Innovative Bicultured Lactic–Acetic Acid Co-fermentation Improves Jujube Puree’s Functionality and Volatile Compounds
Abstract
1. Introduction
2. Experimental Procedures
2.1. Microbial Isolates, Chemicals and Reagents
2.2. Jujube Fruit Sampling and Activation of Bacterial Starter Cultures
2.3. Jujube Puree Preparation and Fermentation Procedure
- A.
- Lacticaseibacillus casei Lc 122-A. pasteurianus Ap-As.1.41 HuNiang 1.01 (JLcAp),
- B.
- Lactobacillus helveticus Lh 43-A. pasteurianus Ap-As.1.41 HuNiang 1.01 (JLhAp), and
- C.
- Lactiplantibacillus plantarum Lp 28-A. pasteurianus Ap-As.1.41 HuNiang 1.01 (JLpAp).
2.4. Functionality of JP
2.4.1. Microbial Profile of JP
2.4.2. Antioxidant Properties
ABTS Radical Scavenging Activity (ABTS-RSA)
DPPH Radical Scavenging Activity (DPPH-RSA)
2.4.3. Color Assessment
2.4.4. Free Amino Acid Profiling
Sample Preparation
Chromatographic Analysis
2.4.5. Phenolic Profile
Sample Preparation
HPLC-UV Detection
2.5. Volatile Analysis
2.5.1. Elucidation of Volatile Compounds Using HS-SPME-GC-MS
2.5.2. Sample Extraction
2.5.3. HS-SPME-GC-MS Column Analysis
2.5.4. Aroma Profile Using Electronic Nose
2.6. Microstructural Analysis with Scanning Electron Microscopy (SEM)
2.7. Statistical Analysis
3. Results and Discussion
3.1. Viable Counts of Lactic and Acetic Acid Bacteria
3.2. Antioxidant Properties
3.3. Effect of Lactic–Acetic Acid Co-Fermentation on Color
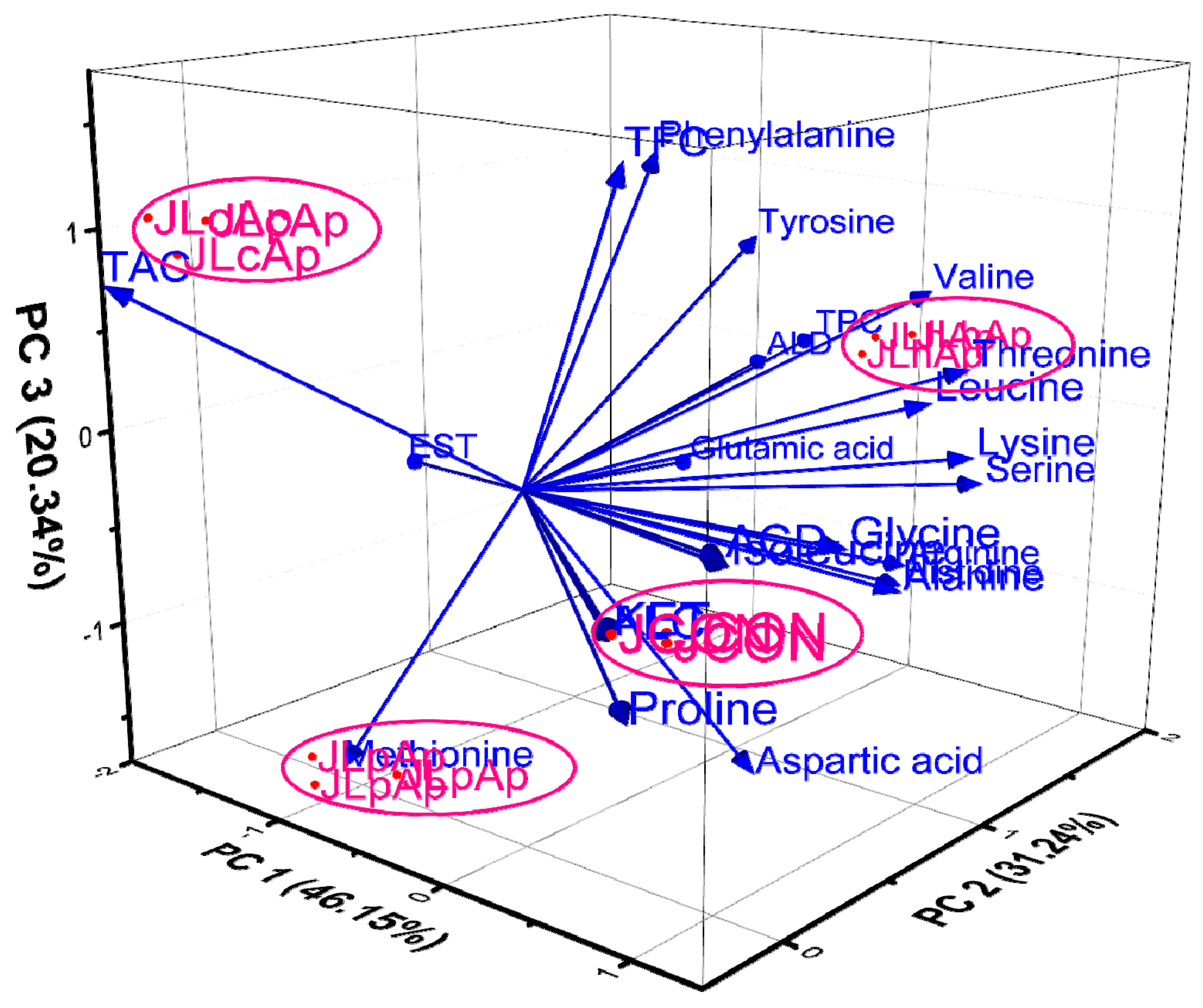
3.4. Assessment of Free Amino Acid Content
3.5. Effect of Lactic–Acetic Acid Co-Fermentation on Phenolics of Bicultured Jujube Purees
3.6. Smell Analysis with Electronic Nose (E-Nose)
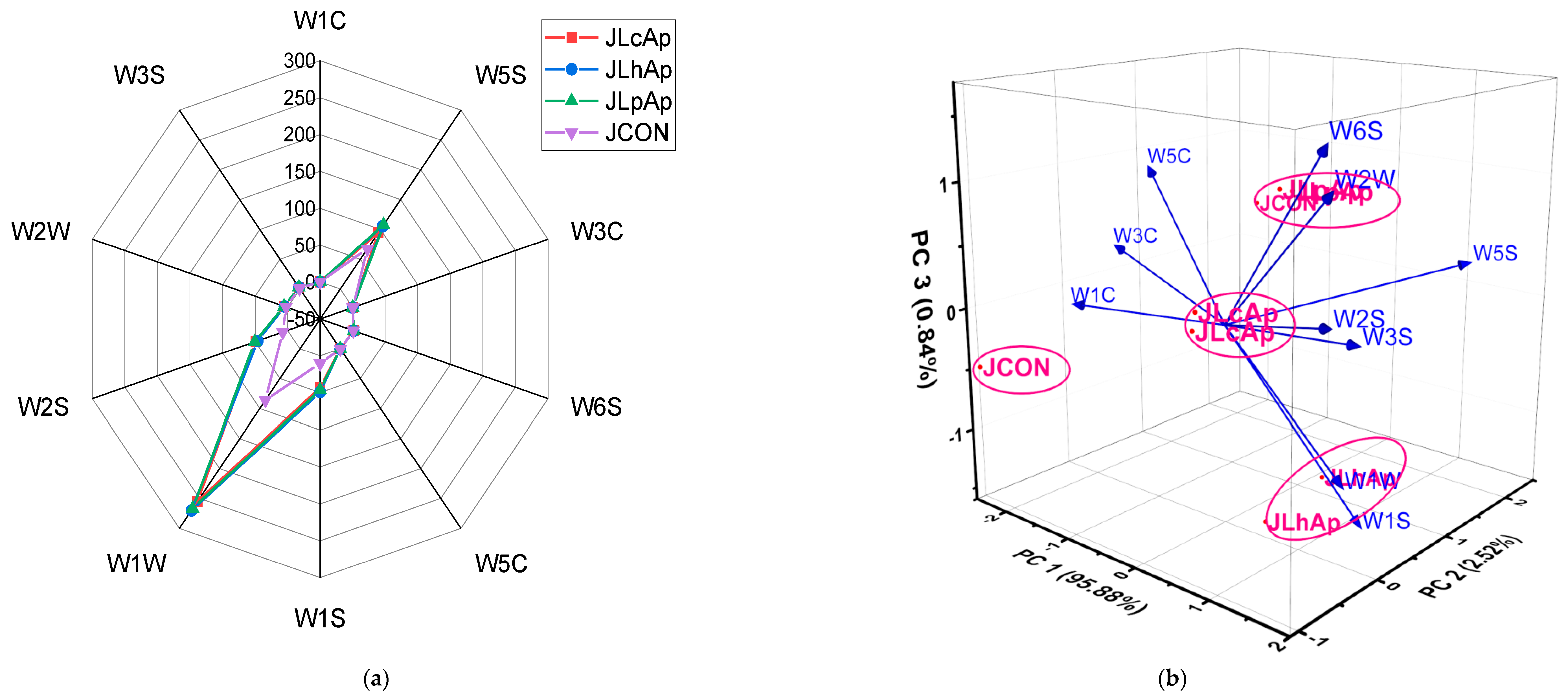
3.7. Volatile Profiling Using HS-SPME/GC–MS
3.8. Effect of Bicultural Fermentation on the Microstructural Changes
3.9. Principal Component Analysis (PCA)
4. Conclusions and Future Studies
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AAB | acetic acid bacteria |
| ABTS-RSA | 2,2-azino-bis-3-ethylbenzothiazoline-6-sulfonic acid radical scavenging activity |
| b* | yellowness-blueness |
| DPPH-RSA | 2,2-diphenyl-1-picrylhydrazyl radical scavenging activity |
| E-nose | electronic nose |
| FAA | free amino acid |
| HPLC | high-performance liquid chromatography |
| HS-SPME/GC–MS | headspace solid-phase microextraction gas chromatography–mass spectrometry |
| JCON | pasteurized puree without inoculation |
| JLcAp | Lacticaseibacillus casei Lc 122-Acetobacter pasteurianus Ap-As.1.41 HuNiang 1.01 puree |
| JLhAp | Lactobacillus helveticus Lh 43-A. pasteurianus Ap-As.1.41 HuNiang 1.01 puree |
| JLpAp | Lactiplantibacillus plantarum Lp 28-A. pasteurianus Ap-As.1.41 HuNiang 1.01 puree |
| JP | bicultured jujube puree |
| LAB | lactic acid bacteria |
| SEM | scanning electron microscopy |
| TAC | total anthocyanin content |
| TFC | total flavonol content |
| TPC | total phenolic content |
References
- Rashwan, A.K.; Karim, N.; Shishir, M.R.I.; Bao, T.; Lu, Y.; Chen, W. Jujube fruit: A potential nutritious fruit for the development of functional food products. J. Funct. Foods 2020, 75, 104205. [Google Scholar] [CrossRef]
- Liu, M.; Wang, J.; Wang, L.; Liu, P.; Zhao, J.; Zhao, Z.; Yao, S.; Stanica, F.; Liu, Z.; Wang, L.; et al. The historical and current research progress on jujube-a superfruit for the future. Hortic. Res. 2020, 7, 119. [Google Scholar] [CrossRef] [PubMed]
- Pu, Y.; Ding, T.; Wang, W.; Xiang, Y.; Ye, X.; Li, M.; Liu, D. Effect of harvest, drying and storage on the bitterness, moisture, sugars, free amino acids and phenolic compounds of jujube fruit (Zizyphus jujuba cv. Junzao). J. Sci. Food Agric. 2018, 98, 628–634. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Zhang, S.; Xu, X.; Lao, F.; Wu, J. Volatile and non-volatile profiles in jujube pulp co-fermented with lactic acid bacteria. LWT 2022, 154, 112772. [Google Scholar] [CrossRef]
