Seeing Colors: A Literature Review on Colorimetric Whole-Cell Biosensors
Abstract
1. Introduction
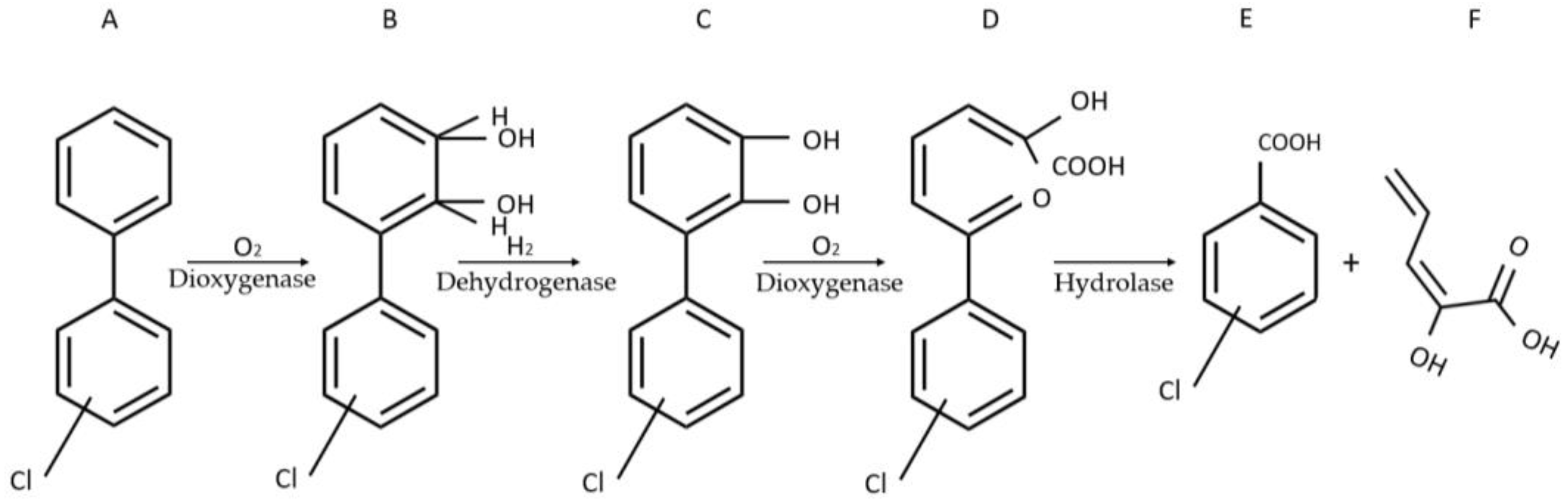
2. Response of WCBs to Synthetic Molecules
3. Detection of Metals by WCBs
3.1. Response of WCBs to Copper
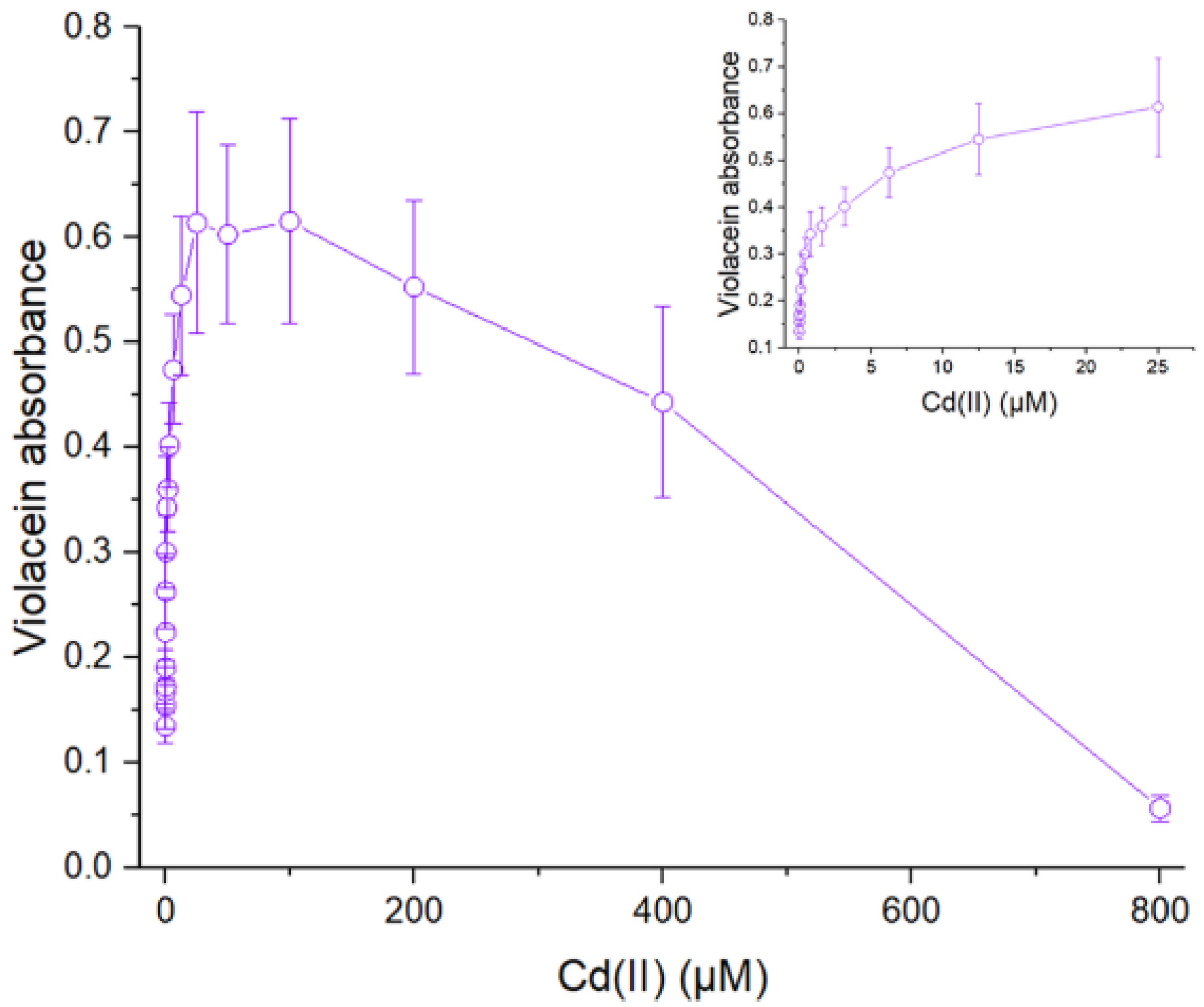
3.2. Response of WCBs to Cadmium
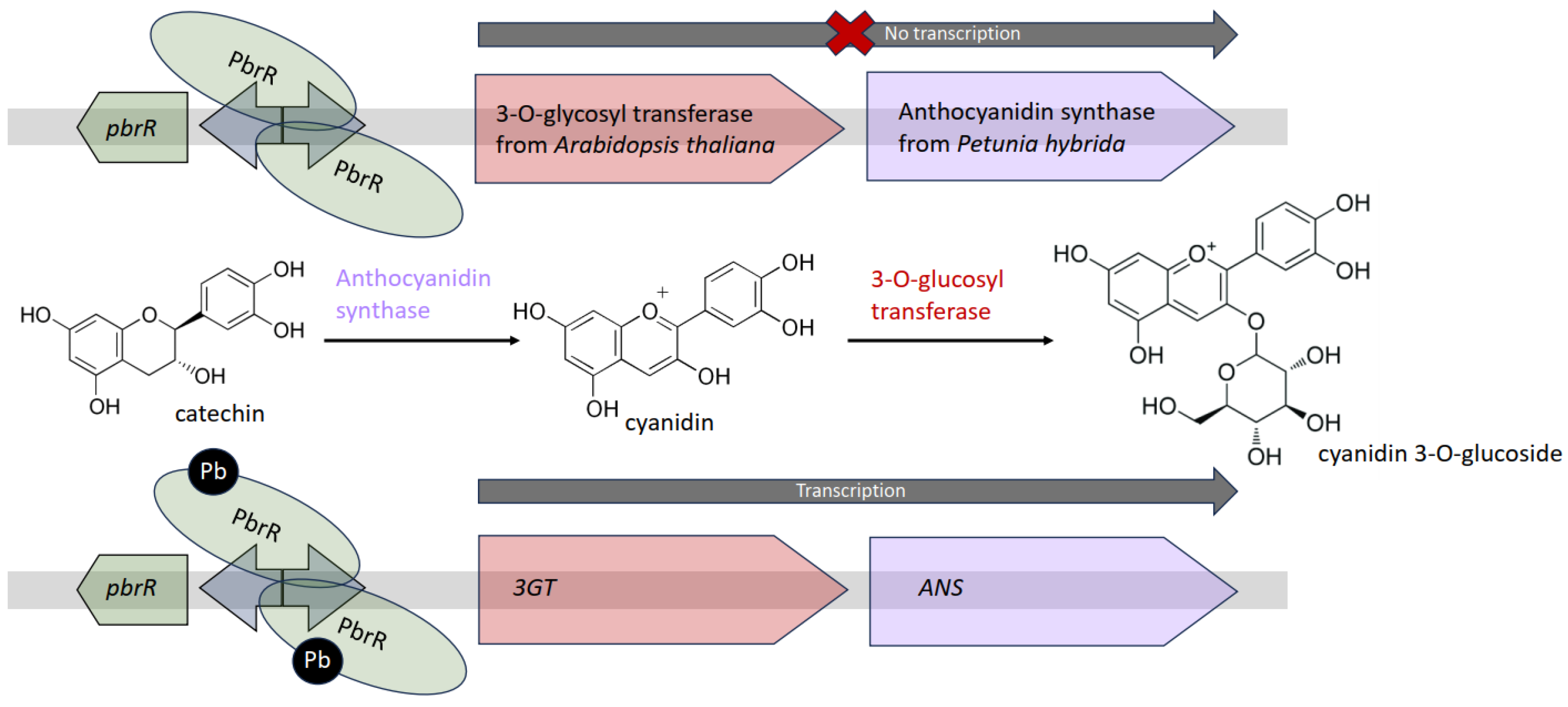
3.3. Response of WCBs to Lead
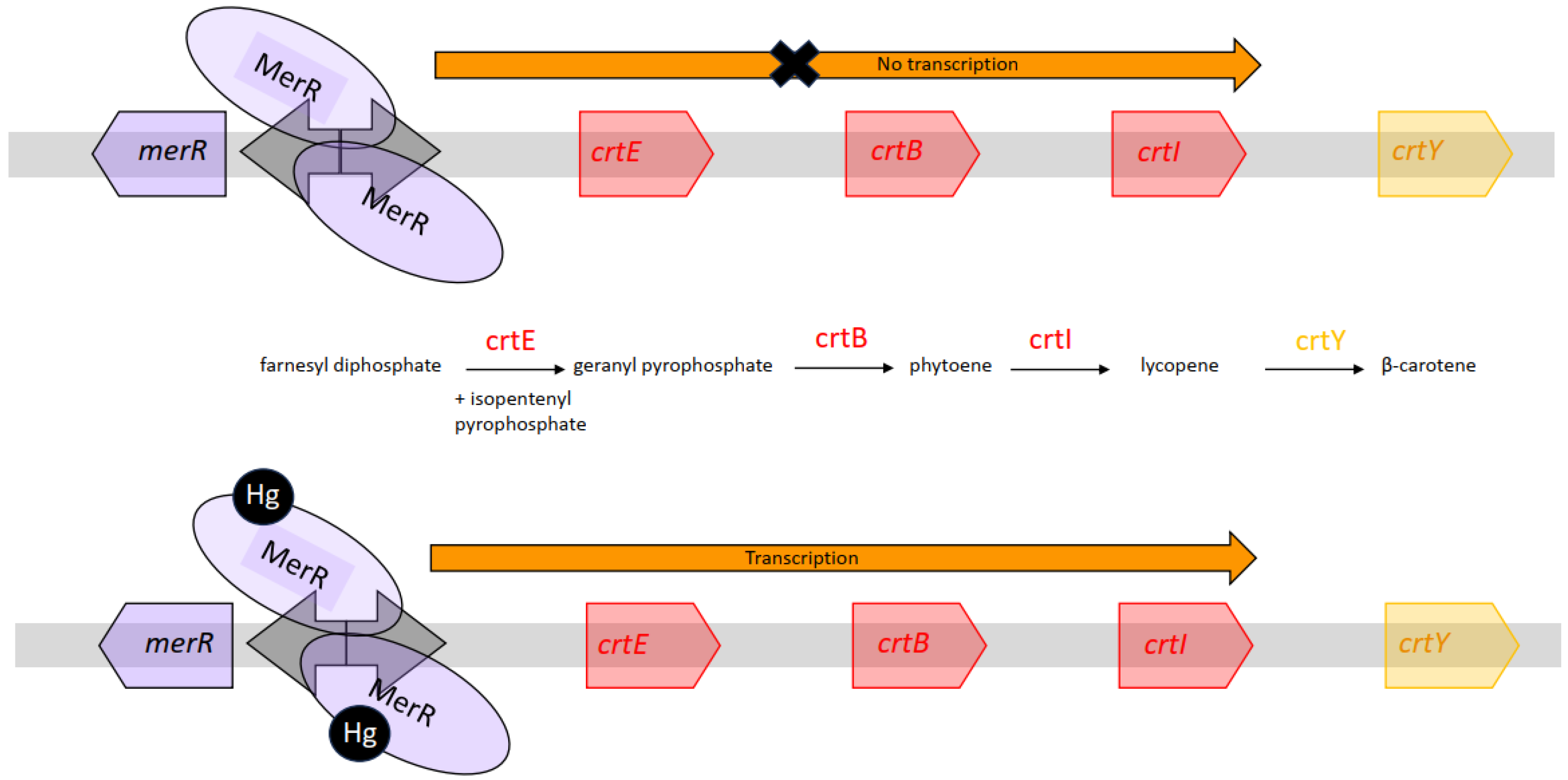
3.4. Response of WCBs to Mercury
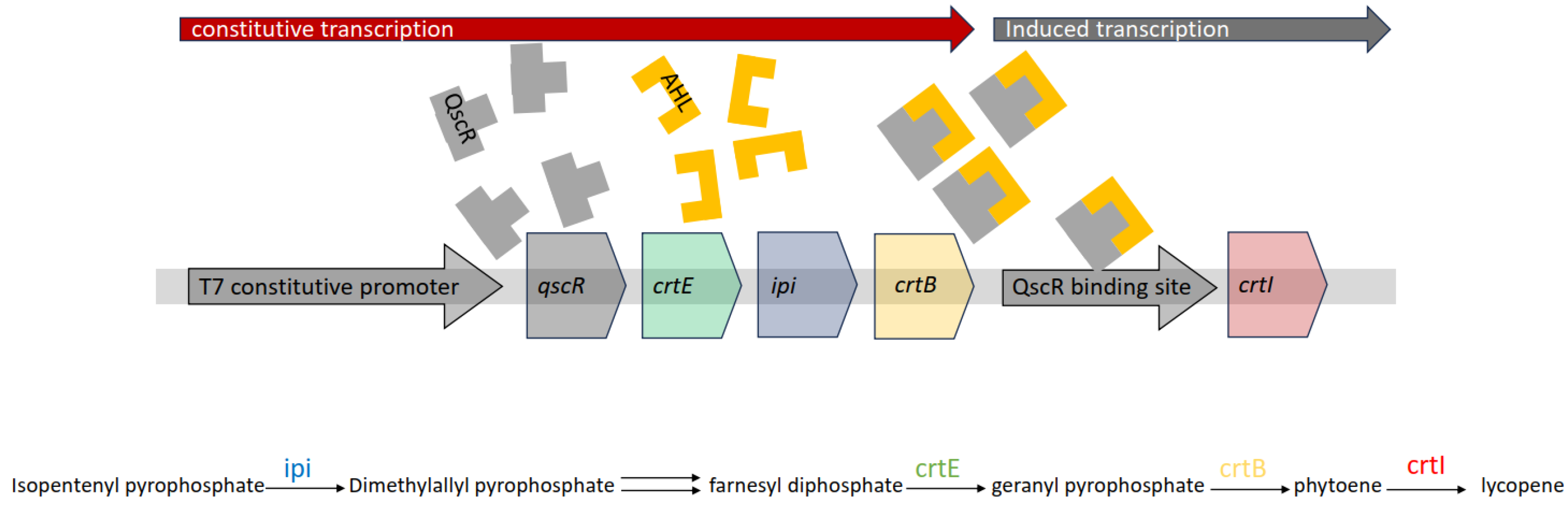
3.5. Response of WCBs to Arsenic
4. Biomonitoring and Control
4.1. High-Level Producer Detection
4.2. Pathogen Detection
4.3. Micronutrient Quantification
5. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bousse, L. Whole Cell Biosensors. Sens. Actuators B Chem. 1996, 34, 270–275. [Google Scholar] [CrossRef]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to Biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Barahona, M.; Buck, M. A Modular Cell-Based Biosensor Using Engineered Genetic Logic Circuits to Detect and Integrate Multiple Environmental Signals. Biosens. Bioelectron. 2013, 40, 368–376. [Google Scholar] [CrossRef]
- Wen, K.Y.; Rutter, J.W.; Barnes, C.P.; Dekker, L. Fundamental Building Blocks of Whole-Cell Biosensor Design. Handb. Cell Biosens. 2020, 1–23. [Google Scholar] [CrossRef]
- Khalil, A.S.; Collins, J.J. Synthetic Biology: Applications Come of Age. Nat. Rev. Genet. 2010, 11, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Hossain, G.S.; Saini, M.; Miyake, R.; Ling, H.; Chang, M.W. Genetic Biosensor Design for Natural Product Biosynthesis in Microorganisms. Trends Biotechnol. 2020, 38, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Serganov, A.; Nudler, E. A Decade of Riboswitches. Cell 2013, 152, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Barahona, M.; Buck, M. Amplification of Small Molecule-Inducible Gene Expression via Tuning of Intracellular Receptor Densities. Nucleic Acids Res. 2015, 43, 1955–1964. [Google Scholar] [CrossRef]
- Hui, C.Y.; Guo, Y.; Gao, C.X.; Li, H.; Lin, Y.R.; Yun, J.P.; Chen, Y.T.; Yi, J. A Tailored Indigoidine-Based Whole-Cell Biosensor for Detecting Toxic Cadmium in Environmental Water Samples. Environ. Technol. Innov. 2022, 27, 102511. [Google Scholar] [CrossRef]
- Joe, M.H.; Lee, K.H.; Lim, S.Y.; Im, S.H.; Song, H.P.; Lee, I.S.; Kim, D.H. Pigment-Based Whole-Cell Biosensor System for Cadmium Detection Using Genetically Engineered Deinococcus Radiodurans. Bioprocess Biosyst. Eng. 2012, 35, 265–272. [Google Scholar] [CrossRef]
- Liu, J.; Xu, J.Z.; Rao, Z.M.; Zhang, W.G. An Enzymatic Colorimetric Whole-Cell Biosensor for High-Throughput Identification of Lysine Overproducers. Biosens. Bioelectron. 2022, 216, 114681. [Google Scholar] [CrossRef] [PubMed]
- Watstein, D.M.; Styczynski, M.P. Development of a Pigment-Based Whole-Cell Zinc Biosensor for Human Serum. ACS Synth. Biol. 2018, 7, 267–275. [Google Scholar] [CrossRef]
- Chong, H.; Ching, C.B. Development of Colorimetric-Based Whole-Cell Biosensor for Organophosphorus Compounds by Engineering Transcription Regulator DmpR. ACS Synth. Biol. 2016, 5, 1290–1298. [Google Scholar] [CrossRef]
- Carpenter, D.O. Polychlorinated Biphenyls (PCBs): Routes of Exposure and Effects on Human Health. Rev. Environ. Health 2006, 21, 1–23. [Google Scholar] [CrossRef]
- Přibyl, J.; Hepel, M.; Skládal, P. Piezoelectric Immunosensors for Polychlorinated Biphenyls Operating in Aqueous and Organic Phases. Sens. Actuators B Chem. 2006, 113, 900–910. [Google Scholar] [CrossRef]
- Shimomura, M.; Nomura, Y.; Zhang, W.; Sakino, M.; Lee, K.H.; Ikebukuro, K.; Karube, I. Simple and Rapid Detection Method Using Surface Plasmon Resonance for Dioxins, Polychlorinated Biphenylx and Atrazine. Anal. Chim. Acta 2001, 434, 223–230. [Google Scholar] [CrossRef]
