Systematic Analysis of Long Non-Coding RNAs in Inflammasome Activation in Monocytes/Macrophages
Abstract
:1. Introduction
2. Results
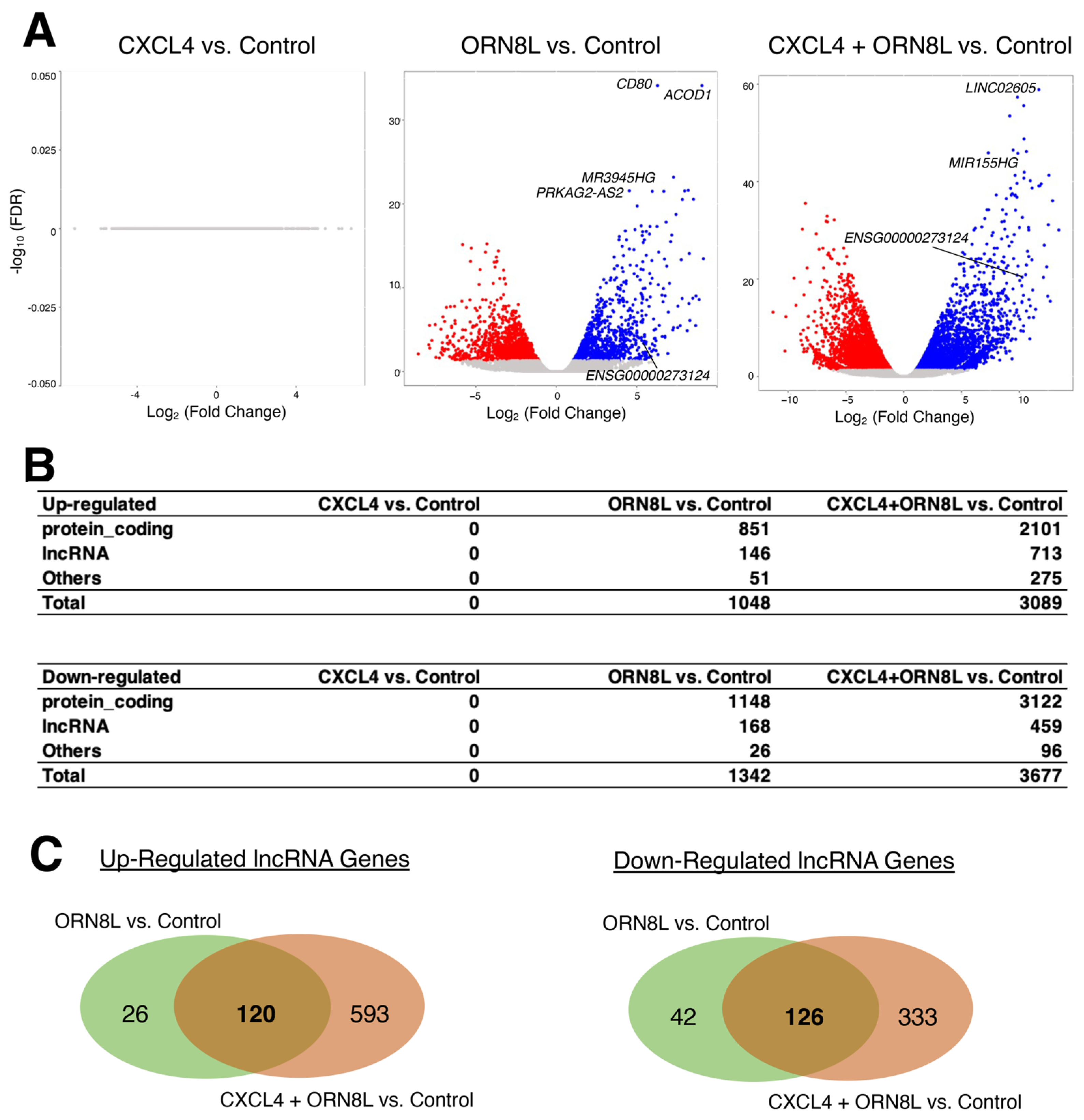
2.1. CXCL4 and TLR8 Co-Stimulation Induces the Expression of lncRNA Genes
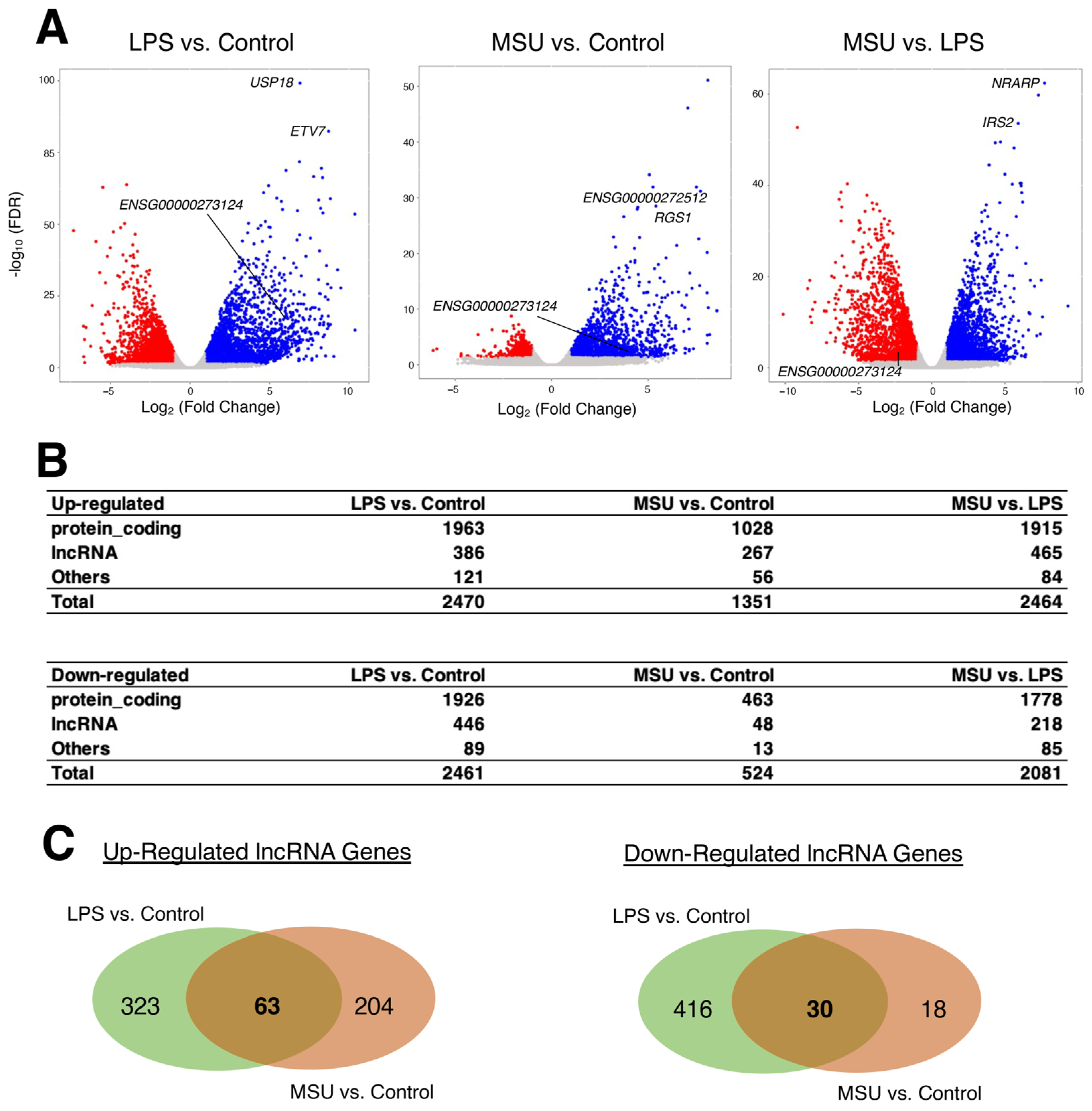
2.2. Distinctive Sets of lncRNA Genes Were Induced by LPS and MSU Treatments in Macrophages
2.3. Expression and Functional Analysis of Inflammasome-Regulated lncRNAs
2.4. Subcellular Localization of lncRNAs May Indicate Their Functionalities
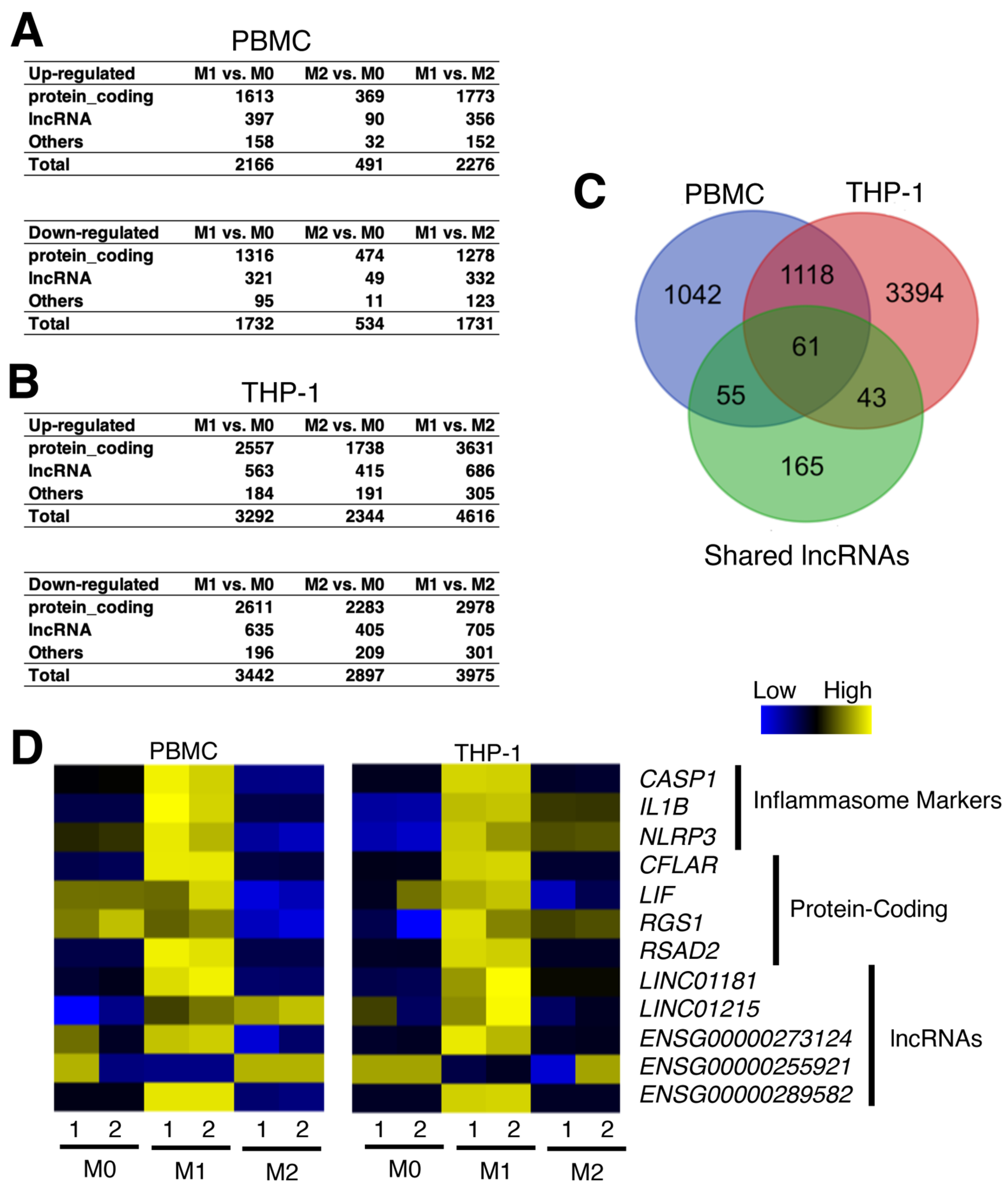
2.5. Polarization of Macrophages to Pro- and Anti-Inflammatory Macrophages
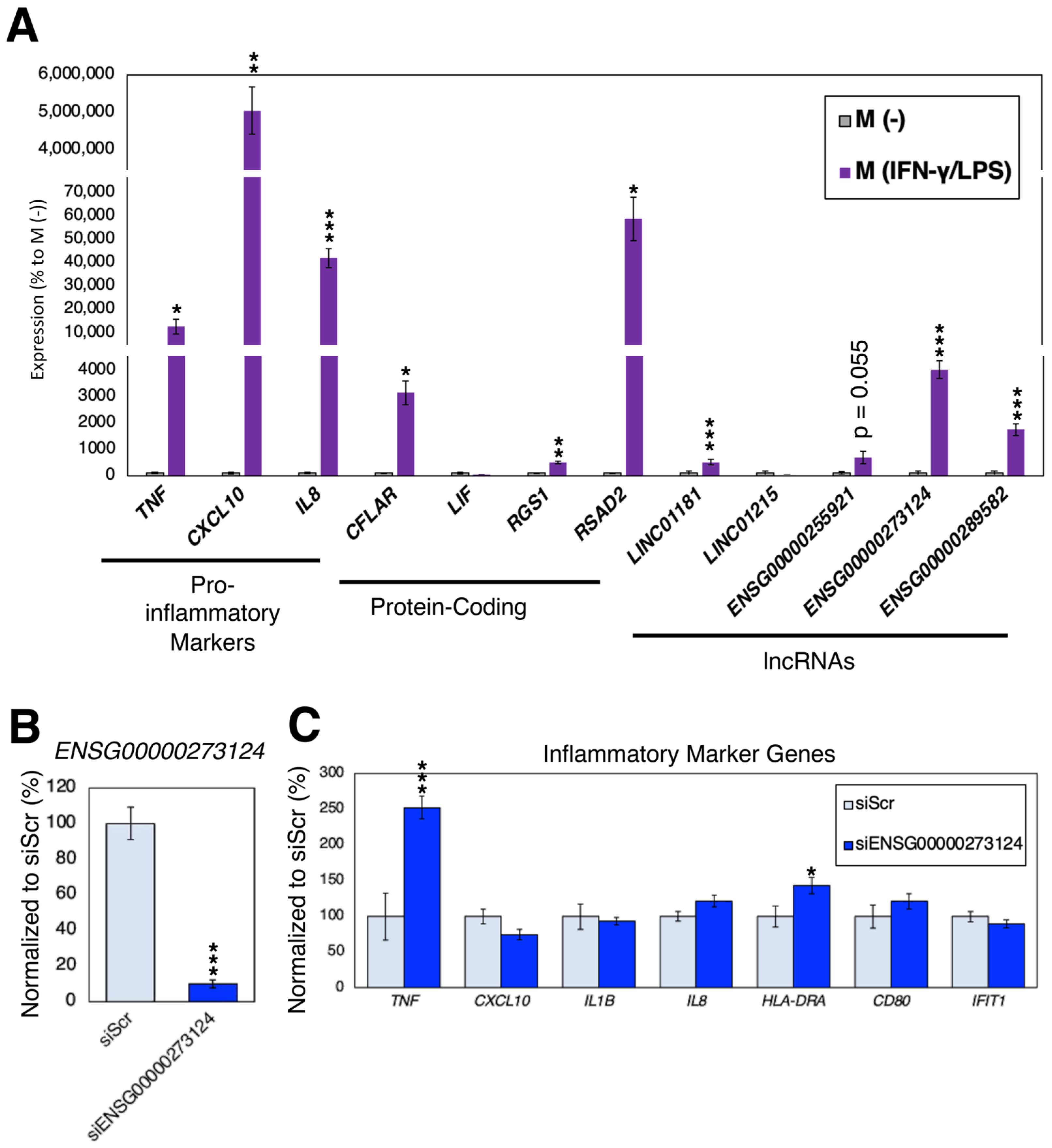
2.6. Loss-of-Function Experiments in Pro-Inflammatory Macrophages
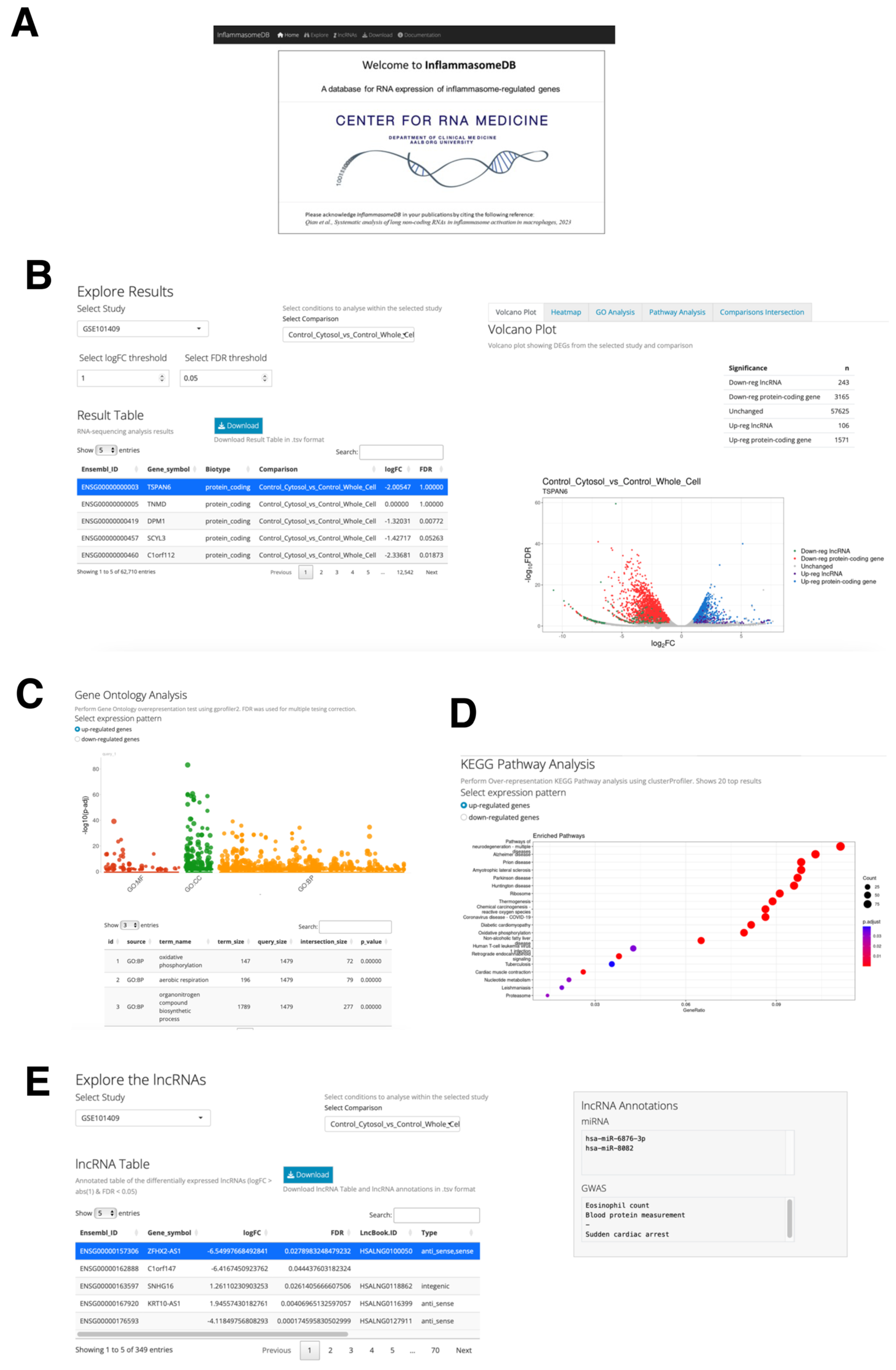
2.7. The Web Database, InflammasomeDB, for Expression Analysis of Inflammasome-Regulated Protein-Coding and lncRNA Genes
3. Discussion
4. Materials and Methods
4.1. RNA-Seq Data Analysis and Visualization
4.2. Cell Culture
4.3. Isolation of Total RNA and RT-PCR
4.4. Inflammasome Web Database
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cassel, S.L.; Sutterwala, F.S. Sterile Inflammatory Responses Mediated by the NLRP3 Inflammasome. Eur. J. Immunol. 2010, 40, 607–611. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, S.; Xiao, Y.; Zhang, W.; Wu, S.; Qin, T.; Yue, Y.; Qian, W.; Li, L. NLRP3 Inflammasome and Inflammatory Diseases. Oxidative Med. Cell. Longev. 2020, 2020, 4063562. [Google Scholar] [CrossRef]
- Mangan, M.S.J.; Olhava, E.J.; Roush, W.R.; Seidel, H.M.; Glick, G.D.; Latz, E. Erratum: Targeting the NLRP3 Inflammasome in Inflammatory Diseases. Nat. Rev. Drug Discov. 2018, 17, 688. [Google Scholar] [CrossRef]