- Rajendran, S.; Silcock, P.; Bremer, P. Flavour Volatiles of Fermented Vegetable and Fruit Substrates: A Review. Molecules 2023, 28, 3236. [Google Scholar] [CrossRef] [PubMed]
- Mollov, P.; Denkova-Kostova, R.; Goranov, B.; Tomova, T.; Yanakieva, V.; Blazheva, D.; Denkova, Z.; Kostov, G.; Ivanov, G.; Mihalev, K.; et al. Investigation of probiotic properties of Lactobacillus helveticus 2/20 isolated from rose blossom of Rosa damascena Mill. In BIO Web of Conferences; EDP Sciences: Les Ulis, France, 2023; Volume 58. [Google Scholar] [CrossRef]
- Song, J.; Wang, J.; Wang, X.; Zhao, H.; Hu, T.; Feng, Z.; Lei, Z.; Li, W.; Zheng, Y.; Wang, M. Improving the Acetic Acid Fermentation of Acetobacter pasteurianus by Enhancing the Energy Metabolism. Front. Bioeng. Biotechnol. 2022, 10, 815614. [Google Scholar] [CrossRef]
- Xia, M.; Zhang, X.; Xiao, Y.; Sheng, Q.; Tu, L.; Chen, F.; Yan, Y.; Zheng, Y.; Wang, M. Interaction of acetic acid bacteria and lactic acid bacteria in multispecies solid-state fermentation of traditional Chinese cereal vinegar. Front. Microbiol. 2022, 13, 964855. [Google Scholar] [CrossRef]
- Fu, L.; Yang, J.; Shang, H.; Song, J. Changes of characteristic sugar, fatty acid, organic acid and amino acid in jujubes at different dry mature stages. J. Food Compos. Anal. 2021, 104, 104104. [Google Scholar] [CrossRef]
- Yan, M.; Wang, Y.; Watharkar, R.B.; Pu, Y.; Wu, C.; Lin, M.; Lu, D.; Liu, M.; Bao, J.; Xia, Y. Physicochemical and antioxidant activity of fruit harvested from eight jujube (Ziziphus jujuba Mill.) cultivars at different development stages. Sci. Rep. 2022, 12, 2272. [Google Scholar] [CrossRef]
- Song, L.; Zheng, J.; Zhang, L.; Yan, S.; Huang, W.; He, J.; Liu, P. Phytochemical Profiling and Fingerprint Analysis of Chinese Jujube (Ziziphus jujuba Mill.) Leaves of 66 Cultivars from Xinjiang Province. Molecules 2019, 24, 4528. [Google Scholar] [CrossRef]
- Gao, Q.-H.; Wu, P.-T.; Liu, J.-R.; Wu, C.-S.; Parry, J.W.; Wang, M. Physico-chemical properties and antioxidant capacity of different jujube (Ziziphus jujuba Mill.) cultivars grown in loess plateau of China. Sci. Hortic. 2011, 130, 67–72. [Google Scholar] [CrossRef]
- Kwaw, E.; Ma, Y.; Tchabo, W.; Apaliya, M.T.; Wu, M.; Sackey, A.S.; Xiao, L.; Tahir, H.E. Effect of lactobacillus strains on phenolic profile, color attributes and antioxidant activities of lactic-acid-fermented mulberry juice. Food Chem. 2018, 250, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Maoz, I.; Lewinsohn, E.; Gonda, I. Amino acids metabolism as a source for aroma volatiles biosynthesis. Curr. Opin. Plant Biol. 2022, 67, 102221. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Bi, J.; Chen, Q.; Wu, X.; Lyu, Y.; Meng, X. Assessment of sugar content, fatty acids, free amino acids, and volatile profiles in jujube fruits at different ripening stages. Food Chem. 2019, 270, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Boateng, I.D.; Zhang, W.; Li, Y.Y.; Saalia, F.K.; Yang, X.M. Non-thermal pretreatment affects Ginkgo biloba L. seed’s product qualities, sensory, and physicochemical properties. J. Food Sci. 2022, 87, 94–111. [Google Scholar] [CrossRef]
- Li, T.; Jiang, T.; Liu, N.; Wu, C.; Xu, H.; Lei, H. Biotransformation of phenolic profiles and improvement of antioxidant capacities in jujube juice by select lactic acid bacteria. Food Chem. 2021, 339, 127859. [Google Scholar] [CrossRef]
- Pan, X.; Li, J.; Lao, F.; Hou, X.; Gao, L.; Wu, J. Phenolic characterization of fermented jujube puree by HPLC-MS/MS and their release during in vitro dynamic digestion. Food Chem. 2023, 413, 135630. [Google Scholar] [CrossRef]