- Gavlasova, P.; Kuncova, G.; Kochankova, L.; Mackova, M. Whole Cell Biosensor for Polychlorinated Biphenyl Analysis Based on Optical Detection. Int. Biodeterior. Biodegrad. 2008, 62, 304–312. [Google Scholar] [CrossRef]
- Dercová, K.; Baláž, Š.; Vrana, B.; Tandlich, R. Aerobic Biodegradation of Polychlorinated Biphenyls (PCBs). In The Utilization of Bioremediation to Reduce Soil Contamination: Problems and Solutions; Springer: Dordrecht, The Netherlands, 2003; pp. 95–113. [Google Scholar] [CrossRef]
- Nováková, H.; Vošahlíková, M.; Pazlarová, J.; Macková, M.; Burkhard, J.; Demnerová, K. PCB Metabolism by Pseudomonas Sp. P2. Int. Biodeterior. Biodegrad. 2002, 50, 47–54. [Google Scholar] [CrossRef]
- Kuncova, G.; Berkova, D.; Burkhard, J.; Demnerova, K.; Pazlarova, J.; Triska, J.; Vrchotova, N. Optical Detection of Polychiorinated Biphenyls. In Environmental Monitoring and Remediation Technologies II; Vo-Dinh, T., Spellicy, R.L., Eds.; SPIE: Bellingham, WA, USA, 1999; Volume 3853, pp. 72–80. [Google Scholar]
- Mulchandani, A.; Rajesh. Microbial Biosensors for Organophosphate Pesticides. Appl. Biochem. Biotechnol. 2011, 165, 687–699. [Google Scholar] [CrossRef]
- Kumar, J.; Jha, S.K.; D’Souza, S.F. Optical Microbial Biosensor for Detection of Methyl Parathion Pesticide Using Flavobacterium Sp. Whole Cells Adsorbed on Glass Fiber Filters as Disposable Biocomponent. Biosens. Bioelectron. 2006, 21, 2100–2105. [Google Scholar] [CrossRef]
- Wise, A.A.; Kuske, C.R. Generation of Novel Bacterial Regulatory Proteins That Detect Priority Pollutant Phenols. Appl. Environ. Microbiol. 2000, 66, 163–169. [Google Scholar] [CrossRef]
- Pavel, H.; Forsman, M.; Shingler, V. An Aromatic Effector Specificity Mutant of the Transcriptional Regulator DmpR Overcomes the Growth Constraints of Pseudomonas Sp. Strain CF600 on Para-Substituted Methylphenols. J. Bacteriol. 1994, 176, 7550–7557. [Google Scholar] [CrossRef]
- Campos, V.L.; Zaror, C.A.; Mondaca, M.A. Detection of Chlorinated Phenols in Kraft Pulp Bleaching Effluents Using DmpR Mutant Strains. Bull. Environ. Contam. Toxicol. 2004, 73, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Eger, A.; Koele, N.; Caspari, T.; Poggio, M.; Kumar, K.; Burge, O.R. Quantifying the Importance of Soil-Forming Factors Using Multivariate Soil Data at Landscape Scale. J. Geophys. Res. Earth Surf. 2021, 126, 1–19. [Google Scholar] [CrossRef]
- Baumhardt, R.L.; Stewart, B.A.; Sainju, U.M. North American Soil Degradation: Processes, Practices, and Mitigating Strategies. Sustainability 2015, 7, 2936–2960. [Google Scholar] [CrossRef]
- Turdean, G.L. Design and Development of Biosensors for the Detection of Heavy Metal Toxicity. Int. J. Electrochem. 2011, 2011, 1–15. [Google Scholar] [CrossRef]
- Fei, X.; Lou, Z.; Xiao, R.; Ren, Z.; Lv, X. Contamination Assessment and Source Apportionment of Heavy Metals in Agricultural Soil through the Synthesis of PMF and GeogDetector Models. Sci. Total Environ. 2020, 747, 141293. [Google Scholar] [CrossRef]
- Nies, D.H. Heavy Metal-Resistant Bacteria as Extremophiles: Molecular Physiology and Biotechnological Use of Ralstonia Sp. CH34. Extremophiles 2000, 4, 77–82. [Google Scholar] [CrossRef]
- Knight, S.A.B.; Tamai, K.T.; Kosman, D.J.; Thiele, D.J. Identification and Analysis of a Saccharomyces Cerevisiae Copper Homeostasis Gene Encoding a Homeodomain Protein. Mol. Cell. Biol. 1994, 14, 7792–7804. [Google Scholar] [CrossRef]
- Vopálenská, I.; Váchová, L.; Palková, Z. New Biosensor for Detection of Copper Ions in Water Based on Immobilized Genetically Modified Yeast Cells. Biosens. Bioelectron. 2015, 72, 160–167. [Google Scholar] [CrossRef]
- Henikoff, S. The Saccharomyces Cerevisiae ADE5,7 Protein Is Homologous to Overlapping Drosophila Melanogaster Gart Polypeptides. J. Mol. Biol. 1986, 190, 519–528. [Google Scholar] [CrossRef]