- Lu, A.; Magupalli, V.G.; Ruan, J.; Yin, Q.; Atianand, M.K.; Vos, M.R.; Schröder, G.F.; Fitzgerald, K.A.; Wu, H.; Egelman, E.H. Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes. Cell 2014, 156, 1193–1206. [Google Scholar] [CrossRef]
- Próchnicki, T.; Latz, E. Inflammasomes on the Crossroads of Innate Immune Recognition and Metabolic Control. Cell Metab. 2017, 26, 71–93. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 Catalog of Human Long Noncoding RNAs: Analysis of Their Gene Structure, Evolution, and Expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Sommerauer, C.; Kutter, C. Noncoding RNAs and RNA-Binding Proteins: Emerging Governors of Liver Physiology and Metabolic Diseases. Am. J. Physiol.-Cell Physiol. 2022, 323, C1003–C1017. [Google Scholar] [CrossRef]
- Pisignano, G.; Ladomery, M. Epigenetic Regulation of Alternative Splicing: How LncRNAs Tailor the Message. ncRNA 2021, 7, 21. [Google Scholar] [CrossRef]
- Bhatti, G.K.; Khullar, N.; Sidhu, I.S.; Navik, U.S.; Reddy, A.P.; Reddy, P.H.; Bhatti, J.S. Emerging Role of Non-coding RNA in Health and Disease. Metab. Brain Dis. 2021, 36, 1119–1134. [Google Scholar] [CrossRef]
- López-Jiménez, E.; Andrés-León, E. The Implications of NcRNAs in the Development of Human Diseases. ncRNA 2021, 7, 17. [Google Scholar] [CrossRef]
- Al-Hawary, S.I.S.; Jasim, S.A.; Romero-Parra, R.M.; Bustani, G.S.; Hjazi, A.; Alghamdi, M.; Kareem, A.K.; Alwaily, E.R.; Zabibah, R.S.; Gupta, J.; et al. NLRP3 Inflammasome Pathway in Atherosclerosis: Focusing on the Therapeutic Potential of Non-Coding RNAs. Pathol. Res. Pract. 2023, 246, 154490. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, Y.; Li, N.; Jiang, Z.; Li, X. Role of Mitochondrial Stress and the NLRP3 Inflammasome in Lung Diseases. Inflamm. Res. 2023, 72, 829–846. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Tan, Q.; Ma, J.; Zhang, J.; Yu, P. Emerging Role of LncRNA Regulation for NLRP3 Inflammasome in Diabetes Complications. Front. Cell Dev. Biol. 2022, 9, 792401. [Google Scholar] [CrossRef] [PubMed]
- Austermann, J.; Roth, J.; Barczyk-Kahlert, K. The Good and the Bad: Monocytes’ and Macrophages’ Diverse Functions in Inflammation. Cells 2022, 11, 1979. [Google Scholar] [CrossRef] [PubMed]
- Chiu, S.; Bharat, A. Role of Monocytes and Macrophages in Regulating Immune Response Following Lung Transplantation. Curr. Opin. Organ Transplant. 2016, 21, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Affandi, A.J.; Carvalheiro, T.; Ottria, A.; De Haan, J.J.; Brans, M.A.D.; Brandt, M.M.; Tieland, R.G.; Lopes, A.P.; Fernández, B.M.; Bekker, C.P.J.; et al. CXCL4 Drives Fibrosis by Promoting Several Key Cellular and Molecular Processes. Cell Rep. 2022, 38, 110189. [Google Scholar] [CrossRef]
- Yang, C.; Bachu, M.; Du, Y.; Brauner, C.; Yuan, R.; Ah Kioon, M.D.; Chesi, G.; Barrat, F.J.; Ivashkiv, L.B. CXCL4 Synergizes with TLR8 for TBK1-IRF5 Activation, Epigenomic Remodeling and Inflammatory Response in Human Monocytes. Nat. Commun. 2022, 13, 3426. [Google Scholar] [CrossRef]
- Guilliams, M.; Svedberg, F.R. Does Tissue Imprinting Restrict Macrophage Plasticity? Nat. Immunol. 2021, 22, 118–127. [Google Scholar] [CrossRef]
- Das, A.; Sinha, M.; Datta, S.; Abas, M.; Chaffee, S.; Sen, C.K.; Roy, S. Monocyte and Macrophage Plasticity in Tissue Repair and Regeneration. Am. J. Pathol. 2015, 185, 2596–2606. [Google Scholar] [CrossRef]
