Long Non-Coding RNA Signatures in Lymphopoiesis and Lymphoid Malignancies
Abstract
1. Introduction: A Glance into Long Non-Coding RNAs
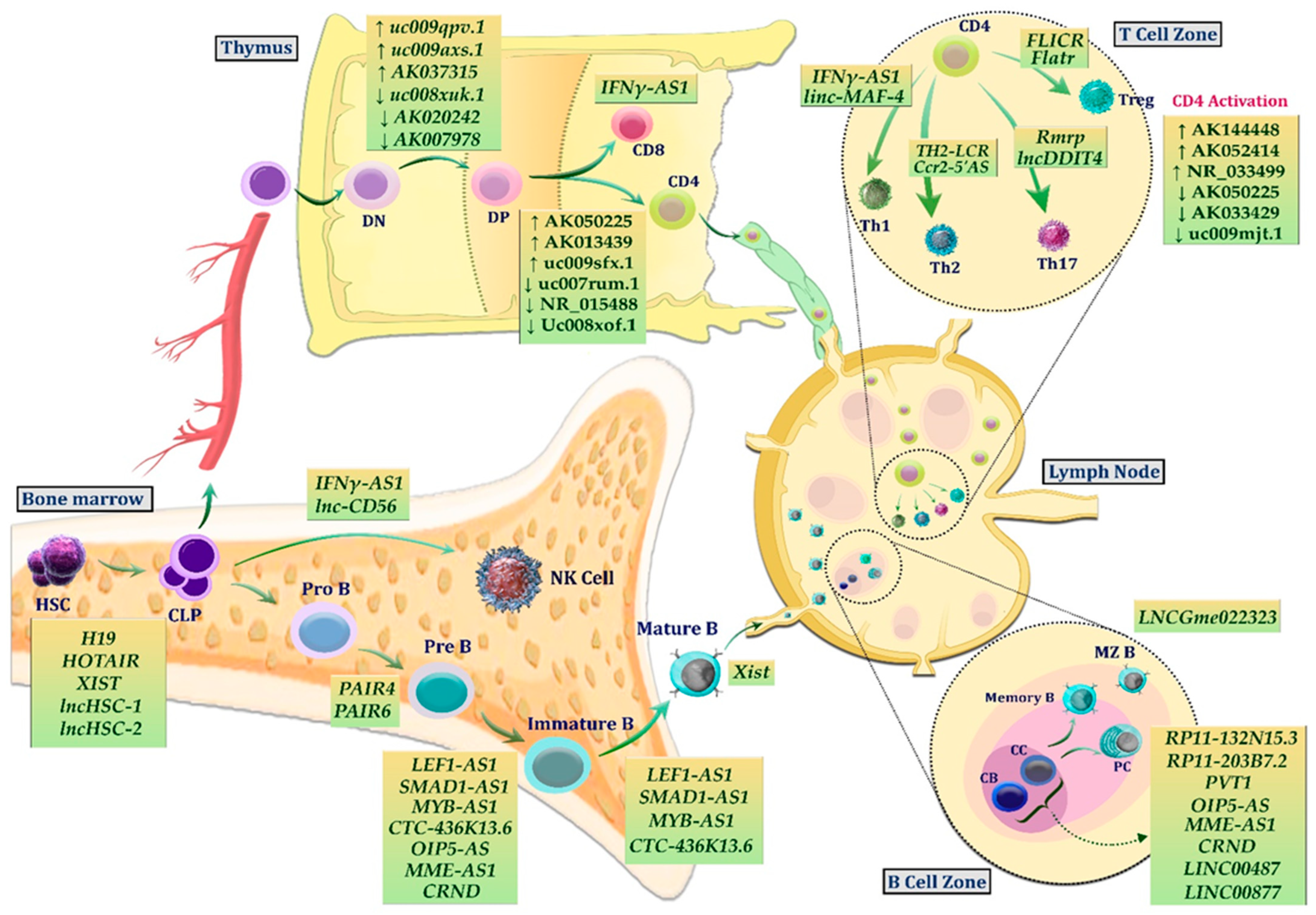
2. LncRNAs in Normal Lymphopoiesis
2.1. LncRNAs in Normal B Lymphopoiesis
2.2. LncRNAs in Normal T Lymphopoiesis
2.3. LncRNAs in Normal NK Lymphopoiesis
3. Alterations in lncRNAs in LYMPHOID Malignancy
3.1. Alterations in lncRNAs in Acute Lymphocytic Leukemia (ALL)
3.1.1. Alterations in lncRNAs in B-ALL
3.1.2. Alterations in lncRNAs in T-ALL
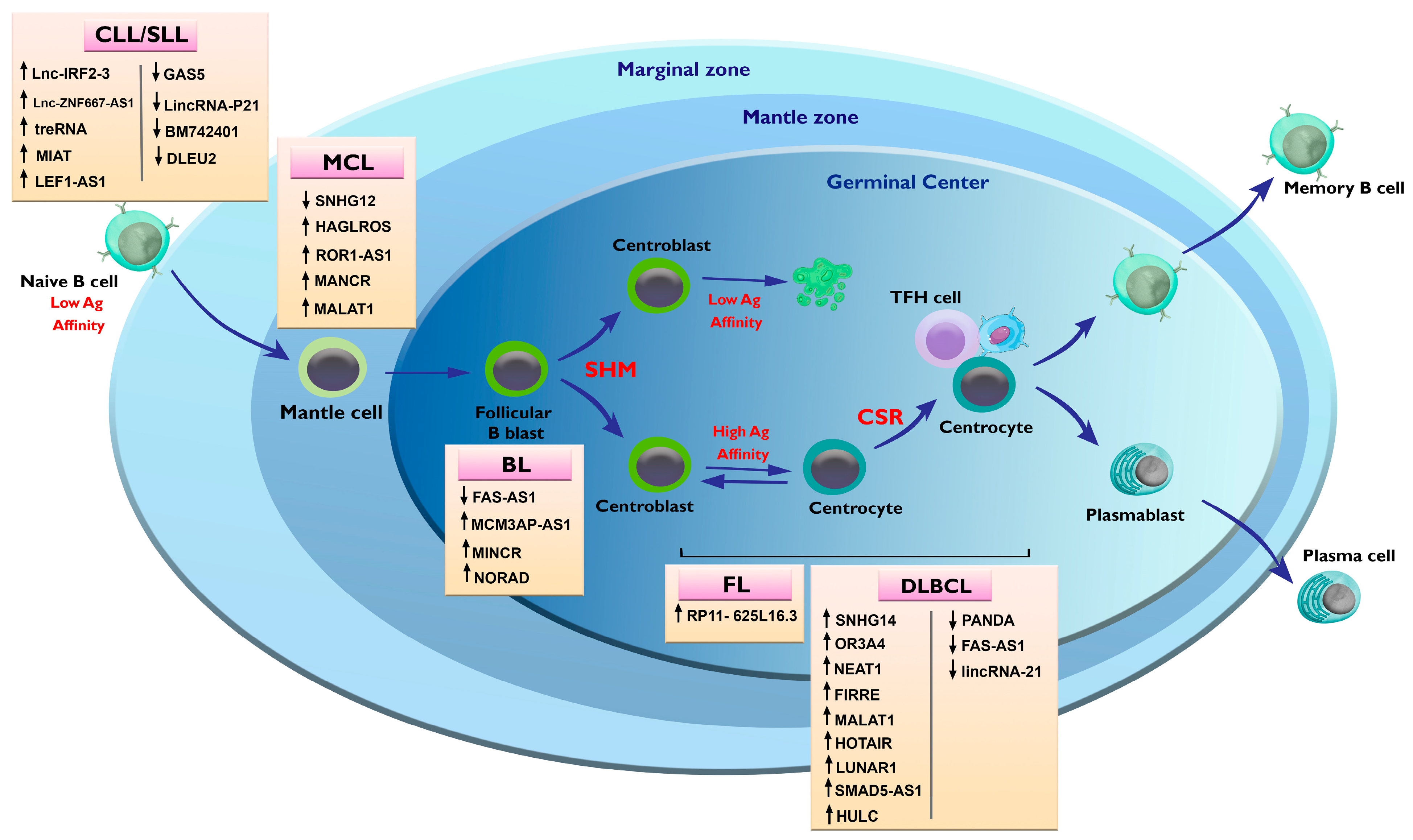
3.2. Alterations in lncRNAs in Chronic Lymphocytic Leukemia (CLL)
4. Alterations in lncRNAs in Non-Hodgkin Lymphoma
4.1. Alterations in lncRNAs in B-Cell Lymphomas
4.2. Alterations in lncRNAs in T/NK-Cell Lymphomas
5. Future Prospectives and Clinical Applications of lncRNAs
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALL | Acute lymphocytic leukemia |
| BL | Burkitt’s lymphoma |
| CASC15 | Cancer susceptibility candidate 15 |
| CLL | Chronic lymphocytic leukemia |
| DDX5 | DEAD-box RNA helicase |
| DLBCL | Diffuse large B-cell lymphoma |
| FLICR | Foxp3 long intergenic noncoding RNA |
| Flatr | Foxp3-specific lncRNA anticipatory of Tregs |
| FL | Follicular lymphoma |
| GAS5 | Growth arrest-specific 5 |
| HSC | Hematopoietic stem cells |
| HOTAIR | Hox transcript antisense intergenic RNA |
| HOTTIP | HOXA transcript at the distal tip |
| HOXA-AS2 | HOXA cluster antisense RNA 2 |
| IFNγ-AS1 | Interferon gamma antisense RNA 1 |
| LncRNAs | Long non-coding RNAs |
| LEF1-AS1 | Lymphoid enhancer-binding factor 1-antisense RNA 1 |
| MALAT1 | Metastasis associated lung adenocarcinoma transcript 1 |
| MEG3 | Maternally expressed gene 3 |
| MYB-AS1 | MYB antisense RNA 1 |
| MIAT | Myocardial infarction associated transcript |
| MCL | Mantle-cell lymphoma |
| mTOR | Mammalian target of rapamycin |
| NcRNAs | Non-coding RNAs |
| NK cell | Natural killer cell |
| NEAT1 | Nuclear enriched abundant transcript 1 |
| NHL | Non-Hodgkin lymphoma |
| NKTCL | Natural killer/T-cell lymphoma |
| PAIR | PAX5-activated intergenic repeat |
| PANDA | P21-associated ncRNA DNA damage-activated |
| PRC2 | Polycomb repressive complex 2 |
| PI3K | Phosphatidylinositol 3-kinase |
| PiRNAs | PIWI-interacting RNAs |
| RNASeq | RNA-sequencing |
| RT-PCR | Reverse transcription polymerase chain reaction |
| SncRNAs | Small non-coding RNAs |
| SiRNAs | Small interfering RNAs |
| SMAD1-AS1 | SMAD1 antisense RNA 1 |
| ShRNA | Short hairpin RNA |
| XIST | X-inactive specific transcript |
References
- Saltanatpour, Z.; Heidari, F.; Sabet, M.N.; Heidari, R.; Hamidieh, A.A. Menstrual Blood-Derived Mesenchymal Stem Cell Therapy for Severe COVID-19 Patients. Curr. Stem Cell Res. Ther. 2023, 18. [Google Scholar] [CrossRef]
- Boehm, T. Design principles of adaptive immune systems. Nat. Rev. Immunol. 2011, 11, 307–317. [Google Scholar] [CrossRef]