- Shetty, R.S.; Deo, S.K.; Liu, Y.; Daunert, S. Fluorescence-Based Sensing System for Copper Using Genetically Engineered Living Yeast Cells. Biotechnol. Bioeng. 2004, 88, 664–670. [Google Scholar] [CrossRef]
- Chen, P.H.; Lin, C.; Guo, K.H.; Yeh, Y.C. Development of a Pigment-Based Whole-Cell Biosensor for the Analysis of Environmental Copper. RSC Adv. 2017, 7, 29302–29305. [Google Scholar] [CrossRef]
- Kumar, A.; Subrahmanyam, G.; Mondal, R.; Cabral-Pinto, M.M.S.; Shabnam, A.A.; Jigyasu, D.K.; Malyan, S.K.; Fagodiya, R.K.; Khan, S.A.; Yu, Z.G. Bio-Remediation Approaches for Alleviation of Cadmium Contamination in Natural Resources. Chemosphere 2021, 268, 128855. [Google Scholar] [CrossRef]
- Hui, C.Y.; Hu, S.Y.; Li, L.M.; Yun, J.P.; Zhang, Y.F.; Yi, J.; Zhang, N.X.; Guo, Y. Metabolic Engineering of the Carotenoid Biosynthetic Pathway toward a Specific and Sensitive Inorganic Mercury Biosensor. RSC Adv. 2022, 12, 36142–36148. [Google Scholar] [CrossRef]
- Meima, R.; Lindstrom, M.E. Characterization of the Minimal Replicon of a Cryptic Deinococcus Radiodurans SARK Plasmid and Development of Versatile Escherichia Coli-D. Radiodurans Shuttle Vectors. Appl. Environ. Microbiol. 2000, 66, 3856–3867. [Google Scholar] [CrossRef] [PubMed]
- Joe, M.H.; Jung, S.W.; Im, S.H.; Lim, S.Y.; Song, H.P.; Kwon, O.; Kim, D.H. Genome-Wide Response of Deinococcus Radiodurans on Cadmium Toxicity. J. Microbiol. Biotechnol. 2011, 21, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Hui, C.Y.; Guo, Y.; Li, H.; Gao, C.X.; Yi, J. Detection of Environmental Pollutant Cadmium in Water Using a Visual Bacterial Biosensor. Sci. Rep. 2022, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Glickmann, E.; Cooksey, D.A. Chromosomal Locus for Cadmium Resistance in Pseudomonas Putida Consisting of a Cadmium-Transporting ATPase and a MerR Family Response Regulator. Appl. Environ. Microbiol. 2001, 67, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- August, P.R.; Grossman, T.H.; Minor, C.; Draper, M.P.; MacNeil, I.A.; Pemberton, J.M.; Call, K.M.; Holt, D.; Osbourne, S. Sequence Analysis and Functional Characterization of the Violacein Biosynthetic Pathway from Chromobacterium Violaceum. J. Mol. Microbiol. Biotechnol. 2000, 2, 513–519. [Google Scholar] [PubMed]
- Hui, C.Y.; Guo, Y.; Liu, L.; Zhang, N.X.; Gao, C.X.; Yang, X.Q.; Yi, J. Genetic Control of Violacein Biosynthesis to Enable a Pigment-Based Whole-Cell Lead Biosensor. RSC Adv. 2020, 10, 28106–28113. [Google Scholar] [CrossRef]
- Shahid, M.; Dumat, C.; Khalid, S.; Niazi, N.K.; Antunes, P.M.C. Cadmium Bioavailability, Uptake, Toxicity and Detoxification in Soil-Plant System. In How to Recruit Voluntary Donors in the Third World? Springer: Cham, Switzerland, 2016; Volume 238, pp. 73–137. [Google Scholar]
- Brocklehurst, K.R.; Megit, S.J.; Morby, A.P. Characterisation of CadR from Pseudomonas Aeruginosa: A Cd(II)-Responsive MerR Homologue. Biochem. Biophys. Res. Commun. 2003, 308, 234–239. [Google Scholar] [CrossRef]
- Liu, X.; Hu, Q.; Yang, J.; Huang, S.; Wei, T.; Chen, W.; He, Y.; Wang, D.; Liu, Z.; Wang, K.; et al. Selective Cadmium Regulation Mediated by a Cooperative Binding Mechanism in CadR. Proc. Natl. Acad. Sci. USA 2019, 116, 20398–20403. [Google Scholar] [CrossRef]
- Hui, C.Y.; Guo, Y.; Li, L.M.; Liu, L.; Chen, Y.T.; Yi, J.; Zhang, N.X. Indigoidine Biosynthesis Triggered by the Heavy Metal-Responsive Transcription Regulator: A Visual Whole-Cell Biosensor. Appl. Microbiol. Biotechnol. 2021, 105, 6087–6102. [Google Scholar] [CrossRef]
- Hui, C.Y.; Guo, Y.; Wu, J.; Liu, L.; Yang, X.Q.; Guo, X.; Xie, Y.; Yi, J. Detection of Bioavailable Cadmium by Double-Color Fluorescence Based on a Dual-Sensing Bioreporter System. Front. Microbiol. 2021, 12, 1–15. [Google Scholar] [CrossRef]