- Meng, F.; Lowell, C.A. Lipopolysaccharide (LPS)-Induced Macrophage Activation and Signal Transduction in the Absence of Src-Family Kinases Hck, Fgr, and Lyn. J. Exp. Med. 1997, 185, 1661–1670. [Google Scholar] [CrossRef]
- Cobo, I.; Cheng, A.; Murillo-Saich, J.; Coras, R.; Torres, A.; Abe, Y.; Lana, A.J.; Schlachetzki, J.; Liu-Bryan, R.; Terkeltaub, R.; et al. Monosodium Urate Crystals Regulate a Unique JNK-Dependent Macrophage Metabolic and Inflammatory Response. Cell Rep. 2022, 38, 110489. [Google Scholar] [CrossRef] [PubMed]
- Bukhari, I.; Khan, M.R.; Li, F.; Swiatczak, B.; Thorne, R.F.; Zheng, P.; Mi, Y. Clinical Implications of LncRNA LINC-PINT in Cancer. Front. Mol. Biosci. 2023, 10, 1097694. [Google Scholar] [CrossRef] [PubMed]
- Ou, C.; He, X.; Liu, Y.; Zhang, X. LncRNA Cytoskeleton Regulator RNA (CYTOR): Diverse Functions in Metabolism, Inflammation and Tumorigenesis, and Potential Applications in Precision Oncology. Genes Dis. 2023, 10, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yang, Y.; Wei, J.; Yue, J.; Wang, Y.; Zhang, Q.; Jin, M.; Wang, R.; Yang, X.; Zhang, J.; et al. LNCGM1082-Mediated NLRC4 Activation Drives Resistance to Bacterial Infection. Cell. Mol. Immunol. 2023, 20, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhu, Y.; Dutta, S.; Almuntashiri, S.; Wang, X.; Zhang, D. A Proinflammatory Long Noncoding RNA Lncenc1 Regulates Inflammasome Activation in Macrophage. Am. J. Physiol. Lung Cell. Mol. Physiol. 2023, 324, L584–L595. [Google Scholar] [CrossRef]
- Zou, W.; Zhang, J.; Zhang, K.; Peng, Z.; Xin, R.; Wang, L.; Li, J. Asiatic Acid Attenuates Inflammation Induced by Salmonella via Upregulating LncRNA TVX1 in Microglia. IJMS 2022, 23, 10978. [Google Scholar] [CrossRef]