- Lejman, M.; Chałupnik, A.; Chilimoniuk, Z.; Dobosz, M. Genetic Biomarkers and Their Clinical Implications in B-Cell Acute Lymphoblastic Leukemia in Children. Int. J. Mol. Sci. 2022, 23, 2755. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Heidari, R.; Akbariqomi, M.; Asgari, Y.; Ebrahimi, D.; Alinejad-Rokny, H. A systematic review of long non-coding RNAs with a potential role in breast cancer. Mutat. Res. Mol. Mech. Mutagen. 2021, 787, 108375. [Google Scholar] [CrossRef] [PubMed]
- Mirmazhari, S.P.; Abdi, M.J.; Mosadeghrad, A.H.; Bonab, R.A.; Babadi, A.A.; Rigi, G.; Shahrokhi, S.Z.; Chamanara, M.; Amodio, N.; Heidari, R. Unraveling the Complex Interplay between lncRNAs and Myc in Breast Cancer. Preprints 2023, 2023071901. [Google Scholar] [CrossRef]
- Jafari-Raddani, F.; Davoodi-Moghaddam, Z.; Yousefi, A.; Ghaffari, S.H.; Bashash, D. An overview of long noncoding RNAs: Biology, functions, therapeutics, analysis methods, and bioinformatics tools. Cell Biochem. Funct. 2022, 40, 800–825. [Google Scholar] [CrossRef]
- Plasek, L.M.; Valadkhan, S. lncRNAs in T lymphocytes: RNA regulation at the heart of the immune response. Am. J. Physiol. Physiol. 2021, 320, C415–C427. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Liu, J.; Wan, L.; Wang, F. Long noncoding RNA/circular RNA regulates competitive endogenous RNA networks in rheumatoid arthritis: Molecular mechanisms and traditional Chinese medicine therapeutic significances. Ann. Med. 2023, 55, 973–989. [Google Scholar] [CrossRef]
- Zhang, T.-N.; Wang, W.; Huang, X.-M.; Gao, S.-Y. Non-Coding RNAs and Extracellular Vehicles: Their Role in the Pathogenesis of Gestational Diabetes Mellitus. Front. Endocrinol. 2021, 12, 664287. [Google Scholar] [CrossRef]
- Vajari, M.K.; Moradinasab, S.; Yousefi, A.M.; Bashash, D. Noncoding RNAs in diagnosis and prognosis of graft-versus-host disease (GVHD). J. Cell. Physiol. 2022, 237, 3480–3495. [Google Scholar] [CrossRef] [PubMed]
- Mosallaei, M.; Siri, G.; Alani, B.; Khomartash, M.S.; Naghoosi, H.; Pourghazi, F.; Heidari, R.; Sabet, M.N.; Behroozi, J. Differential methylation of DNA promoter sequences in peripheral blood mononuclear cells as promising diagnostic biomarkers for colorectal cancer. J. Cancer Res. Ther. 2023, 9. [Google Scholar] [CrossRef]
- Giuliani, B.; Tordonato, C.; Nicassio, F. Mechanisms of Long Non-Coding RNA in Breast Cancer. Int. J. Mol. Sci. 2023, 24, 4538. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, L.; Chen, L.-L. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet. 2017, 33, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Song, X.; Glass, C.K.; Rosenfeld, M.G. The Long Arm of Long Noncoding RNAs: Roles as Sensors Regulating Gene Transcriptional Programs. Cold Spring Harb. Perspect. Biol. 2010, 3, a003756. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Cruz-Miranda, G.M.; Hidalgo-Miranda, A.; Bárcenas-López, D.A.; Núñez-Enríquez, J.C.; Ramírez-Bello, J.; Mejía-Aranguré, J.M.; Jiménez-Morales, S. Long non-coding RNA and acute leukemia. Int. J. Mol. Sci. 2019, 20, 735. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Dominguez, J.R.; Lodish, H.F. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood 2017, 130, 1965–1975. [Google Scholar] [CrossRef]
- Paralkar, V.R.; Weiss, M.J. Long noncoding RNAs in biology and hematopoiesis. Blood J. Am. Soc. Hematol. 2013, 121, 4842–4846. [Google Scholar] [CrossRef]
- Venkatraman, A.; He, X.C.; Thorvaldsen, J.L.; Sugimura, R.; Perry, J.M.; Tao, F.; Zhao, M.; Christenson, M.K.; Sanchez, R.; Yu, J.Y.; et al. Maternal imprinting at the H19–Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature 2013, 500, 345–349. [Google Scholar] [CrossRef]
- Gao, S.; Zhou, B.; Li, H.; Huang, X.; Wu, Y.; Xing, C.; Yu, X.; Ji, Y. Long noncoding RNA HOTAIR promotes the self-renewal of leukemia stem cells through epigenetic silencing of p15. Exp. Hematol. 2018, 67, 32–40.e3. [Google Scholar] [CrossRef] [PubMed]
- Sommerkamp, P.; Renders, S.; Ladel, L.; Hotz-Wagenblatt, A.; Schönberger, K.; Zeisberger, P.; Przybylla, A.; Sohn, M.; Zhou, Y.; Klibanski, A.; et al. The long non-coding RNA Meg3 is dispensable for hematopoietic stem cells. Sci. Rep. 2019, 9, 2110. [Google Scholar] [CrossRef]
- Luo, H.; Zhu, G.; Xu, J.; Lai, Q.; Yan, B.; Guo, Y.; Fung, T.K.; Zeisig, B.B.; Cui, Y.; Zha, J.; et al. HOTTIP lncRNA Promotes Hematopoietic Stem Cell Self-Renewal Leading to AML-like Disease in Mice. Cancer Cell 2019, 36, 645–659.e8. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.-Y.; Wang, J.-H.; Wang, J.-L.; Ma, C.X.; Wang, X.-C.; Liu, F.-S. Malat1 as an evolutionarily conserved lncRNA, plays a positive role in regulating proliferation and maintaining undifferentiated status of early-stage hematopoietic cells. BMC Genom. 2015, 16, 676. [Google Scholar] [CrossRef]
- Casero, D.; Sandoval, S.; Seet, C.S.; Scholes, J.; Zhu, Y.; Ha, V.L.; Luong, A.; Parekh, C.; Crooks, G.M. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat. Immunol. 2015, 16, 1282–1291. [Google Scholar] [CrossRef] [PubMed]
- Petri, A.; Dybkær, K.; Bøgsted, M.; Thrue, C.A.; Hagedorn, P.H.; Schmitz, A.; Bødker, J.S.; Johnsen, H.E.; Kauppinen, S. Long Noncoding RNA Expression during Human B-Cell Development. PLoS ONE 2015, 10, e0138236. [Google Scholar] [CrossRef]
- Bonnal, R.J.; Ranzani, V.; Arrigoni, A.; Curti, S.; Panzeri, I.; Gruarin, P.; Abrignani, S.; Rossetti, G.; Pagani, M. De novo transcriptome profiling of highly purified human lymphocytes primary cells. Sci. Data 2015, 2, 150051. [Google Scholar] [CrossRef]