- Tian, H.; Jiao, L.; Dong, D. Rapid Determination of Trace Cadmium in Drinking Water Using Laser-Induced Breakdown Spectroscopy Coupled with Chelating Resin Enrichment. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.; Wu, P.; Zhang, X.; Hou, X.; Jones, B. Determination of Cadmium in Biological Samples. Appl. Spectrosc. Rev. 2006, 41, 35–75. [Google Scholar] [CrossRef]
- Büyükpınar, Ç.; Bodur, S.; Yazıcı, E.; Tekin, Z.; San, N.; Tarık Komesli, O.; Bakırdere, S. An Accurate Analytical Method for the Determination of Cadmium: Ultraviolet Based Photochemical Vapor Generation-Slotted Quartz Tube Based Atom Trap-Flame Atomic Absorption Spectrophotometry. Meas. J. Int. Meas. Confed. 2021, 176, 109192. [Google Scholar] [CrossRef]
- Wei, W.; Liu, X.; Sun, P.; Wang, X.; Zhu, H.; Hong, M.; Mao, Z.W.; Zhao, J. Simple Whole-Cell Biodetection and Bioremediation of Heavy Metals Based on an Engineered Lead-Specific Operon. Environ. Sci. Technol. 2014, 48, 3363–3371. [Google Scholar] [CrossRef] [PubMed]
- Shetty, R.S.; Deo, S.K.; Shah, P.; Sun, Y.; Rosen, B.P.; Daunert, S. Luminescence-Based Whole-Cell-Sensing Systems for Cadmium and Lead Using Genetically Engineered Bacteria. Anal. Bioanal. Chem. 2003, 376, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Borremans, B.; Hobman, J.L.; Provoost, A.; Brown, N.L.; Van Der Lelie, D. Cloning and Functional Analysis of the Pbr Lead Resistance Determinant of Ralstonia Metallidurans CH34. J. Bacteriol. 2001, 183, 5651–5658. [Google Scholar] [CrossRef] [PubMed]
- Hui, C.; Ma, B.; Wang, Y.; Yang, X.; Cai, J. Designed Bacteria Based on Natural Pbr Operons for Detecting and Detoxifying Environmental Lead: A Mini-Review. Ecotoxicol. Environ. Saf. 2023, 267, 115662. [Google Scholar] [CrossRef] [PubMed]
- Bereza-Malcolm, L.; Aracic, S.; Franks, A.E. Development and Application of a Synthetically-Derived Lead Biosensor Construct for Use in Gram-Negative Bacteria. Sensors 2016, 16, 2174. [Google Scholar] [CrossRef] [PubMed]
- Nourmohammadi, E.; Hosseinkhani, S.; Nedaeinia, R.; Khoshdel-Sarkarizi, H.; Nedaeinia, M.; Ranjbar, M.; Ebrahimi, N.; Farjami, Z.; Nourmohammadi, M.; Mahmoudi, A.; et al. Construction of a Sensitive and Specific Lead Biosensor Using a Genetically Engineered Bacterial System with a Luciferase Gene Reporter Controlled by Pbr and CadA Promoters. Biomed. Eng. Online 2020, 19, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Zhao, T.; Liu, Y.; Bu, R.; Wu, K. Gene Circuit Engineering to Improve the Performance of a Whole-Cell Lead Biosensor. FEMS Microbiol. Lett. 2018, 365, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Hui, C.Y.; Liu, L.; Zheng, H.Q.; Wu, H.M. Improved Monitoring of Low-Level Transcription in Escherichia Coliby a β-Galactosidase α-Complementation System. Front. Microbiol. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Hui, C.Y.; Guo, Y.; Zhu, D.L.; Li, L.M.; Yi, J.; Zhang, N.X. Metabolic Engineering of the Violacein Biosynthetic Pathway toward a Low-Cost, Minimal-Equipment Lead Biosensor. Biosens. Bioelectron. 2022, 214, 114531. [Google Scholar] [CrossRef]
- Zhao, E.M.; Suek, N.; Wilson, M.Z.; Dine, E.; Pannucci, N.L.; Gitai, Z.; Avalos, J.L.; Toettcher, J.E. Light-Based Control of Metabolic Flux through Assembly of Synthetic Organelles. Nat. Chem. Biol. 2019, 15, 589–597. [Google Scholar] [CrossRef]
- Guo, Y.; Huang, Z.L.; Zhu, D.L.; Hu, S.Y.; Li, H.; Hui, C.Y. Anthocyanin Biosynthetic Pathway Switched by Metalloregulator PbrR to Enable a Biosensor for the Detection of Lead Toxicity. Front. Microbiol. 2022, 13, 1–10. [Google Scholar] [CrossRef]