- Yao, Q.; Xie, Y.; Xu, D.; Qu, Z.; Wu, J.; Zhou, Y.; Wei, Y.; Xiong, H.; Zhang, X.-L. Lnc-EST12, Which Is Negatively Regulated by Mycobacterial EST12, Suppresses Antimycobacterial Innate Immunity through Its Interaction with FUBP3. Cell. Mol. Immunol. 2022, 19, 883–897. [Google Scholar] [CrossRef]
- Yuan, J.; Zhu, Q.; Zhang, X.; Wen, Z.; Zhang, G.; Li, N.; Pei, Y.; Wang, Y.; Pei, S.; Xu, J.; et al. Ezh2 Competes with P53 to License LncRNA Neat1 Transcription for Inflammasome Activation. Cell Death Differ. 2022, 29, 2009–2023. [Google Scholar] [CrossRef]
- Shen, M.; Pan, X.; Gao, Y.; Ye, H.; Zhang, J.; Chen, Y.; Pan, M.; Huang, W.; Xu, X.; Zhao, Y.; et al. LncRNA CRNDE Exacerbates IgA Nephropathy Progression by Promoting NLRP3 Inflammasome Activation in Macrophages. Immunol. Investig. 2022, 51, 1515–1527. [Google Scholar] [CrossRef]
- Zou, L.; Yu, Q.; Zhang, L.; Yuan, X.; Fang, F.; Xu, F. Identification of Inflammation Related LncRNAs and Gm33647 as a Potential Regulator in Septic Acute Lung Injury. Life Sci. 2021, 282, 119814. [Google Scholar] [CrossRef]
- Wang, Z.; Kun, Y.; Lei, Z.; Dawei, W.; Lin, P.; Jibo, W. LncRNA MIAT Downregulates IL-1β, TNF-α to Suppress Macrophage Inflammation but Is Suppressed by ATP-Induced NLRP3 Inflammasome Activation. Cell Cycle 2021, 20, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Guo, J.; Sun, Z.; Liu, L.; Zhao, T.; Li, J.; Tang, G.; Zhang, H.; Wang, W.; Cao, S.; et al. Brucella-Induced Downregulation of LncRNA Gm28309 Triggers Macrophages Inflammatory Response Through the MiR-3068-5p/NF-ΚB Pathway. Front. Immunol. 2020, 11, 581517. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Zhang, S.; Qiao, J. Silencing of Long Non-Coding RNA MEG3 Alleviates Lipopolysaccharide-Induced Acute Lung Injury by Acting as a Molecular Sponge of MicroRNA-7b to Modulate NLRP3. Aging 2020, 12, 20198–20211. [Google Scholar] [CrossRef]
