Fine-Tuning the Expression of the Glycolate Biosynthetic Pathway in Escherichia coli Using Synthetic Promoters
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Genetic Manipulations and Plasmid Construction
2.3. Fluorescence Analysis
2.4. Real-Time Quantitative PCR (RT-qPCR)
2.5. Batch and Fed-Batch Cultivation
2.6. Analytical Methods
2.7. Data Availability
3. Result
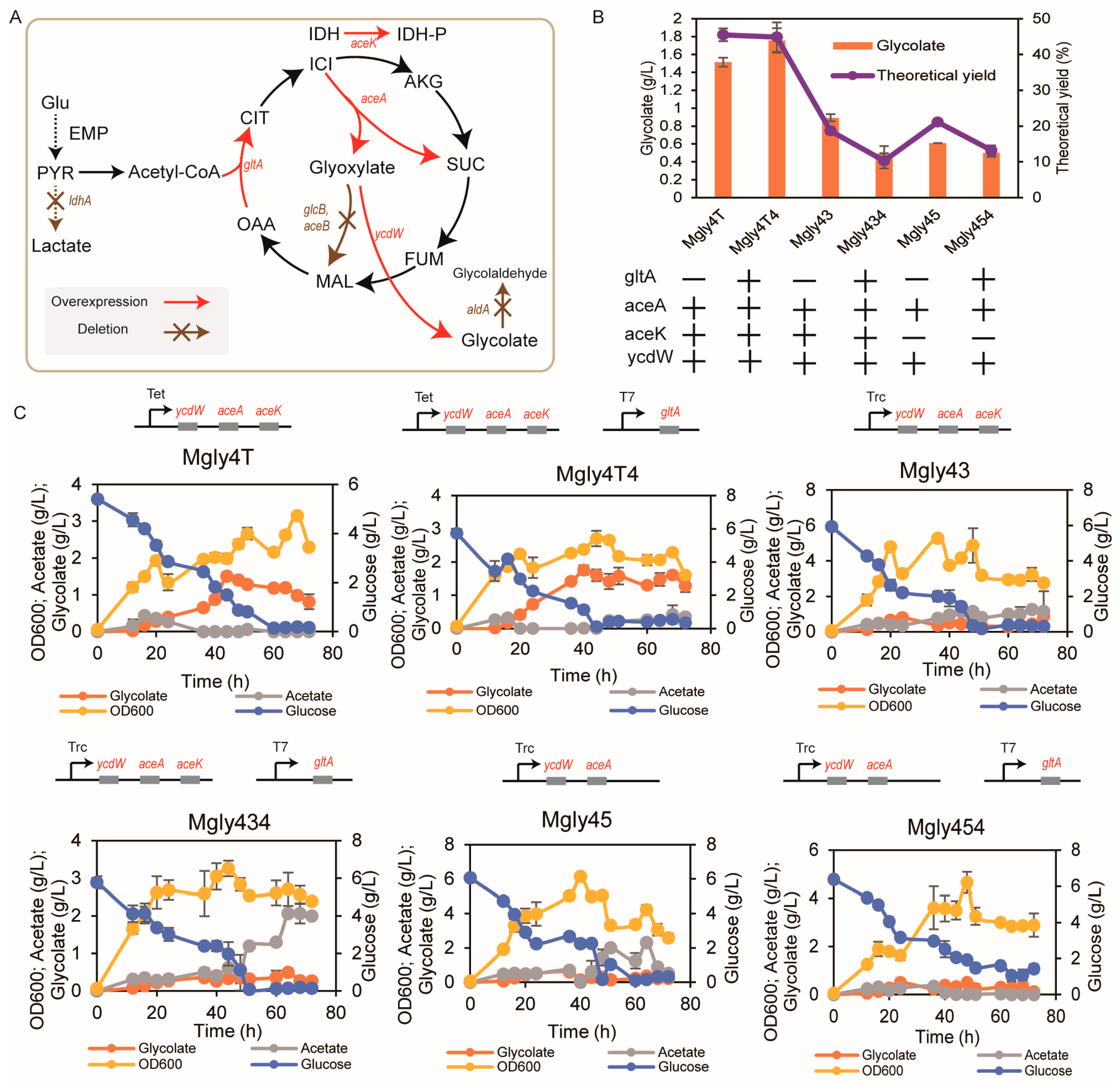
3.1. Gene Expression and Construction of the Modified Glycolate Pathway
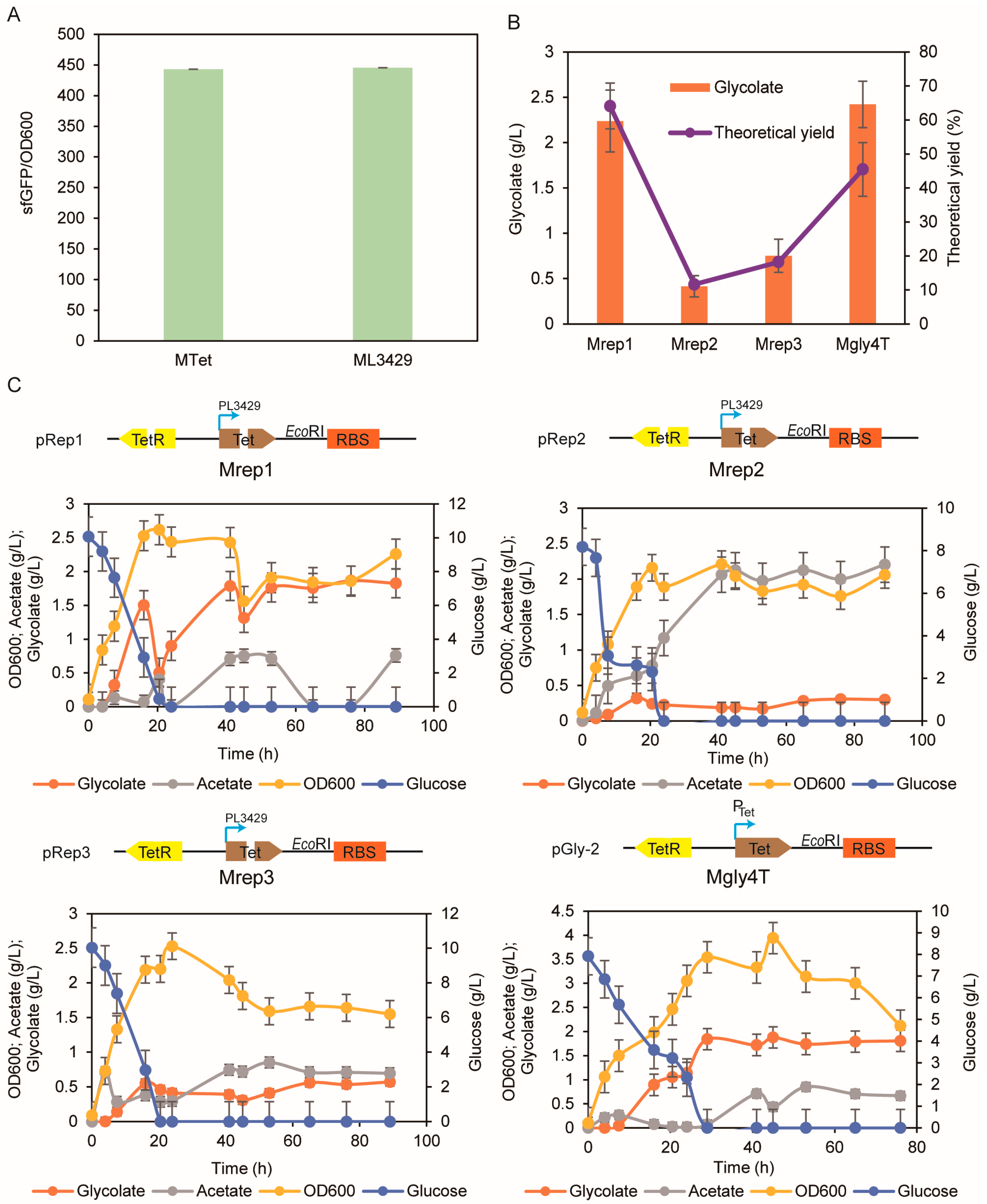
3.2. Replacement of PTet with the Synthetic Promoter PL3429
3.3. Optimizing the Different Synthetic Promoters for Glycolate Production
3.4. Production of Glycolate in 5-L Bioreactor
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Salusjarvi, L.; Havukainen, S.; Koivistoinen, O.; Toivari, M. Biotechnological production of glycolic acid and ethylene glycol: Current state and perspectives. Appl. Microbiol. Biotechnol. 2019, 103, 2525–2535. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.S.; Park, J.; Heo, Y.B.; Woo, H.M. Case study of xylose conversion to glycolate in Corynebacterium glutamicum: Current limitation and future perspective of the CRISPR-Cas systems. Enzyme Microb. Technol. 2020, 132, 109395–109399. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Zhang, L.; Zhou, S.; Deng, Y. Biosensor-based multigene pathway optimization for enhancing the production of glycolate. Appl. Microbiol. Biotechnol. 2021, 87, e00113-21. [Google Scholar] [CrossRef]
- Schad, A.; Wagner, H.; Wilhelm, C. Optimising biotechnological glycolate production in Chlamydomonas reinhardtii by improving carbon allocation towards the product. Chem. Eng. J. 2023, 459, 141432. [Google Scholar] [CrossRef]
- Choi, S.Y.; Park, S.J.; Kim, W.J.; Yang, J.E.; Lee, H.; Shin, J.; Lee, S.Y. One-step fermentative production of poly(lactate-co-glycolate) from carbohydrates in Escherichia coli. Nat. Biotechnol. 2016, 34, 435. [Google Scholar] [CrossRef]