- Ranzani, V.; Rossetti, G.; Panzeri, I.; Arrigoni, A.; Bonnal, R.J.P.; Curti, S.; Gruarin, P.; Provasi, E.; Sugliano, E.; Marconi, M.; et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat. Immunol. 2015, 16, 318–325. [Google Scholar] [CrossRef]
- Tayari, M.M.; Winkle, M.; Kortman, G.; Sietzema, J.; de Jong, D.; Terpstra, M.; Mestdagh, P.; Kroese, F.G.; Visser, L.; Diepstra, A.; et al. Long Noncoding RNA Expression Profiling in Normal B-Cell Subsets and Hodgkin Lymphoma Reveals Hodgkin and Reed-Sternberg Cell–Specific Long Noncoding RNAs. Am. J. Pathol. 2016, 186, 2462–2472. [Google Scholar] [CrossRef]
- Brazão, T.F.; Johnson, J.S.; Müller-Winkler, J.; Heger, A.; Ponting, C.P.; Tybulewicz, V.L.J. Long noncoding RNAs in B-cell development and activation. Blood 2016, 128, e10–e19. [Google Scholar] [CrossRef]
- Verma-Gaur, J.; Torkamani, A.; Schaffer, L.; Head, S.R.; Schork, N.J.; Feeney, A.J. Noncoding transcription within the Igh distal VH region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc. Natl. Acad. Sci. USA 2012, 109, 17004–17009. [Google Scholar] [CrossRef]
- Syrett, C.M.; Sindhava, V.; Hodawadekar, S.; Myles, A.; Liang, G.; Zhang, Y.; Nandi, S.; Cancro, M.; Atchison, M.; Anguera, M.C. Loss of Xist RNA from the inactive X during B cell development is restored in a dynamic YY1-dependent two-step process in activated B cells. PLoS Genet. 2017, 13, e1007050. [Google Scholar] [CrossRef]
- Meng, Y.-B.; He, X.; Huang, Y.-F.; Wu, Q.-N.; Zhou, Y.-C.; Hao, D.-J. Long Noncoding RNA CRNDE Promotes Multiple Myeloma Cell Growth by Suppressing miR-451. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2017, 25, 1207–1214. [Google Scholar] [CrossRef] [PubMed]
- Shao, K.; Shi, T.; Yang, Y.; Wang, X.; Xu, D.; Zhou, P. Highly expressed lncRNA CRNDE promotes cell proliferation through Wnt/β-catenin signaling in renal cell carcinoma. Tumor Biol. 2016, 37, 15997–16004. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Han, L.-M.; Gao, Q.; Sun, Y. Long non-coding RNA CRNDE promotes tumor growth in medulloblastoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2588–2597. [Google Scholar] [PubMed]
- Clark, E.A.; Giltiay, N.V. CD22: A Regulator of Innate and Adaptive B Cell Responses and Autoimmunity. Front. Immunol. 2018, 9, 2235. [Google Scholar] [CrossRef]
- Chung, E.Y.; Dews, M.; Cozma, D.; Yu, D.; Wentzel, E.A.; Chang, T.-C.; Schelter, J.M.; Cleary, M.A.; Mendell, J.T.; Thomas-Tikhonenko, A. c-Myb oncoprotein is an essential target of the dleu2 tumor suppressor microRNA cluster. Cancer Biol. Ther. 2008, 7, 1758–1764. [Google Scholar] [CrossRef]
- Tschumper, R.C.; Hoelzinger, D.B.; Walters, D.K.; Davila, J.I.; Osborne, C.A.; Jelinek, D.F. Stage-Specific Non-Coding RNA Expression Patterns during In Vitro Human B Cell Differentiation into Antibody Secreting Plasma Cells. Non-Coding RNA 2022, 8, 15. [Google Scholar] [CrossRef]