- Zhang, P.; Cao, L.; Zhou, R.; Yang, X.; Wu, M. The LncRNA Neat1 Promotes Activation of Inflammasomes in Macrophages. Nat. Commun. 2019, 10, 1495. [Google Scholar] [CrossRef]
- Xue, Z.; Zhang, Z.; Liu, H.; Li, W.; Guo, X.; Zhang, Z.; Liu, Y.; Jia, L.; Li, Y.; Ren, Y.; et al. LincRNA-Cox2 Regulates NLRP3 Inflammasome and Autophagy Mediated Neuroinflammation. Cell Death Differ. 2019, 26, 130–145. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Liu, Q.; Lv, H.; Wang, F.; Liu, R.; Zeng, N. Effect of Pulegone on the NLPR3 Inflammasome during Inflammatory Activation of THP-1 Cells. Exp. Ther. Med. 2019, 19, 1304–1312. [Google Scholar] [CrossRef]
- Yeom, K.-H.; Pan, Z.; Lin, C.-H.; Lim, H.Y.; Xiao, W.; Xing, Y.; Black, D.L. Tracking Pre-MRNA Maturation across Subcellular Compartments Identifies Developmental Gene Regulation through Intron Retention and Nuclear Anchoring. Genome Res. 2021, 31, 1106–1119. [Google Scholar] [CrossRef]
- Carlevaro-Fita, J.; Johnson, R. Global Positioning System: Understanding Long Noncoding RNAs through Subcellular Localization. Mol. Cell 2019, 73, 869–883. [Google Scholar] [CrossRef]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: LncRNA Localization and Function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef]
- Mas-Ponte, D.; Carlevaro-Fita, J.; Palumbo, E.; Hermoso Pulido, T.; Guigo, R.; Johnson, R. LncATLAS Database for Subcellular Localization of Long Noncoding RNAs. RNA 2017, 23, 1080–1087. [Google Scholar] [CrossRef]