- Samantaray, P.K.; Little, A.; Haddleton, D.M.; McNally, T.; Tan, B.; Sun, Z.; Huang, W.; Ji, Y.; Wan, C. Poly(glycolic acid) (PGA): A versatile building block expanding high performance and sustainable bioplastic applications. Green Chem. 2020, 22, 4055–4081. [Google Scholar] [CrossRef]
- Global Glycolic Acid Market by Grade (Cosmetic, Technical), Application (Personal Care & Dermatology, Industrial, Household) and Region (Asia Pacific, North America, Europe, South America, Middle East & Africa)—Forecast to 2027. Available online: https://www.researchandmarkets.com/reports/4846741/global-glycolic-acid-market-by-grade-cosmetic (accessed on 1 May 2022).
- Long, B.H.; Nishiyama, M.; Sato, R.; Tanaka, T.; Ohara, H.; Aso, Y. Production of glyoxylate from glucose in engineered Escherichia coli. Fermentation 2023, 9, 534–546. [Google Scholar] [CrossRef]
- Alkim, C.; Trichez, D.; Cam, Y.; Spina, L.; Francois, J.M.; Walther, T. The synthetic xylulose-1 phosphate pathway increases production of glycolic acid from xylose-rich sugar mixtures. Biotechnol. Biofuels 2016, 9, 201–211. [Google Scholar] [CrossRef]
- Cabulong, R.B.; Valdehuesa, K.N.G.; Ramos, K.R.M.; Nisola, G.M.; Lee, W.K.; Lee, C.R.; Chung, W.J. Enhanced yield of ethylene glycol production from D-xylose by pathway optimization in Escherichia coli. Enzyme Microb. Technol. 2017, 97, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Cam, Y.; Alkim, C.; Trichez, D.; Trebosc, V.; Vax, A.; Bartolo, F.; Besse, P.; Francois, J.M.; Walther, T. Engineering of a synthetic metabolic pathway for the assimilation of (D)-xylose into value-added chemicals. ACS Synth. Biol. 2016, 5, 607–618. [Google Scholar] [CrossRef]
- Jo, H.-J.; Yu, H.; Cheon, H.; Kim, J.-H.; Kim, G.Y.; Kim, B.S.; Kim, J.-S.; Park, J.-B. Multilayer engineering of an Escherichia coli-based biotransformation system to exclusively produce glycolic acid from formaldehyde. ACS Sustain. Chem. Eng. 2023, 11, 1078–1086. [Google Scholar] [CrossRef]
- Dischert, W.; Soucaille, P. Method for Producing High Amount of Glycolic Acid by Fermentation. U.S. Patent 8,945,888, 24 March 2009. [Google Scholar]