- Miyazawa, T.; Nakagawa, K.; Kudo, M.; Muraishi, K.; Someya, K. Direct Intestinal Absorption of Red Fruit Anthocyanins, Cyanidin-3- Glucoside and Cyanidin-3,5-Diglucoside, into Rats and Humans. J. Agric. Food Chem. 1999, 47, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Olivas-Aguirre, F.J.; Rodrigo-García, J.; Martínez-Ruiz, N.D.R.; Cárdenas-Robles, A.I.; Mendoza-Díaz, S.O.; Álvarez-Parrilla, E.; González-Aguilar, G.A.; De La Rosa, L.A.; Ramos-Jiménez, A.; Wall-Medrano, A. Cyanidin-3-O-Glucoside: Physical-Chemistry, Foodomics and Health Effects. Molecules 2016, 21, 1264. [Google Scholar] [CrossRef] [PubMed]
- Masten, S.J.; Davies, S.H.; McElmurry, S.P. Flint Water Crisis: What Happened and Why? J. Am. Water Works Assoc. 2016, 108, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Helmann, J.D.; Ballard, B.T.; Walsh, C.T. The MerR Metalloregulatory Protein Binds Mercuric Ion as a Tricoordinate, Metal-Bridged Dimer. Science 1990, 247, 946–948. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.X.; Guo, Y.; Li, H.; Yang, X.Q.; Gao, C.X.; Hui, C.Y. Versatile Artificial Mer Operons in Escherichia Coli towards Whole Cell Biosensing and Adsorption of Mercury. PLoS ONE 2021, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Hui, C.Y.; Liu, L.; Chen, M.P.; Huang, H.Y. Development of a Bioavailable Hg(II) Sensing System Based on MerR-Regulated Visual Pigment Biosynthesis. Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef]
- Huang, L.; Huang, Y.; Lou, Y.; Qian, H.; Xu, D.; Ma, L.; Jiang, C.; Zhang, D. Pyocyanin-Modifying Genes PhzM and PhzS Regulated the Extracellular Electron Transfer in Microbiologically-Influenced Corrosion of X80 Carbon Steel by Pseudomonas Aeruginosa. Corros. Sci. 2020, 164, 108355. [Google Scholar] [CrossRef]
- Wang, D.; Zheng, Y.; Fan, X.; Xu, L.; Pang, T.; Liu, T.; Liang, L.; Huang, S.; Xiao, Q. Visual Detection of Hg2+ by Manipulation of Pyocyanin Biosynthesis through the Hg2+-Dependent Transcriptional Activator MerR in Microbial Cells. J. Biosci. Bioeng. 2020, 129, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Ketelboeter, L.M.; Bardy, S.L. Methods to Inhibit Bacterial Pyomelanin Production and Determine the Corresponding Increase in Sensitivity to Oxidative Stress. J. Vis. Exp. 2015, 2015, 1–9. [Google Scholar] [CrossRef]
- De Mora, K.; Joshi, N.; Balint, B.L.; Ward, F.B.; Elfick, A.; French, C.E. A PH-Based Biosensor for Detection of Arsenic in Drinking Water. Anal. Bioanal. Chem. 2011, 400, 1031–1039. [Google Scholar] [CrossRef]
- Wu, J.; Rosen, B.P. The ArsR Protein Is a Trans-acting Regulatory Protein. Mol. Microbiol. 1991, 5, 1331–1336. [Google Scholar] [CrossRef]
- Fujimoto, H.; Wakabayashi, M.; Yamashiro, H.; Maeda, I.; Isoda, K.; Kondoh, M.; Kawase, M.; Miyasaka, H.; Yagi, K. Whole-Cell Arsenite Biosensor Using Photosynthetic Bacterium Rhodovulum Sulfidophilum: Rhodovulum Sulfidophilum as an Arsenite Biosensor. Appl. Microbiol. Biotechnol. 2006, 73, 332–338. [Google Scholar] [CrossRef]
- Yoshida, K.; Inoue, K.; Takahashi, Y.; Ueda, S.; Isoda, K.; Yagi, K.; Maeda, I. Novel Carotenoid-Based Biosensor for Simple Visual Detection of Arsenite: Characterization and Preliminary Evaluation for Environmental Application. Appl. Environ. Microbiol. 2008, 74, 6730–6738. [Google Scholar] [CrossRef]
- Yoshida, K.; Yoshioka, D.; Inoue, K.; Takaichi, S.; Maeda, I. Evaluation of Colors in Green Mutants Isolated from Purple Bacteria as a Host for Colorimetric Whole-Cell Biosensors. Appl. Microbiol. Biotechnol. 2007, 76, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Zaynab, M.; Al-Yahyai, R.; Ameen, A.; Sharif, Y.; Ali, L.; Fatima, M.; Khan, K.A.; Li, S. Health and Environmental Effects of Heavy Metals. J. King Saud Univ.-Sci. 2022, 34, 101653. [Google Scholar] [CrossRef]
- Ali, H.; Khan, E. Trophic Transfer, Bioaccumulation, and Biomagnification of Non-Essential Hazardous Heavy Metals and Metalloids in Food Chains/Webs—Concepts and Implications for Wildlife and Human Health. Hum. Ecol. Risk Assess. 2019, 25, 1353–1376. [Google Scholar] [CrossRef]