- Aznaourova, M.; Janga, H.; Sefried, S.; Kaufmann, A.; Dorna, J.; Volkers, S.M.; Georg, P.; Lechner, M.; Hoppe, J.; Dökel, S.; et al. Noncoding RNA MaIL1 Is an Integral Component of the TLR4–TRIF Pathway. Proc. Natl. Acad. Sci. USA 2020, 117, 9042–9053. [Google Scholar] [CrossRef] [PubMed]
- Yunna, C.; Mengru, H.; Lei, W.; Weidong, C. Macrophage M1/M2 Polarization. Eur. J. Pharmacol. 2020, 877, 173090. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, X.; Wan, C.; Liu, Y.; Wang, Y.; Meng, C.; Zhang, Y.; Jiang, C. NLRP3 Inflammasome Mediates M1 Macrophage Polarization and IL-1β Production in Inflammatory Root Resorption. J. Clin. Periodontol. 2020, 47, 451–460. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Jhong, J.-H.; Chen, Q.; Huang, K.-Y.; Strittmatter, K.; Kreuzer, J.; De Ran, M.; Wu, X.; Lee, T.-Y.; Slavov, N.; et al. Global Characterization of Macrophage Polarization Mechanisms and Identification of M2-Type Polarization Inhibitors. Cell Rep. 2021, 37, 109955. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, L.; Feng, C.; Qin, Y.; Xiao, J.; Zhang, Z.; Ma, L. LncBook 2.0: Integrating Human Long Non-Coding RNAs with Multi-Omics Annotations. Nucleic Acids Res. 2023, 51, D186–D191. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR : A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis (Use R!), 2nd ed.; Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics Enrichment Tools: Paths toward the Comprehensive Functional Analysis of Large Gene Lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New Capabilities and Interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Cheng, J.; Allaire, J.J.; Sievert, C.; Schloerke, B.; Xie, Y.; Allen, J.; McPherson, J.; Dipert, A.; Borges, B. Shiny: Web Application Framework for R; CRAN: Wien, Austria, 2023. [Google Scholar]