- Li, J.; Zhao, W.; Pan, X.; Lao, F.; Liao, X.; Shi, Y.; Wu, J. Improvement of antioxidant properties of jujube puree by biotransformation of polyphenols via Streptococcus thermophilus fermentation. Food Chem. X 2022, 13, 100214. [Google Scholar] [CrossRef]
- Boasiako, T.A.; Yinka, A.A.; Yuqing, X.; Boateng, I.D.; Ma, Y. Tri-cultured lactic-acetic acid co-fermentation improves stored jujube puree functionality, physicochemical, volatile compounds, and sensory characteristics. Food Biosci. 2024, 57, 103534. [Google Scholar] [CrossRef]
- Boateng, I.D.; Yang, X.-M.; Tahany, A.A.A.; Li, Y.-Y.; Yolandani. Drying methods affect organoleptic and physicochemical properties of rehydrated ginkgo seed slices. Ind. Crops Prod. 2021, 160, 113166. [Google Scholar] [CrossRef]
- Dou, Z.; Chen, C.; Fu, X.; Liu, R. A dynamic view on the chemical composition and bioactive properties of mulberry fruit using an in vitro digestion and fermentation model. Food Funct. 2022, 13, 4142–4157. [Google Scholar] [CrossRef] [PubMed]
- Kwaw, E.; Ma, Y.; Tchabo, W.; Sackey, A.S.; Apaliya, M.T.; Xiao, L.; Wu, M.; Sarpong, F. Ultrasonication effects on the phytochemical, volatile and sensorial characteristics of lactic acid fermented mulberry juice. Food Biosci. 2018, 24, 17–25. [Google Scholar] [CrossRef]
- Chen, Y.; Wei, Z.; Zhang, T.; Ng, K.H.; Ye, J.; He, W.; Lozano, J. Physicochemical, Electronic Nose and Tongue, Sensory Evaluation Determination Combined with Chemometrics to Characterize Ficus hirta Vahl. (Moraceae) Beer. J. Food Qual. 2022, 2022, 8948603. [Google Scholar] [CrossRef]
- Wahia, H.; Zhou, C.; Mustapha, A.T.; Amanor-Atiemoh, R.; Mo, L.; Fakayode, O.A.; Ma, H. Storage effects on the quality quartet of orange juice submitted to moderate thermosonication: Predictive modeling and odor fingerprinting approach. Ultrason. Sonochem. 2020, 64, 104982. [Google Scholar] [CrossRef] [PubMed]
- Bonah, E.; Huang, X.; Yi, R.; Aheto, J.H.; Osae, R.; Golly, M. Electronic nose classification and differentiation of bacterial foodborne pathogens based on support vector machine optimized with particle swarm optimization algorithm. J. Food Process Eng. 2019, 42, 13236. [Google Scholar] [CrossRef]
- Dinu, L.; Lordache, O.; Vamanu, E. Scanning Electron Microscopy Study on the Biodeterioration of Natural Fiber Materials Compared to Disposable Hygiene and Sanitary Products. Fermentation 2022, 8, 287. [Google Scholar] [CrossRef]
- Mustapha, A.T.; Zhou, C. Novel assisted/unassisted ultrasound treatment: Effect on respiration rate, ethylene production, enzymes activity, volatile composition, and odor of cherry tomato. LWT 2021, 149, 111779. [Google Scholar] [CrossRef]
- Boateng, I.D.; Yang, X.-M. Do non-thermal pretreatments followed by intermediate-wave infrared drying affect toxicity, allergenicity, bioactives, functional groups, and flavor components of Ginkgo biloba seed? A case study. Ind. Crops Prod. 2021, 165, 113421. [Google Scholar] [CrossRef]
- Liu, G.; Wu, M.; Li, Y.; Qayyum, N.; Li, X.; Zhang, J.; Wang, C. The effect of different pretreatment methods on jujube juice and lactic acid bacteria-fermented jujube juice. LWT 2023, 181, 114692. [Google Scholar] [CrossRef]
- Ekumah, J.-N.; Ma, Y.; Akpabli-Tsigbe, N.D.K.; Kwaw, E.; Jie, H.; Quaisie, J.; Manqing, X.; Johnson Nkuma, N.A. Effect of selenium supplementation on yeast growth, fermentation efficiency, phytochemical and antioxidant activities of mulberry wine. LWT 2021, 146, 111425. [Google Scholar] [CrossRef]