- Li, C.; Zhou, K.; Qin, W.; Tian, C.; Qi, M.; Yan, X.; Han, W. A Review on Heavy Metals Contamination in Soil: Effects, Sources, and Remediation Techniques. Soil Sediment Contam. 2019, 28, 380–394. [Google Scholar] [CrossRef]
- Gupta, A.; Sharma, V.; Sharma, K.; Kumar, V.; Choudhary, S.; Mankotia, P.; Kumar, B.; Mishra, H.; Moulick, A.; Ekielski, A.; et al. A Review of Adsorbents for Heavy Metal Decontamination: Growing Approach to Wastewater Treatment. Materials 2021, 14, 4702. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Rout, C.; Singh, J.; Saharan, Y.; Goyat, R.; Umar, A.; Akbar, S.; Baskoutas, S. A Review on the Clean-up Technologies for Heavy Metal Ions Contaminated Soil Samples. Heliyon 2023, 9, e15472. [Google Scholar] [CrossRef] [PubMed]
- Félix, F.K. do C.; Letti, L.A.J.; Vinícius de Melo Pereira, G.; Bonfim, P.G.B.; Soccol, V.T.; Soccol, C.R. L-Lysine Production Improvement: A Review of the State of the Art and Patent Landscape Focusing on Strain Development and Fermentation Technologies. Crit. Rev. Biotechnol. 2019, 39, 1031–1055. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Chen, P.; Song, A.; Wang, D.; Wang, Q. Expanding Lysine Industry: Industrial Biomanufacturing of Lysine and Its Derivatives. J. Ind. Microbiol. Biotechnol. 2018, 45, 719–734. [Google Scholar] [CrossRef]
- Ikeda, M. Lysine Fermentation: History and Genome Breeding. In Advances in Biochemical Engineering/Biotechnology; Springer: Tokyo, Japan, 2016; Volume 123, pp. 73–102. [Google Scholar]
- Bellman, A.; Vrljić, M.; Pátek, M.; Sahm, H.; Krämer, R.; Eggeling, L. Expression Control and Specificity of the Basic Amino Acid Exporter LysE of Corynebacterium Glutamicum. Microbiology 2001, 147, 1765–1774. [Google Scholar] [CrossRef]
- Henke, N.A.; Wendisch, V.F. Improved Astaxanthin Production with Corynebacterium Glutamicum by Application of a Membrane Fusion Protein. Mar. Drugs 2019, 17, 621. [Google Scholar] [CrossRef]
- Kumari, A.; Pasini, P.; Daunert, S. Detection of Bacterial Quorum Sensing N-Acyl Homoserine Lactones in Clinical Samples. Anal. Bioanal. Chem. 2008, 391, 1619–1627. [Google Scholar] [CrossRef]
- Yeon, K.M.; Cheong, W.S.; Oh, H.S.; Lee, W.N.; Hwang, B.K.; Lee, C.H.; Beyenal, H.; Lewandowski, Z. Quorum Sensing: A New Biofouling Control Paradigm in a Membrane Bioreactor for Advanced Wastewater Treatment. Environ. Sci. Technol. 2009, 43, 380–385. [Google Scholar] [CrossRef]
- Valle, A.; Bailey, M.J.; Whiteley, A.S.; Manefield, M. N-Acyl-L-Homoserine Lactones (AHLs) Affect Microbial Community Composition and Function in Activated Sludge. Environ. Microbiol. 2004, 6, 424–433. [Google Scholar] [CrossRef]
- Song, S.; Jia, Z.; Xu, J.; Zhang, Z.; Bian, Z. N-Butyryl-Homoserine Lactone, a Bacterial Quorum-Sensing Signaling Molecule, Induces Intracellular Calcium Elevation in Arabidopsis Root Cells. Biochem. Biophys. Res. Commun. 2011, 414, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Yong, Y.C.; Zhong, J.J. A Genetically Engineered Whole-Cell Pigment-Based Bacterial Biosensing System for Quantification of N-Butyryl Homoserine Lactone Quorum Sensing Signal. Biosens. Bioelectron. 2009, 25, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.X.; Zhou, W.Q.; Zhong, J.J. Organotin Decomposition by Pyochelin, Secreted by Pseudomonas Aeruginosa Even in an Iron-Sufficient Environment. Appl. Environ. Microbiol. 2006, 72, 6411–6413. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, C.W.; Wang, D.; Wei, N. A Whole-Cell Biosensor for Point-of-Care Detection of Waterborne Bacterial Pathogens. ACS Synth. Biol. 2021, 10, 333–344. [Google Scholar] [CrossRef] [PubMed]