- Distefano, R.; Ilieva, M.; Madsen, J.H.; Rennie, S.; Uchida, S. DoxoDB: A Database for the Expression Analysis of Doxorubicin-Induced LncRNA Genes. ncRNA 2023, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Distefano, R.; Ilieva, M.; Madsen, J.H.; Ishii, H.; Aikawa, M.; Rennie, S.; Uchida, S. T2DB: A Web Database for Long Non-Coding RNA Genes in Type II Diabetes. ncRNA 2023, 9, 30. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Yu, G. Enrichplot: Visualization of Functional Enrichment Result. Available online: https://github.com/YuLab-SMU/enrichplot (accessed on 5 August 2023).
- Chen, H.; Boutros, P.C. VennDiagram: A Package for the Generation of Highly-Customizable Venn and Euler Diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, N.; Distefano, R.; Ilieva, M.; Madsen, J.H.; Rennie, S.; Uchida, S. Systematic Analysis of Long Non-Coding RNAs in Inflammasome Activation in Monocytes/Macrophages. Non-Coding RNA 2023, 9, 50. https://doi.org/10.3390/ncrna9050050
Qian N, Distefano R, Ilieva M, Madsen JH, Rennie S, Uchida S. Systematic Analysis of Long Non-Coding RNAs in Inflammasome Activation in Monocytes/Macrophages. Non-Coding RNA. 2023; 9(5):50. https://doi.org/10.3390/ncrna9050050
Chicago/Turabian StyleQian, Na, Rebecca Distefano, Mirolyuba Ilieva, Jens Hedelund Madsen, Sarah Rennie, and Shizuka Uchida. 2023. "Systematic Analysis of Long Non-Coding RNAs in Inflammasome Activation in Monocytes/Macrophages" Non-Coding RNA 9, no. 5: 50. https://doi.org/10.3390/ncrna9050050
APA StyleQian, N., Distefano, R., Ilieva, M., Madsen, J. H., Rennie, S., & Uchida, S. (2023). Systematic Analysis of Long Non-Coding RNAs in Inflammasome Activation in Monocytes/Macrophages. Non-Coding RNA, 9(5), 50. https://doi.org/10.3390/ncrna9050050