- Deng, Y.; Ma, N.; Zhu, K.J.; Mao, Y.; Wei, X.T.; Zhao, Y.Y. Balancing the carbon flux distributions between the TCA cycle and glyoxylate shunt to produce glycolate at high yield and titer in Escherichia coli. Metab. Eng. 2018, 46, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Koivistoinen, O.M.; Kuivanen, J.; Barth, D.; Turkia, H.; Pitkanen, J.P.; Penttila, M.; Richard, P. Glycolic acid production in the engineered yeasts Saccharomyces cerevisiae and Kluyveromyces lactis. Microb. Cell Factories 2013, 12, 82–97. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Yan, Q.; Jones, J.A.; Tang, Y.J.; Fong, S.S.; Koffas, M.A.G. Metabolic burden: Cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol. 2016, 34, 652–664. [Google Scholar] [CrossRef]
- Jervis, A.J.; Carbonell, P.; Taylor, S.; Sung, R.; Dunstan, M.S.; Robinson, C.J.; Breitling, R.; Takano, E.; Scrutton, N.S. SelProm: A queryable and predictive expression vector selection tool for Escherichia coli. ACS Synth. Biol. 2019, 8, 1478. [Google Scholar] [CrossRef]
- Marbach, A.; Bettenbrock, K. Lac operon induction in Escherichia coli: Systematic comparison of IPTG and TMG induction and influence of the transacetylase LacA. J. Biotechnol. 2012, 157, 82–88. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhao, M.; Zhou, S.H.; Zhao, Y.Y.; Li, G.H.; Deng, Y. Biosynthesis of adipic acid by a highly e fficient induction -free system in Escherichia coli. J. Biotechnol. 2020, 314, 8–13. [Google Scholar] [CrossRef]
- Leavitt, J.M.; Alper, H.S. Advances and current limitations in transcript-level control of gene expression. Curr. Opin. Biotechnol. 2015, 34, 98–104. [Google Scholar] [CrossRef]
- Bergman, D.T.; Jones, T.R.; Liu, V.; Ray, J.; Jagoda, E.; Siraj, L.; Kang, H.Y.; Nasser, J.; Kane, M.; Rios, A.; et al. Compatibility rules of human enhancer and promoter sequences. Nature 2022, 607, 176–184. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, H.; Xu, H.; Wei, L.; Liu, L.; Hu, Z.; Wang, X. Deep flanking sequence engineering for efficient promoter design using DeepSEED. Nat. Commun. 2023, 14, 6309. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Wei, L.; Li, S.; Liu, L.; Wang, X. Synthetic promoter design in Escherichia coli based on a deep generative network. Nucleic Acids Res. 2020, 48, 6403–6412. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.; Li, P.; Xie, X.; Li, J.; Liao, Y.; Ma, X.; Cai, D.; Chen, S. Construction and characterization of a gradient strength promoter library for fine-tuned gene expression in Bacillus licheniformis. ACS Synth. Biol. 2021, 10, 2331–2339. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Yuan, Z.; Wu, L.; Zhou, S.; Deng, Y. Precise prediction of promoter strength based on a de novo synthetic promoter library coupled with machine learning. ACS Synth. Biol. 2022, 11, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Pédelacq, J.-D.; Cabantous, S.; Tran, T.; Terwilliger, T.C.; Waldo, G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006, 24, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.H.; Dhamankar, H.; Tseng, H.-C.; Sheppard, M.J.; Reisch, C.R.; Prather, K.L.J. A platform pathway for production of 3-hydroxyacids provides a biosynthetic route to 3-hydroxy-γ-butyrolactone. Nat. Commun. 2013, 4, 1414–1422. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.J.; Li, G.H.; Wei, R.; Mao, Y.; Zhao, Y.Y.; He, A.Y.; Bai, Z.H.; Deng, Y. Systematic analysis of the effects of different nitrogen source and ICDH knockout on glycolate synthesis in Escherichia coli. J. Med. Biol. Eng. 2019, 13, 30–42. [Google Scholar] [CrossRef]
- Miyazaki, K. Creating random mutagenesis libraries by megaprimer PCR of whole plasmid (MEGAWHOP). Methods Mol. Biol. 2003, 231, 23–28. [Google Scholar]
- Liu, D.Y.; Mao, Z.T.; Guo, J.X.; Wei, L.Y.; Ma, H.W.; Tang, Y.J.; Chen, T.; Wang, Z.W.; Zhao, X.M. Construction, model-based analysis, and characterization of a promoter library for fine-tuned gene expression in Bacillus subtilis. ACS Synth. Biol. 2018, 7, 1785–1797. [Google Scholar] [CrossRef]