- Winkle, M.; Kluiver, J.; Diepstra, A.; Berg, A.V.D. Emerging roles for long noncoding RNAs in B-cell development and malignancy. Crit. Rev. Oncol. 2017, 120, 77–85. [Google Scholar] [CrossRef]
- Xia, F.; Dong, F.; Yang, Y.; Huang, A.; Chen, S.; Sun, D.; Xiong, S.; Zhang, J. Dynamic Transcription of Long Non-Coding RNA Genes during CD4+ T Cell Development and Activation. PLoS ONE 2014, 9, e101588. [Google Scholar] [CrossRef]
- Collier, S.P.; Collins, P.L.; Williams, C.L.; Boothby, M.R.; Aune, T.M. Cutting edge: Influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J. Immunol. 2012, 189, 2084–2088. [Google Scholar] [CrossRef] [PubMed]
- Vigneau, S.; Rohrlich, P.S.; Brahic, M.; Bureau, J.F. Tmevpg1, a candidate gene for the control of Theiler′s virus persistence, could be implicated in the regulation of gamma interferon. J. Virol. 2003, 77, 5632–5638. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Liu, G.; Wei, C.; Gao, C.; Hao, J. Linc-MAF-4 regulates Th1/Th2 differentiation and is associated with the pathogenesis of multiple sclerosis by targeting MAF. FASEB J. 2017, 31, 519–525. [Google Scholar] [CrossRef]
- Gibbons, H.R.; Shaginurova, G.; Kim, L.C.; Chapman, N.; Spurlock, C.F.I.; Aune, T.M. Divergent lncRNA GATA3-AS1 Regulates GATA3 Transcription in T-Helper 2 Cells. Front. Immunol. 2018, 9, 2512. [Google Scholar] [CrossRef] [PubMed]
- Kanduri, K.; Tripathi, S.; Larjo, A.; Mannerström, H.; Ullah, U.; Lund, R.; Hawkins, R.D.; Ren, B.; Lähdesmäki, H.; Lahesmaa, R. Identification of global regulators of T-helper cell lineage specification. Genome Med. 2015, 7, 122. [Google Scholar] [CrossRef]
- Spurlock, C.F.; Tossberg, J.T.; Guo, Y.; Collier, S.P.; Crooke, P.S.; Aune, T.M. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat. Commun. 2015, 6, 6932. [Google Scholar] [CrossRef]
- Koh, B.H.; Hwang, S.S.; Kim, J.Y.; Lee, W.; Kang, M.-J.; Lee, C.G.; Park, J.-W.; Flavell, R.A.; Lee, G.R. Th2 LCR is essential for regulation of Th2 cytokine genes and for pathogenesis of allergic asthma. Proc. Natl. Acad. Sci. USA 2010, 107, 10614–10619. [Google Scholar] [CrossRef]
- Nie, J.; Zhao, Q. Lnc-ITSN1-2, derived from RNA sequencing, correlates with increased disease risk, activity and promotes CD4+ T cell activation, proliferation and Th1/Th17 cell differentiation by serving as a ceRNA for IL-23R via sponging miR-125a in inflammatory bowel disease. Front. Immunol. 2020, 11, 852. [Google Scholar]
- Hu, G.; Tang, Q.; Sharma, S.; Yu, F.; Escobar, T.M.; Muljo, S.A.; Zhu, J.; Zhao, K. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol. 2013, 14, 1190–1198. [Google Scholar] [CrossRef]
- Huang, W.; Thomas, B.; Flynn, R.A.; Gavzy, S.J.; Wu, L.; Kim, S.V.; Hall, J.A.; Miraldi, E.R.; Ng, C.P.; Rigo, F.; et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature 2015, 528, 517–522. [Google Scholar] [CrossRef]