- Dzah, C.S.; Duan, Y.; Zhang, H.; Ma, H. Effects of pretreatment and type of hydrolysis on the composition, antioxidant potential and HepG2 cytotoxicity of bound polyphenols from Tartary buckwheat (Fagopyrum tataricum L. Gaerth) hulls. Food Res. Int. 2021, 142, 110187. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Lee, M.J.; Choi, Y.; Park, S.J.; Lee, M.; Min, S.G.; Park, S.; Seo, H.; Yun, Y. Free Amino Acid and Volatile Compound Profiles of Jeotgal Alternatives and Its Application to Kimchi. Foods 2021, 10, 423. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, Y.; Wang, W.; Zheng, F.; Chen, F. Comparison of volatile compositions of 15 different varieties of Chinese jujube (Ziziphus jujuba Mill.). J. Food Sci. Technol. 2019, 56, 1631–1640. [Google Scholar] [CrossRef] [PubMed]
- Alenyorege, E.A.; Ma, H.; Aheto, J.H.; Agyekum, A.A.; Zhou, C. Effect of sequential multi-frequency ultrasound washing processes on quality attributes and volatile compounds profiling of fresh-cut Chinese cabbage. LWT 2020, 117, 108666. [Google Scholar] [CrossRef]
- Chen, K.; Fan, D.; Fu, B.; Zhou, J.; Li, H. Comparison of physical and chemical composition of three chinese jujube (Ziziphus jujuba Mill.) cultivars cultivated in four districts of Xinjiang region in China. Food Sci. Technol. 2019, 39, 912–921. [Google Scholar] [CrossRef]
- Krakowska-Sieprawska, A.; Rafinska, K.; Walczak-Skierska, J.; Buszewski, B. The Influence of Plant Material Enzymatic Hydrolysis and Extraction Conditions on the Polyphenolic Profiles and Antioxidant Activity of Extracts: A Green and Efficient Approach. Molecules 2020, 25, 2074. [Google Scholar] [CrossRef] [PubMed]
- Degrain, A.; Manhivi, V.; Remize, F.; Garcia, C.; Sivakumar, D. Effect of Lactic Acid Fermentation on Color, Phenolic Compounds and Antioxidant Activity in African Nightshade. Microorganisms 2020, 8, 1324. [Google Scholar] [CrossRef]
- Ye, M.; Yue, T.; Yuan, Y. Evolution of polyphenols and organic acids during the fermentation of apple cider. J. Sci. Food Agric. 2014, 94, 2951–2957. [Google Scholar] [CrossRef]
- Acosta-Estrada, B.A.; Gutierrez-Uribe, J.A.; Serna-Saldivar, S.O. Bound phenolics in foods, a review. Food Chem. 2014, 152, 46–55. [Google Scholar] [CrossRef]
- Xue, Y.; Zhang, Y.; Grace, S.; He, Q. Functional expression of an Arabidopsis p450 enzyme, p -coumarate-3-hydroxylase, in the cyanobacterium Synechocystis PCC 6803 for the biosynthesis of caffeic acid. Appl. Phycol. 2014, 26, 219–226. [Google Scholar] [CrossRef]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, E.A.; Bai, J.; Plotto, A.; Dea, S. Electronic Noses and Tongues: Applications for the Food and Pharmaceutical Industries. Sensors 2011, 11, 4744–4766. [Google Scholar] [CrossRef] [PubMed]
- Shishir, M.R.I.; Karim, N.; Gowd, V.; Xie, J.; Zheng, X.; Chen, W. Pectin-chitosan conjugated nanoliposome as a promising delivery system for neohesperidin: Characterization, release behavior, cellular uptake, and antioxidant property. Food Hydrocoll. 2019, 95, 432–444. [Google Scholar] [CrossRef]
- Rauf, A.; Imran, M.; Abu-Izneid, T.; Iahtisham Ul, H.; Patel, S.; Pan, X.; Naz, S.; Sanches Silva, A.; Saeed, F.; Rasul Suleria, H.A. Proanthocyanidins: A comprehensive review. Biomed. Pharmacother. 2019, 116, 108999. [Google Scholar] [CrossRef]
- Liang, J.R.; Deng, H.; Hu, C.Y.; Zhao, P.T.; Meng, Y.H. Vitality, fermentation, aroma profile, and digestive tolerance of the newly selected Lactiplantibacillus plantarum and Lacticaseibacillus paracasei in fermented apple juice. Front. Nutr. 2022, 9, 1045347. [Google Scholar] [CrossRef]
- Chen, Q.; Song, J.; Bi, J.; Meng, X.; Wu, X. Characterization of volatile profile from ten different varieties of Chinese jujubes by HS-SPME/GC-MS coupled with E-nose. Food Res. Int. 2018, 105, 605–615. [Google Scholar] [CrossRef]
- Boateng, I.D.; Yang, X.-M. Thermal and non-thermal processing affect Maillard reaction products, flavor, and phytochemical profiles of Ginkgo biloba seed. Food Biosci. 2021, 41, 101044. [Google Scholar] [CrossRef]

| Parameters | JP | |||