- D’Agata, E.M.C.; Dupont-Rouzeyrol, M.; Magal, P.; Olivier, D.; Ruan, S. The Impact of Different Antibiotic Regimens on the Emergence of Antimicrobial-Resistant Bacteria. PLoS ONE 2008, 3, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.; Song, F.; Guo, F.; Zhou, Z.; Zhu, C.; Xiang, J.; Huan, J. Acinetobacter Baumannii Quorum-Sensing Signalling Molecule Induces the Expression of Drug-Resistance Genes. Mol. Med. Rep. 2017, 15, 4061–4068. [Google Scholar] [CrossRef]
- Saipriya, K.; Swathi, C.H.; Ratnakar, K.S.; Sritharan, V. Quorum-Sensing System in Acinetobacter Baumannii: A Potential Target for New Drug Development. J. Appl. Microbiol. 2020, 128, 15–27. [Google Scholar] [CrossRef]
- Haque, S.; Yadav, D.K.; Bisht, S.C.; Yadav, N.; Singh, V.; Dubey, K.K.; Jawed, A.; Wahid, M.; Dar, S.A. Quorum Sensing Pathways in Gram-Positive and -Negative Bacteria: Potential of Their Interruption in Abating Drug Resistance. J. Chemother. 2019, 31, 161–187. [Google Scholar] [CrossRef]
- Zhao, X.; Yu, Z.; Ding, T. Quorum-Sensing Regulation of Antimicrobial Resistance in Bacteria. Microorganisms 2020, 8, 425. [Google Scholar] [CrossRef]
- Stock, F.; Cirri, E.; Nuwanthi, S.G.L.I.; Stock, W.; Ueberschaar, N.; Mangelinckx, S.; Pohnert, G.; Vyverman, W. Sampling, Separation, and Quantification of N-Acyl Homoserine Lactones from Marine Intertidal Sediments. Limnol. Oceanogr. Methods 2021, 19, 145–157. [Google Scholar] [CrossRef]
- Hoang, T.P.T.; Barthélemy, M.; Lami, R.; Stien, D.; Eparvier, V.; Touboul, D. Annotation and Quantification of N-Acyl Homoserine Lactones Implied in Bacterial Quorum Sensing by Supercritical-Fluid Chromatography Coupled with High-Resolution Mass Spectrometry. Anal. Bioanal. Chem. 2020, 412, 2261–2276. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.M.S.; Lami, R.; Escoubeyrou, K.; Intertaglia, L.; Mazurek, C.; Doberva, M.; Pérez-Ferrer, P.; Stien, D. Straightforward N -Acyl Homoserine Lactone Discovery and Annotation by LC–MS/MS-Based Molecular Networking. J. Proteome Res. 2022, 21, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Stevens, G.A.; Beal, T.; Mbuya, M.N.N.; Luo, H.; Neufeld, L.M.; Addo, O.Y.; Adu-Afarwuah, S.; Alayón, S.; Bhutta, Z.; Brown, K.H.; et al. Micronutrient Deficiencies among Preschool-Aged Children and Women of Reproductive Age Worldwide: A Pooled Analysis of Individual-Level Data from Population-Representative Surveys. Lancet Glob. Health 2022, 10, e1590–e1599. [Google Scholar] [CrossRef]
- Wessells, K.R.; Brown, K.H. Estimating the Global Prevalence of Zinc Deficiency: Results Based on Zinc Availability in National Food Supplies and the Prevalence of Stunting. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- McNerney, M.P.; Michel, C.L.; Kishore, K.; Standeven, J.; Styczynski, M.P. Dynamic and Tunable Metabolite Control for Robust Minimal-Equipment Assessment of Serum Zinc. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Reyes, S.; Le, N.; Fuentes, M.D.; Upegui, J.; Dikici, E.; Broyles, D.; Quinto, E.; Daunert, S.; Deo, S.K. An Intact Cell Bioluminescence-Based Assay for the Simple and Rapid Diagnosis of Urinary Tract Infection. Int. J. Mol. Sci. 2020, 21, 5015. [Google Scholar] [CrossRef] [PubMed]
- Lubkowicz, D.; Ho, C.L.; Hwang, I.Y.; Yew, W.S.; Lee, Y.S.; Chang, M.W. Reprogramming Probiotic Lactobacillus Reuteri as a Biosensor for Staphylococcus Aureus Derived AIP-I Detection. ACS Synth. Biol. 2018, 7, 1229–1237. [Google Scholar] [CrossRef] [PubMed]
- Raut, N.; Pasini, P.; Daunert, S. Deciphering Bacterial Universal Language by Detecting the Quorum Sensing Signal, Autoinducer-2, with a Whole-Cell Sensing System. Anal. Chem. 2013, 85, 9604–9609. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.G.; Moon, S.J.; Kim, S.K.; Kim, T.H.; Lim, H.S.; Yeon, G.H.; Sung, B.H.; Lee, C.H.; Lee, S.G.; Hwang, J.H.; et al. A Designed Whole-Cell Biosensor for Live Diagnosis of Gut Inflammation through Nitrate Sensing. Biosens. Bioelectron. 2020, 168, 112523. [Google Scholar] [CrossRef]