- Enayati, H.; Ayatollahi, H.; Keramati, M.R.; Sheikhi, M.; Bagheri, H.; Shams, S.F.; Sadeghian, M.H. Expression of ROR1 gene in patients with acute lymphoblastic leukemia. Iran. J. Blood Cancer 2019, 11, 57–62. [Google Scholar]
- Bonafé, L.; Dermitzakis, E.T.; Unger, S.; Greenberg, C.R.; Campos-Xavier, B.A.; Zankl, A.; Ucla, C.; Antonarakis, S.E.; Superti-Furga, A.; Reymond, A. Evolutionary Comparison Provides Evidence for Pathogenicity of RMRP Mutations. PLoS Genet. 2005, 1, e47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Liu, G.; Li, D.; Wei, C.; Hao, J. DDIT4 and associated lncDDIT4 modulate Th17 differentiation through the DDIT4/TSC/mTOR pathway. J. Immunol. 2018, 200, 1618–1626. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Lee, J.; Krummey, S.; Lu, W.; Cai, H.; Lenardo, M.J. The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat. Immunol. 2011, 12, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Zemmour, D.; Pratama, A.; Loughhead, S.M.; Mathis, D.; Benoist, C. Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proc. Natl. Acad. Sci. USA 2017, 114, E3472–E3480. [Google Scholar] [CrossRef]
- Brajic, A.; Franckaert, D.; Burton, O.; Bornschein, S.; Calvanese, A.L.; Demeyer, S.; Cools, J.; Dooley, J.; Schlenner, S.; Liston, A. The Long Non-coding RNA Flatr Anticipates Foxp3 Expression in Regulatory T Cells. Front. Immunol. 2018, 9, 1989. [Google Scholar] [CrossRef]
- Kanbar, J.N.; Ma, S.; Kim, E.S.; Kurd, N.S.; Tsai, M.S.; Tysl, T.; Widjaja, C.E.; Limary, A.E.; Yee, B.; He, Z.; et al. The long noncoding RNA Malat1 regulates CD8+ T cell differentiation by mediating epigenetic repression. J. Exp. Med. 2022, 219, e20211756. [Google Scholar] [CrossRef]
- Cheng, S.; Li, F.; Qin, H.; Ping, Y.; Zhao, Q.; Gao, Q.; Song, M.; Qu, J.; Shan, J.; Zhang, K.; et al. Long Noncoding RNA lncNDEPD1 Regulates PD-1 Expression via miR-3619-5p in CD8+ T Cells. J. Immunol. 2022, 208, 1483–1492. [Google Scholar] [CrossRef]
- Zhang, R.; Ni, F.; Fu, B.; Wu, Y.; Sun, R.; Tian, Z.; Wei, H. A long noncoding RNA positively regulates CD56 in human natural killer cells. Oncotarget 2016, 7, 72546–72558. [Google Scholar] [CrossRef]
- Stein, N.; Berhani, O.; Schmiedel, D.; Duev-Cohen, A.; Seidel, E.; Kol, I.; Tsukerman, P.; Hecht, M.; Reches, A.; Gamliel, M.; et al. IFNG-AS1 Enhances Interferon Gamma Production in Human Natural Killer Cells. iScience 2019, 11, 466–473. [Google Scholar] [CrossRef]
- Collier, S.P.; Henderson, M.A.; Tossberg, J.T.; Aune, T.M. Regulation of the Th1 Genomic Locus from Ifng through Tmevpg1 by T-bet. J. Immunol. 2014, 193, 3959–3965. [Google Scholar] [CrossRef] [PubMed]
- Gasic, V.; Karan-Djurasevic, T.; Pavlovic, D.; Zukic, B.; Pavlovic, S.; Tosic, N. Diagnostic and Therapeutic Implications of Long Non-Coding RNAs in Leukemia. Life 2022, 12, 1770. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Bian, H.; Cao, Y.; Juan, C.; Cao, Q.; Zhou, G.; Fang, Y. Identification of novel lncRNAs involved in the pathogenesis of childhood acute lymphoblastic leukemia. Oncol. Lett. 2018, 17, 2081–2090. [Google Scholar] [CrossRef] [PubMed]