|---|---|---|---|---|
| JLcAp | JLhAp | JLpAp | JCON | |
| Cell count ( CFU/mL): | ||||
| Before fermentation: LAB | 8.41 ± 0.19 a,b | 8.83 ± 0.14 a | 8.85 ± 0.23 a | ˂1 c |
| AAB: | 8.87 ± 0.31 a | 8.87 ± 0.31 a | 8.87 ± 0.31 a | ˂1 b |
| After fermentation: LAB | 11.10 ± 0.11 b | 12.20 ± 0.17 a | 11.39 ± 0.42 b | ˂1 c |
| AAB | 11.81 ± 0.07 b | 12.07 ± 0.15 a,b | 12.26 ± 0.19 a | ˂1 c |
| Antioxidant properties: | ||||
| ABTS-RSA (mg AAE/100 g FW) | 33.41 ± 0.13 c | 34.52 ± 0.07 b | 34.61 ± 0.04 b | 36.70 ± 0.07 a |
| DPPH-RSA (mg AAE/100 g FW) | 34.70 ± 0.04 d | 35.68 ± 0.04 c | 35.96 ± 0.04 b | 38.15 ± 0.04 a |
| Colorimetric: | ||||
| ΔE | 4.99 ± 2.41 b | 9.66 ± 1.69 a | 3.07 ± 0.62 b,c | - |
| Amino Acids | JP (mg/100 g) | |||
|---|---|---|---|---|
| JLcAp | JLhAp | JLpAp | JCON | |
| L-Aspartic acid | 23.35 ± 0.05 d | 26.45 ± 0.11 c | 27.46 ± 0.11 a | 27.10 ± 0.10 b |
| L-Threonine | 2.00 ± 0.05 b | 2.41 ± 0.05 a | 1.93 ± 0.05 b | 2.30 ± 0.10 a |
| L-Serine | 2.85 ± 0.11 b | 3.47 ± 0.00 a | 3.03 ± 0.09 b | 3.30 ± 0.10 a |
| L-Glutamic acid | 7.57 ± 0.05 c | 8.76 ± 0.11 a | 8.37 ± 0.14 b | 6.93 ± 0.06 d |
| L-Glycine | 2.03 ± 0.05 c | 2.57 ± 0.11 b | 2.09 ± 0.09 c | 3.20 ± 0.10 a |
| L-Alanine | 3.82 ± 0.05 c | 4.23 ± 0.09 a,b | 4.01 ± 0.14 b,c | 4.40 ± 0.10 a |
| L-Valine | 2.60 ± 0.11 b | 4.82 ± 0.11 a | 1.99 ± 0.09 c | 2.83 ± 0.06 b |
| L-Methionine | nd | nd | 0.85 ± 0.09 a | nd |
| L-Isoleucine | 1.41 ± 0.19 c | 1.84 ± 0.07 b | 1.23 ± 0.09 c | 3.70 ± 0.10 a |
| L-Leucine | 2.32 ± 0.05 b | 2.82 ± 0.00 a | 2.15 ± 0.05 c | 2.93 ± 0.06 a |
| L-Tyrosine | 1.00 ± 0.05 a,b | 1.15 ± 0.10 a | 0.85 ± 0.01 b | 0.83 ± 0.06 b |
| L-Phenylalanine | 2.57 ± 0.11 a | 2.66 ± 0.05 a | 0.76 ± 0.00 c | 1.17 ± 0.06 b |
| L-Histidine | 5.51 ± 0.05 c | 6.23 ± 0.11 a | 6.00 ± 0.14 a,b | 5.93 ± 0.06 b |
| L-Lysine | 2.75 ± 0.05 c | 5.10 ± 0.05 a | 2.91 ± 0.05 b | 5.17 ± 0.06 a |
| L-Arginine | 2.19 ± 0.05 c | 2.82 ± 0.09 a | 2.59 ± 0.05 b | 2.53 ± 0.06 b |
| L-Proline | 77.65 ± 0.09 d | 81.85 ± 0.05 c | 86.89 ± 0.55 b | 111.70 ± 0.10 a |
| Total free amino acids | 139.62 ± 1.15 d | 157.17 ± 1.12 b | 153.11 ± 1.80 c | 184.03 ± 1.16 a |
| Phenolic Compounds | MF | RT (min) | Concentration (mg/100 g) | |||
|---|---|---|---|---|---|---|
| JLcAp | JLhAp | JLpAp | JCON | |||
| Phenolic acids | ||||||
| Chlorogenic acid | C16H18O9 | 11.72 | 0.427 ± 0.000 c | 0.973 ± 0.000 b | 0.385 ± 0.000 d | 1.293 ± 0.000 a |
| Sinapic acid | C11H12O5 | 19.61 | 2.202 ± 0.000 a | 2.011 ± 0.000 b | 1.938 ± 0.000 c | 0.845 ± 0.000 d |
| Ferulic acid | C10H10O4 | 19.66 | 0.731 ± 0.000 a | 0.702 ± 0.000 b | 0.679 ± 0.000 c | 0.335 ± 0.000 d |
| Gallic acid | C7H6O5 | 6.45 | 0.615 ± 0.000 c | 0.633 ± 0.000 b | 0.639 ± 0.000 a | n.d |
| Neochlorogenic acid | C16H18O9 | 9.63 | 0.357 ± 0.000 d | 2.646 ± 0.000 a | 1.306 ± 0.000 b | 0.747 ± 0.000 c |
| Caffeic acid | C9H8O4 | 14.47 | 0.513 ± 0.000 b | 0.898 ± 0.000 a | n.d | 0.235 ± 0.000 c |
| Protocatechuic acid | C7H6O4 | 9.85 | n.d | 0.091 ± 0.000 a | 0.034 ± 0.000 b | n.d |
| Syringic acid | C9H10O5 | 15.19 | 0.023 ± 0.000 b | 0.268 ± 0.000 a | n.d | n.d |
| 2,3,4-trihydroxybenzoic acid | C7H6O5 | 9.10 | 0.147 ± 0.000 c | 0.231 ± 0.000 a | 0.132 ± 0.000 d | 0.168 ± 0.000 b |
| P-coumaric acid | C9H8O3 | 18.56 | n.d | n.d | n.d | 0.057 ± 0.000 a |
| Total phenolic acids | 5.016 ± 0.001 b | 8.453 ± 0.000 a | 5.115 ± 0.000 b | 3.681 ± 0.000 c | ||
| Flavonols | ||||||
| Morin | C15H10O7 | 23.51 | 1.088 ± 0.000 a | n.d | n.d | 0.721 ± 0.000 b |
| Quercetin | C15H10O7 | 23.65 | 1.560 ± 0.000 a | n.d | n.d | 1.293 ± 0.000 b |
| Catechin | C15H14O6 | 12.57 | 0.779 ± 0.000 d | 2.983 ± 0.000 a | 1.139 ± 0.000 c | 1.600 ± 0.000 b |
| Rutin | C27H30O16 | 18.37 | 0.987 ± 0.000 a | 0.950 ± 0.000 b | 0.949 ± 0.000 c | 0.402 ± 0.000 d |