- Moraskie, M.; Roshid, H.; Connor, G.O.; Dikici, E.; Zingg, J.; Deo, S.; Daunert, S. Microbial Whole-Cell Biosensors: Current Applications, Challenges, and Future Perspectives. Biosens. Bioelectron. 2022, 191, 113359. [Google Scholar] [CrossRef]
- Berepiki, A.; Kent, R.; MacHado, L.F.M.; Dixon, N. Development of High-Performance Whole Cell Biosensors Aided by Statistical Modeling. ACS Synth. Biol. 2020, 9, 576–589. [Google Scholar] [CrossRef]
| Application | Sensory Module | Reporter Module | Pigment | Target Molecule | Range of Detection | Ref. |
|---|---|---|---|---|---|---|
| Quantification of PCBs | Biphenyl degradation pathway (unspecified genes) | 2-hydroxy-6-oxo-6-phenyl-2,4-hexadienoic acid | Trichlorobiphenyl mixtures | 0.5 mg D103 l-1 and 0.2 mg 2,4,40CB l-1 | [17] | |
| Detection and quantification of organophosphate pesticides. | DmpR—DM12 | mRFP1 | RFP | parathion | 10 µM | [13] |
| DmpR—E135K | mRFP1 | RFP | fenitrothion | 50 µM | [13] | |
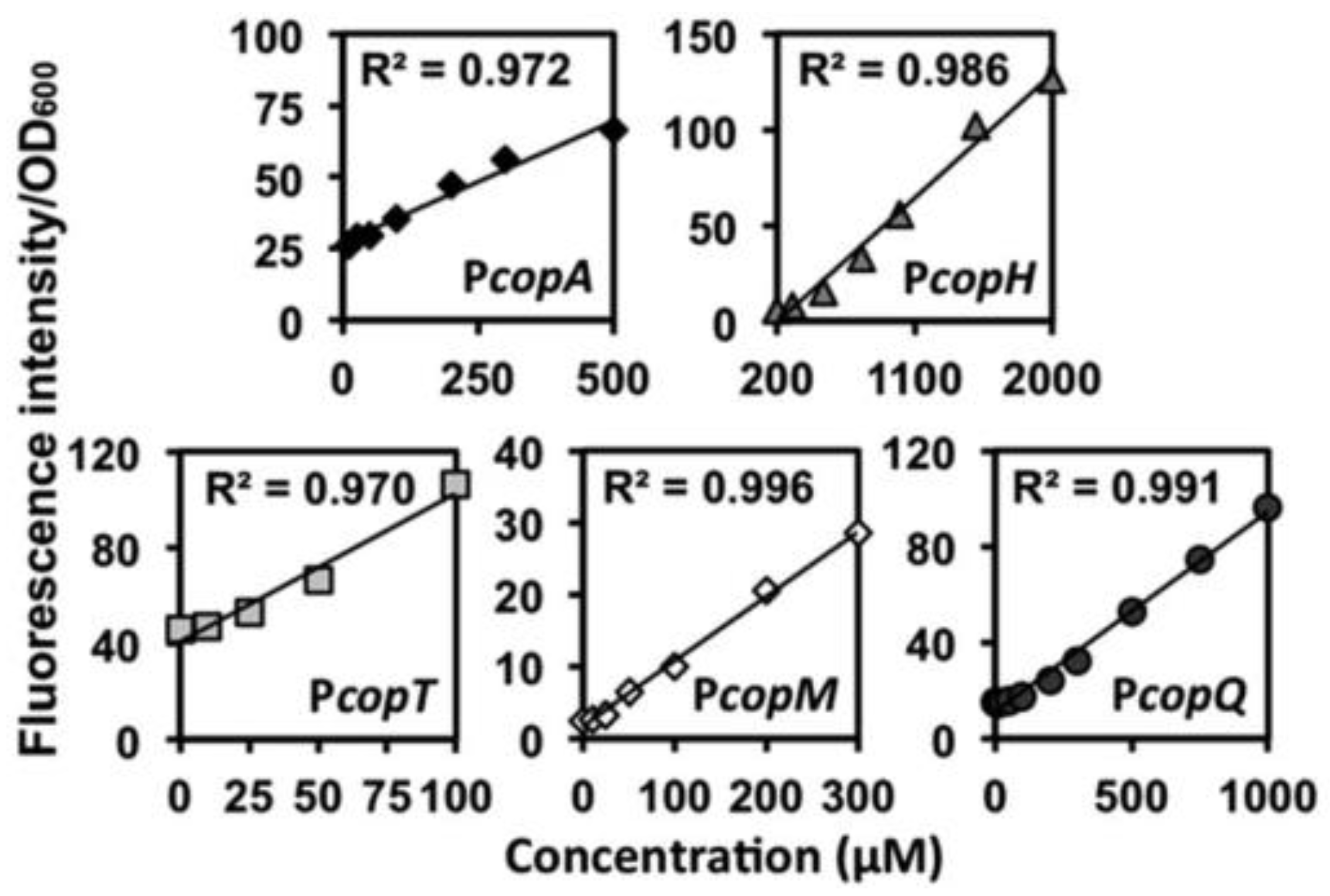
| Detection and quantification of heavy metal ions in water | PCUP1 | AMP pathway ∆ADE2 | Unspecified red pigment | Cu(II) | 1–100 µM | [32] |
| PcopQ | Mjdod | Betaxanthin | Cu(II) | 0–1000 µM | [35] | |
| P0659-1 | crtI | Deinoxanthin | Cd(II) | 50 nM–1 mM | [10] | |
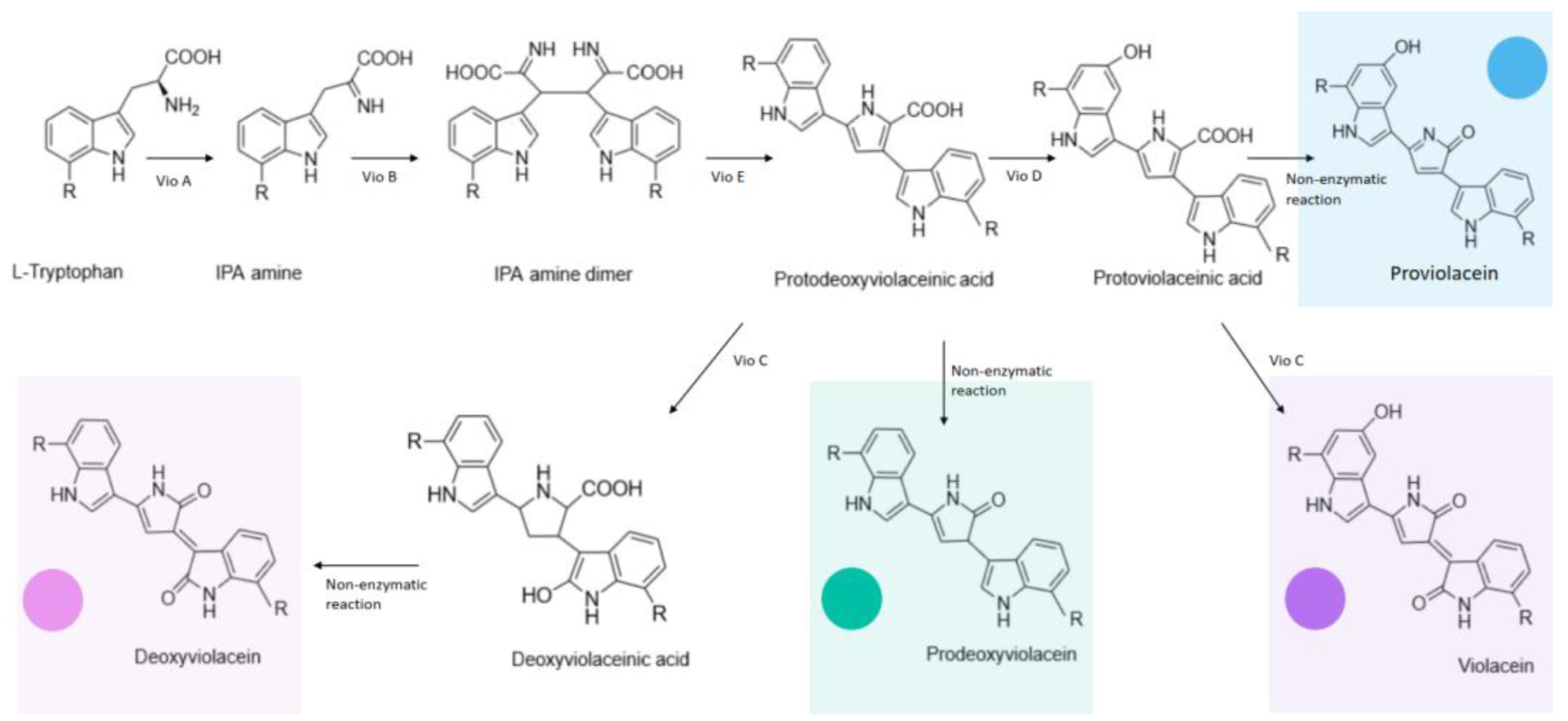
| cadR | vioABCDE | Violacein | Cd(II) | 0–25 µM | [40] | |
| cadR | bpsA, pcpS | Indigoidine | Cd(II) | 0–200 µM | [9] | |
| PbrR | bpsA, pcpS | Indigoidine | Pb(II) | 0.26–8.3 µM | [47] | |
| PbrR | vioABCDE | Violacein | Pb(II) | 0.1875–1.5 mM | [43] | |
| PbrR | vioABCE | Deoxyviolacein | Pb(II) | 2.93–750 nM | [60] | |
| PbrR | 3GT, ANS | Cyanidin 3-O-glucoside | Pb(II) | 0.012–3.125 µM | [62] | |
| MerR | bpsA, pcpS | indigoidine | Hg(II) | 0.008 µM–0.52 µM | [47] | |
| MerR | vioABCDE | Violacein | Hg(II) | 0–0.12 µM | [68] | |
| MerR | phzM and phzS | Pyomelanin | Hg(II) | 25 and 1000 nM | [70] | |
| MerR | crtEBI and crtY | β-carotene | Hg(II) | 12 to 195 nM | [37] | |
| OPars | crtF and crtA | Spheroiden and spheroidenone | Arsenite | 2–8 µg/L | [74] | |
| OPars | CrtI | Lycopene | Arsenite | 1–10 µg/L | [75] | |
| Identification of overproducers | lysG | crtEBI | Lycopene | Lysine | 80–325 mM | [11] |
| Detection of bacterial contamination | RhiI | Blue-green pigment synthesis pathway native to P. aeruginosa | Unspecified blue-green pigment | N-acyl homoserine lactone | 0.11–49.7 µM | [91] |
| QscR | crtEBI | lycopene | N-3-hydroxydecanoyl homoserine lactone, N-3-oxododecanoyl homoserine lactone, N-decanoyl-L-homoserine lactone | ≥2.0 nM | [93] | |
| Quantification of serum zinc | Zur | VioABCDE, crtEBI, crtY | Violacein, lycopene, β-carotene | Zn(II) | Physiologically relevant range | [12] |
| Zur | VioABCDE, crtEBI, crtY | Violacein, lycopene, β-carotene | Zn(II) | Physiologically relevant range | [104] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nemer, G.; Koubaa, M.; El Chamy, L.; Maroun, R.G.; Louka, N. Seeing Colors: A Literature Review on Colorimetric Whole-Cell Biosensors. Fermentation 2024, 10, 79. https://doi.org/10.3390/fermentation10020079
Nemer G, Koubaa M, El Chamy L, Maroun RG, Louka N. Seeing Colors: A Literature Review on Colorimetric Whole-Cell Biosensors. Fermentation. 2024; 10(2):79. https://doi.org/10.3390/fermentation10020079
Chicago/Turabian StyleNemer, Georgio, Mohamed Koubaa, Laure El Chamy, Richard G. Maroun, and Nicolas Louka. 2024. "Seeing Colors: A Literature Review on Colorimetric Whole-Cell Biosensors" Fermentation 10, no. 2: 79. https://doi.org/10.3390/fermentation10020079
APA StyleNemer, G., Koubaa, M., El Chamy, L., Maroun, R. G., & Louka, N. (2024). Seeing Colors: A Literature Review on Colorimetric Whole-Cell Biosensors. Fermentation, 10(2), 79. https://doi.org/10.3390/fermentation10020079