- Chagas, P.F.D.; Sousa, G.R.D.; Kodama, M.H.; Biagi Junior, C.A.O.D.; Yunes, J.A.; Brandalise, S.R.; Calin, G.A.; Tone, L.G.; Scrideli, C.A.; Oliveira, J.C.D. Ultraconserved long non-coding RNA uc. 112 is highly expressed in childhood T versus B-cell acute lymphoblastic leukemia. Hematol. Transfus. Cell Ther. 2020, 43, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Han, Q.; Gu, Y.; Ma, J.; McGrath, M.; Qiao, F.; Chen, B.; Song, C.; Ge, Z. Circular RNA PVT1 expression and its roles in acute lymphoblastic leukemia. Epigenomics 2018, 10, 723–732. [Google Scholar] [CrossRef]
- Yu, D.; Wang, W.-M.; Wu, F.-F.; Ma, P.; Sun, L.; Wan, D.-M.; Liu, Y.-F.; Xie, X.-S.; Wang, C.; Sun, H. Expression of Long-Chain Non-coding RNA RP11-87C12.5 in Acute Lymphocytic Leukemia and Its Cinical Significance. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2018, 26, 26–31. [Google Scholar]
- Gasic, V.; Stankovic, B.; Zukic, B.; Janic, D.; Dokmanovic, L.; Krstovski, N.; Lazic, J.; Milosevic, G.; Lucafò, M.; Stocco, G.; et al. Expression pattern of long non-coding RNA growth arrest-specific 5 in the remission induction therapy in childhood acute lymphoblastic leukemia. J. Med. Biochem. 2019, 38, 292–298. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhao, S.; Li, J.; Zhang, H.; Qian, C.; Wang, H.; Liu, J.; Zhao, Y. TCF7L2 activated HOXA-AS2 decreased the glucocorticoid sensitivity in acute lymphoblastic leukemia through regulating HOXA3/EGFR/Ras/Raf/MEK/ERK pathway. Biomed. Pharmacother. 2018, 109, 1640–1649. [Google Scholar] [CrossRef]
- Pouyanrad, S.; Rahgozar, S.; Ghodousi, E.S. Dysregulation of miR-335-3p, targeted by NEAT1 and MALAT1 long non-coding RNAs, is associated with poor prognosis in childhood acute lymphoblastic leukemia. Gene 2019, 692, 35–43. [Google Scholar] [CrossRef]
- James, A.R.; Schroeder, M.P.; Neumann, M.; Bastian, L.; Eckert, C.; Gökbuget, N.; Tanchez, J.O.; Schlee, C.; Isaakidis, K.; Schwartz, S.; et al. Long non-coding RNAs defining major subtypes of B cell precursor acute lymphoblastic leukemia. J. Hematol. Oncol. 2019, 12, 8. [Google Scholar] [CrossRef]
- Cuadros, M.; Andrades, Á.; Coira, I.F.; Baliñas, C.; Rodríguez, M.I.; Álvarez-Pérez, J.C.; Peinado, P.; Arenas, A.M.; García, D.J.; Jiménez, P.; et al. Expression of the long non-coding RNA TCL6 is associated with clinical outcome in pediatric B-cell acute lymphoblastic leukemia. Blood Cancer J. 2019, 9, 93. [Google Scholar] [CrossRef]
- Ghazavi, F.; De Moerloose, B.; Van Loocke, W.; Wallaert, A.; Ferster, A.; Bakkus, M.; Plat, G.; Delabesse, E.; Uyttebroeck, A.; Van Nieuwerburgh, F.; et al. Unique long non-coding RNA expression signature in ETV6/RUNX1-driven B-cell precursor acute lymphoblastic leukemia. Blood 2016, 128, 3920. [Google Scholar] [CrossRef]