| Total flavonols | 4.416 ± 0.001 a | 3.933 ± 0.000 c | 2.088 ± 0.000 d | 4.016 ± 0.000 b | ||
| Anthocyanins | ||||||
| Cyanidin-3-O-rutinoside | C27H31O15 | 10.64 | 5.146 ± 0.001 a | 3.876 ± 0.000 d | 4.679 ± 0.000 c | 5.102 ± 0.000 b |
| Peonidin-3-O-glucoside | C22H23O12 | 12.21 | 10.642 ± 0.000 b | 6.703 ± 0.000 d | 15.119 ± 0.000 a | 10.343 ± 0.000 c |
| Peonidin-3,5-diglucoside | C28H33O16 | 9.94 | 34.133 ± 0.000 a | 27.484 ± 0.000 b | 19.851 ± 0.000 d | 25.193 ± 0.000 c |
| Total anthocyanins | 49.921 ± 0.001 a | 38.063 ± 0.000 d | 39.649 ± 0.000 c | 40.638 ± 0.000 b | ||
| Total polyphenol concentration | 59.352 ± 0.002 a | 50.450 ± 0.001 b | 46.851 ± 0.000 d | 48.335 ± 0.000 c | ||
| Volatile Groups | SN | Compound Name | CAS Number | Odor Description | Concentration of JP, ng/100 g FW (Mean ± SD) | |||
|---|---|---|---|---|---|---|---|---|
| JCON | JLcAp | JLhAp | JLpAp | |||||
| Alcohols | AL1 | 1-Decanol | 112-30-1 | Floral, fatty | 22.79 ± 0.03 a | n.d | n.d | n.d |
| AL2 | 1-Dodecanol | 112-53-8 | Sweet, fats, coconut | n.d | n.d | 2.22 ± 0.07 a | 1.66 ± 0.10 b | |
| AL3 | 1-Heptanol | 111-70-6 | Green | n.d | 3.22 ± 0.06 b | n.d | 3.39 ± 0.06 a | |
| AL4 | 1-Hexanol | 111-27-3 | Resin, flower, green | 3.02 ± 0.08 a | 2.34 ± 0.004 b | 2.20 ± 0.09 b,c | 2.15 ± 0.0 c | |
| AL5 | 1-Hexanol, 2-ethyl- | 104-76-7 | Green, rose | 14.51 ± 0.03 a | 1.19 ± 0.004 c | 1.28 ± 0.03 b | 1.10 ± 0.03 d | |
| AL6 | 1-Hexanol, 5-methyl- | 627-98-5 | n.d | n.d | 3.41 ± 0.05 a | n.d | ||
| AL7 | 1-Nonanol | 143-8-8 | Fatty | 1.24 ± 0.04 a | n.d | n.d | n.d | |
| AL8 | 1-Octanol | 111-87-5 | Chemical, metal, burnt | 4.08 ± 0.06 b | n.d | n.d | 6.21 ± 0.07 a | |
| AL9 | 2,3-Butanediol | 24347-58-8 | Buttery, creamy | 264.05 ± 2.54 a | 56.36 ± 0.11 d | 65.74 ± 0.03 b | 61.71 ± 0.05 c | |
| AL10 | 2,4-Di-tert-butylphenol | 96-76-4 | Phenolic | 3.48 ± 0.02 a | 1.14 ± 0.04 d | 1.83 ± 0.05 b | 1.40 ± 0.04 c | |
| AL11 | 2-Butanol, 1-methoxy- | 53778-73-7 | n.d | n.d | 1.36 ± 0.04 a | n.d | ||
| AL12 | 2-Nonanol | 628-99-9 | Orange, rose, mushroom | 1.24 ± 0.04 b | n.d | 2.71 ± 0.04 a | n.d | |
| AL13 | Benzyl alcohol | 100-51-6 | Walnut, nutty | 14.95 ± 0.04 a | 4.95 ± 0.12 c | 5.13 ± 0.05 c | 5.42 ± 0.07 b | |
| AL14 | Phenol | 108-95-2 | Plastic, rubber | n.d | 6.43 ± 0.14 b | n.d | 7.26 ± 0.06 a | |
| AL15 | Phenol, 4-ethyl- | 123-7-9 | Phenol | n.d | 4.88 ± 0.13 a | n.d | 2.96 ± 0.05 b | |
| AL16 | Phenylethyl Alcohol | 60-12-8 | Floral, rosy, honey, spice | 5.09 ± 0.04 d | 83.19 ± 0.22 c | 90.72 ± 0.04 a | 84.93 ± 0.03 b | |
| AL17 | Thymol | 89-83-8 | Herb, pleasant | n.d | 13.75 ± 0.05 a | n.d | n.d | |
| Subtotal | 333.20 ± 2.70 a | 177.43 ± 0.22 b | 176.61 ± 0.06 b | 178.19 ± 0.40 b | ||||
| Acids | ACD1 | 2-Heptenoic acid | 18999-28-5 | Green, fruity | 7.87 ± 0.03 a | n.d | n.d | n.d |
| ACD2 | 2-Octenoic acid | 1470-50-4 | n.d | 1.11 ± 0.03 a | 0.89 ± 0.10 b | n.d | ||
| ACD3 | 3-Decenoic acid, (E)- | 53678-20-9 | n.d | 1.00 ± 0.05 b | 1.36 ± 0.10 a | n.d | ||
| ACD4 | 3-Hexenoic acid, (E)- | 1577-18-0 | Must, fat | 2.09 ± 0.03 a | n.d | n.d | n.d | |
| ACD5 | 3-Octenoic acid, (E)- | 5163-67-7 | 3.23 ± 0.06 a | n.d | 2.15 ± 0.06 b | n.d | ||
| ACD6 | Benzoic acid | 65-85-0 | Leather | 6.33 ± 0.01 b | n.d | 19.86 ± 0.07 a | 6.42 ± 0.07 b | |
| ACD7 | Benzoic acid, p-tert-butyl- | 98-73-7 | 1.48 ± 0.03 a | n.d | n.d | n.d | ||
| ACD8 | Butanoic acid, 2-methyl- | 116-53-0 | Cheesy, sweaty | 2.04 ± 0.03 a | n.d | n.d | n.d | |
| ACD9 | Butanoic acid, 3-methyl- | 503-74-2 | Rancid, cheesy, sweaty | 1.73 ± 0.03 a | n.d | n.d | n.d | |
| ACD10 | Dodecanoic acid | 143-7-7 | Rancid, moldy | 15.03 ± 0.01 a | 11.54 ± 0.03 c | 11.62 ± 0.05 c | 11.83 ± 0.04 b | |