- Geyer, C.N.; Fowler, R.C.; Johnson, J.R.; Johnston, B.; Weissman, S.J.; Hawkey, P.; Hanson, N.D. Evaluation of CTX-M steady-state mRNA, mRNA half-life and protein production in various STs of Escherichia coli. J. Antimicrob. Chemother. 2016, 71, 607–616. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhao, M.; Huang, D.; Zhang, X.; Koffas, M.A.G.; Zhou, J.; Deng, Y. Metabolic engineering of Escherichia coli for producing adipic acid through the reverse adipate-degradation pathway. Metab. Eng. 2018, 47, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Ehrt, S.; Guo, X.Z.V.; Hickey, C.M.; Ryou, M.; Monteleone, M.; Riley, L.W.; Schnappinger, D. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 2005, 33, e21. [Google Scholar] [CrossRef] [PubMed]
- Pereira, B.; Li, Z.J.; De Mey, M.; Lim, C.G.; Zhang, H.R.; Hoeltgen, C.; Stephanopoulos, G. Efficient utilization of pentoses for bioproduction of the renewable two-carbon compounds ethylene glycol and glycolate. Metab. Eng. 2016, 34, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Pereira, B.; Zhang, H.R.; De Mey, M.; Lim, C.G.; Li, Z.J.; Stephanopoulos, G. Engineering a Novel Biosynthetic Pathway in Escherichia coli for Production of Renewable Ethylene Glycol. Biotechnol. Bioeng. 2016, 113, 376–383. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, M.; Shang, J.; Chen, J.; Zabed, H.M.; Qi, X. Fine-Tuning the Expression of the Glycolate Biosynthetic Pathway in Escherichia coli Using Synthetic Promoters. Fermentation 2024, 10, 67. https://doi.org/10.3390/fermentation10010067
Zhao M, Shang J, Chen J, Zabed HM, Qi X. Fine-Tuning the Expression of the Glycolate Biosynthetic Pathway in Escherichia coli Using Synthetic Promoters. Fermentation. 2024; 10(1):67. https://doi.org/10.3390/fermentation10010067
Chicago/Turabian StyleZhao, Mei, Jie Shang, Jiaojiao Chen, Hossain M. Zabed, and Xianghui Qi. 2024. "Fine-Tuning the Expression of the Glycolate Biosynthetic Pathway in Escherichia coli Using Synthetic Promoters" Fermentation 10, no. 1: 67. https://doi.org/10.3390/fermentation10010067
APA StyleZhao, M., Shang, J., Chen, J., Zabed, H. M., & Qi, X. (2024). Fine-Tuning the Expression of the Glycolate Biosynthetic Pathway in Escherichia coli Using Synthetic Promoters. Fermentation, 10(1), 67. https://doi.org/10.3390/fermentation10010067