| ACD11 | Heptanoic acid | 111-14-8 | Rancid, fatty | 4.04 ± 0.03 c | 4.52 ± 0.09 b | 11.54 ± 0.07 a | 3.71 ± 0.03 d | |
| ACD12 | Hexanoic acid | 142-62-1 | Cheesy, fatty | 48.76 ± 0.01 a | 6.63 ± 0.08 b | n.d | n.d | |
| ACD13 | Hydrocinnamic acid | 501-52-0 | Cheesy | 3.96 ± 0.06 b | 3.40 ± 0.05 c | 3.06 ± 0.06 d | 4.25 ± 0.06 a | |
| ACD14 | n-Decanoic acid | 334-48-5 | Fatty, citrus | 43.79 ± 0.04 a | 7.21 ± 0.13 c | 9.16 ± 0.09 b | 5.55 ± 0.06 d | |
| ACD15 | Nonanoic acid | 112-5-0 | Waxy, cheese-like | 14.42 ± 0.02 a | 10.45 ± 0.08 b | n.d | 3.34 ± 0.06 c | |
| ACD16 | Octanoic acid | 124-7-2 | Fatty, rancid | 24.43 ± 0.09 a | 10.98 ± 0.15 c | 22.27 ± 0.07 b | 9.72 ± 0.03 d | |
| ACD17 | Pentanoic acid | 109-52-4 | Sweet | 4.77 ± 0.03 a | 2.17 ± 0.03 b | n.d | n.d | |
| ACD18 | Pentanoic acid, 3-methyl- | 105-43-1 | Cheesy, fruity | 4.51 ± 0.01 a | n.d | n.d | n.d | |
| ACD19 | Tetradecanoic acid | 544-63-8 | Waxy, fatty, coconut | 2.01 ± 0.06 a | n.d | n.d | n.d | |
| Subtotal | 190.49 ± 0.51 a | 59.01 ± 0.39 c | 81.90 ± 0.18 b | 44.82 ± 0.19 d | ||||
| Ketones | KTN1 | 2-Pentanone, 5-methoxy- | 17429-4-8 | 8.66 ± 0.10 a | n.d | n.d | n.d | |
| KTN2 | 2-Hydroxy-3-hexanone | 54073-43-7 | 7.32 ± 0.04 a | n.d | n.d | n.d | ||
| KTN3 | 2-Nonanone | 821-55-6 | Vegetable, moldy | 12.66 ± 0.06 a | n.d | n.d | n.d | |
| Subtotal | 28.64 ± 0.20 a | |||||||
| Aldehydes | ALD1 | 2-Decenal, (E)- | 3913-81-3 | Soap, tallow | n.d | n.d | 1.86 ± 0.06 a | 1.66 ± 0.10 b |
| ALD2 | Benzaldehyde | 100-52-7 | Sweet, almond, cherry | n.d | 17.79 ± 0.09 c | 23.54 ± 0.05 a | 18.21 ± 0.05 b | |
| ALD3 | Benzaldehyde, 2,4-dimethyl- | 15764-16-6 | Cherry, almond, vanilla | 25.95 ± 0.04 a | 2.72 ± 0.05 d | 12.67 ± 0.06 b | 2.97 ± 0.09 c | |
| ALD5 | Benzeneacetaldehyde | 122-78-1 | Rose-like, honey, floral | 1.36 ± 0.04 d | 44.59 ± 0.07 b | 92.56 ± 0.07 a | 40.11 ± 0.09 c | |
| ALD6 | Decanal | 112-31-2 | Soap, tallow | n.d | n.d | n.d | 1.33 ± 0.08 a | |
| ALD7 | Heptanal | 111-71-7 | Fat, citrus, rancid | 1.12 ± 0.03 a | n.d | n.d | n.d | |
| ALD8 | Hexanal | 66-25-1 | Green, sweet | n.d | n.d | n.d | 2.72 ± 0.06 a | |
| ALD9 | Paraldehyde | 123-63-7 | Sweet, pleasant | n.d | n.d | n.d | 7.27 ± 0.07 a | |
| Subtotal | 28.43 ± 0.10 d | 65.10 ± 0.06 c | 130.62 ± 0.03 a | 74.27 ± 0.08 b | ||||
| Esters | EST1 | 1-Butanol, 2-methyl-, acetate | 624-41-9 | Fruity, floral | n.d | 16.90 ± 0.06 b | 16.34 ± 0.04 c | 27.00 ± 0.07 a |
| EST2 | 1-Butanol, 3-methyl-, acetate | 123-92-2 | Fruity, floral, sweet | 0.71 ± 0.04 d | 59.18 ± 0.06 b | 54.99 ± 0.04 c | 91.51 ± 0.07 a | |
| EST3 | 1-Methoxy-2-propyl acetate | 108-65-6 | Fruity | n.d | 1.88 ± 0.09 a | 1.92 ± 0.05 a | n.d | |
| EST4 | 2-Butanol, 3-methyl-, acetate | 5343-96-4 | Fruity | n.d | n.d | 1.91 ± 0.05 a | n.d | |
| EST5 | 3-Hexenoic acid, ethyl ester | 2396-83-0 | Fruity, brandy, wine-like | n.d | 1.43 ± 0.11 b | 1.91 ± 0.04 a | 1.27 ± 0.10 b | |
| EST6 | 3-Methyl-3-buten-1-ol, acetate | 5205-07-2 | Fruity | n.d | 2.39 ± 0.09 b | n.d | 2.82 ± 0.06 a | |
| EST7 | 7-Octenoic acid, ethyl ester | 35194-38-8 | Fruity | n.d | 2.66 ± 0.06 b | n.d | 2.84 ± 0.06 a | |
| EST8 | Acetic acid, 2-phenylethyl ester | 103-45-7 | Fruity, rose | n.d | 65.99 ± 0.06 c | 76.05 ± 0.05 b | 82.18 ± 0.06 a | |
| EST9 | Acetic acid, butyl ester | 123-86-4 | Fruity, sweet, solvent | n.d | n.d | n.d | 2.18 ± 0.03 a | |
| EST10 | Acetic acid, pentyl ester | 628-63-7 | Fruity, apple | n.d | n.d | n.d | 0.77 ± 0.10 a | |
| EST11 | Acetic acid, phenylmethyl ester | 140-11-4 | Fruity | n.d | 4.99 ± 0.08 b | 4.98 ± 0.05 b | 7.03 ± 0.05 a | |
| EST12 | Acetoin acetate | 4906-24-5 | Fruity | n.d | 7.82 ± 0.08 a | n.d | n.d | |
| EST13 | Benzeneacetic acid, ethyl ester | 101-97-3 | Fruity | n.d | n.d | n.d | 1.45 ± 0.03 a | |
| EST14 | Benzenepropanoic acid, ethyl ester | 2021-28-5 | Fruity | n.d | 16.82 ± 0.09 a | n.d | 16.17 ± 0.06 b | |
| EST15 | Benzoic acid, ethyl ester | 93-89-0 | Fruity, floral | 2.64 ± 0.03 d | 6.64 ± 0.04 c | 10.97 ± 0.10 a | 8.00 ± 0.06 b | |
| EST16 | cis-9-Tetradecenoic acid, propyl ester | Fruity | n.d | 1.25 ± 0.10 a | 0.98 ± 0.07 b | 0.86 ± 0.07 b | ||
| EST17 | Dodecanoic acid, ethyl ester | 106-33-2 | Fruity | n.d | 5.56 ± 0.03 b | 6.33 ± 0.08 a | 4.04 ± 0.04 c | |
| EST18 | Dodecyl tiglate | 1231959-17-3 | Fruity | n.d | n.d | n.d | 1.05 ± 0.03 a | |
| EST19 | Ethyl 4-acetoxybutanoate | 25560-91-2 | Fruity | n.d | 0.93 ± 0.04 b | n.d | 1.05 ± 0.03 a | |
| EST20 | Ethyl 9-hexadecenoate | 54546-22-4 | Fruity | n.d | 0.91 ± 0.09 a | n.d | n.d | |
| EST21 | Ethyl Acetate | 141-78-6 | Pineapple, fruity | n.d | n.d | 0.99 ± 0.03 a | 0.80 ± 0.06 b | |
| EST22 | Heptanoic acid, ethyl ester | 106-30-9 | Wine-like, fruity, brandy | n.d | n.d | 1.58 ± 0.09 a | n.d | |
| EST23 | Hexadecanoic acid, ethyl ester | 628-97-7 | Fruity | n.d | 2.87 ± 0.04 a | 1.92 ± 0.06 b | 1.37 ± 0.07 c | |
| EST24 | Hexanoic acid, ethyl ester | 123-66-0 | Fruity, apple, banana | 16.70 ± 0.03 b | 8.24 ± 0.09 d | 20.41 ± 0.06 a | 9.75 ± 0.06 c | |
| EST25 | Isoamyl lactate | 19329-89-6 | Fruity, banana, pear | n.d | 1.31 ± 0.10 a | n.d | 0.79 ± 0.11 b | |
| EST26 | Methyl salicylate | 119-36-8 | Fruity, peppermint | 2.69 ± 0.02 a | n.d | n.d | n.d | |
| EST27 | Octanoic acid, ethyl ester | 106-32-1 | Brandy, pear, musty | n.d | 3.01 ± 0.09 b | 5.31 ± 0.03 a | 2.92 ± 0.03 b | |
| EST28 | Pentanoic acid, ethyl ester | 539-82-2 | Fruity, berry | n.d | 2.17 ± 0.03 c | 2.28 ± 0.06 b | 2.43 ± 0.06 a | |
| EST29 | Propanoic acid, 2-hydroxy-, ethyl ester | 97-64-3 | Fruity | n.d | 2.95 ± 0.10 a | n.d | 0.96 ± 0.06 b | |
| EST30 | Propanoic acid, 2-methyl-, 3-hydroxy-2,2,4-trimethylpentyl ester | 77-68-9 | Fruity | n.d | 1.67 ± 0.10 b | 1.86 ± 0.07 a | 1.98 ± 0.05 a | |
| EST31 | Tetradecanoic acid, ethyl ester | 124-6-1 | Soap, mild, polish | n.d | 1.35 ± 0.07 a | n.d | n.d | |
| Subtotal | 22.75 ± 0.09 d | 220.86 ± 0.67 b | 210.71 ± 0.24 c | 271.22 ± 0.68 a | ||||
| Other(s) | NAT1 | Naphthalene, 1,2,3,4-tetrahydro-1,1,6-trimethyl- | 475-3-6 | tar | 7.57 ± 0.03 a | n.d | n.d | n.d |
| Total of volatile compounds | 611.07 ± 3.60 a | 522.38 ± 0.77 d | 599.85 ± 0.33 b | 568.49 ± 0.55 c | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boasiako, T.A.; Xiong, Y.; Boateng, I.D.; Appiagyei, J.; Li, Y.; Clark, K.; Aregbe, A.Y.; Yaqoob, S.; Ma, Y. Innovative Bicultured Lactic–Acetic Acid Co-fermentation Improves Jujube Puree’s Functionality and Volatile Compounds. Fermentation 2024, 10, 71. https://doi.org/10.3390/fermentation10010071
Boasiako TA, Xiong Y, Boateng ID, Appiagyei J, Li Y, Clark K, Aregbe AY, Yaqoob S, Ma Y. Innovative Bicultured Lactic–Acetic Acid Co-fermentation Improves Jujube Puree’s Functionality and Volatile Compounds. Fermentation. 2024; 10(1):71. https://doi.org/10.3390/fermentation10010071
Chicago/Turabian StyleBoasiako, Turkson Antwi, Yuqing Xiong, Isaac Duah Boateng, Jeffrey Appiagyei, Yanshu Li, Kerry Clark, Afusat Yinka Aregbe, Sanabil Yaqoob, and Yongkun Ma. 2024. "Innovative Bicultured Lactic–Acetic Acid Co-fermentation Improves Jujube Puree’s Functionality and Volatile Compounds" Fermentation 10, no. 1: 71. https://doi.org/10.3390/fermentation10010071
APA StyleBoasiako, T. A., Xiong, Y., Boateng, I. D., Appiagyei, J., Li, Y., Clark, K., Aregbe, A. Y., Yaqoob, S., & Ma, Y. (2024). Innovative Bicultured Lactic–Acetic Acid Co-fermentation Improves Jujube Puree’s Functionality and Volatile Compounds. Fermentation, 10(1), 71. https://doi.org/10.3390/fermentation10010071