- Lajoie, M.; Drouin, S.; Caron, M.; St-Onge, P.; Ouimet, M.; Gioia, R.; Lafond, M.-H.; Vidal, R.; Richer, C.; Oualkacha, K.; et al. Specific expression of novel long non-coding RNAs in high-hyperdiploid childhood acute lymphoblastic leukemia. PLoS ONE 2017, 12, e0174124. [Google Scholar] [CrossRef] [PubMed]
- Fernando, T.R.; Rodriguez-Malave, N.I.; Waters, E.V.; Yan, W.; Casero, D.; Basso, G.; Pigazzi, M.; Rao, D.S. LncRNA Expression Discriminates Karyotype and Predicts Survival in B-Lymphoblastic Leukemia. Mol. Cancer Res. 2015, 13, 839–851. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, J.; Guo, G.; Feng, R.; Chen, K.; Liao, Y.; Zhang, L.; Sun, L.; Huang, S.; Chen, J.L. Novel lncRNA-IUR suppresses Bcr-Abl-induced tumorigenesis through regulation of STAT5-CD71 pathway. Mol. Cancer 2019, 18, 84. [Google Scholar] [CrossRef]
- Huang, M.; Zheng, J.; Ren, Y.; Zhu, J.; Kou, L.; Nie, J. LINC00221 suppresses the malignancy of children acute lymphoblastic leukemia. Biosci. Rep. 2020, 40, BSR20194070. [Google Scholar] [CrossRef]
- Gioia, R.; Drouin, S.; Ouimet, M.; Caron, M.; St-Onge, P.; Richer, C.; Sinnett, D. LncRNAs downregulated in childhood acute lymphoblastic leukemia modulate apoptosis, cell migration, and DNA damage response. Oncotarget 2017, 8, 80645–80650. [Google Scholar] [CrossRef]
- Fernando, T.R.; Contreras, J.R.; Zampini, M.; Rodriguez-Malave, N.I.; Alberti, M.O.; Anguiano, J.; Tran, T.M.; Palanichamy, J.K.; Gajeton, J.; Ung, N.M.; et al. The lncRNA CASC15 regulates SOX4 expression in RUNX1-rearranged acute leukemia. Mol. Cancer 2017, 16, 126. [Google Scholar] [CrossRef]
- Grasedieck, S.; Ruess, C.; Krowiorz, K.; Lux, S.; Pochert, N.; Schwarzer, A.; Klusmann, J.-H.; Jongen-Lavrencic, M.; Herold, T.; Bullinger, L.; et al. The long non-coding RNA Cancer Susceptibility 15 (CASC15) is induced by isocitrate dehydrogenase (IDH) mutations and maintains an immature phenotype in adult acute myeloid leukemia. Haematologica 2020, 105, e448–e453. [Google Scholar] [CrossRef]
- Ouimet, M.; Drouin, S.; Lajoie, M.; Caron, M.; St-Onge, P.; Gioia, R.; Richer, C.; Sinnett, D. A childhood acute lymphoblastic leukemia-specific lncRNA implicated in prednisolone resistance, cell proliferation, and migration. Oncotarget 2016, 8, 7477–7488. [Google Scholar] [CrossRef]
- Bárcenas-López, D.A.; Núñez-Enríquez, J.C.; Hidalgo-Miranda, A.; Beltrán-Anaya, F.O.; May-Hau, D.I.; Jiménez-Hernández, E.; Bekker-Méndez, V.C.; Flores-Lujano, J.; Medina-Sansón, A.; Tamez-Gómez, E.L.; et al. Transcriptome Analysis Identifies LINC00152 as a Biomarker of Early Relapse and Mortality in Acute Lymphoblastic Leukemia. Genes 2020, 11, 302. [Google Scholar] [CrossRef] [PubMed]
- Fang, K.; Han, B.-W.; Chen, Z.-H.; Lin, K.; Zeng, C.-W.; Li, X.-J.; Li, J.; Luo, X.-Q.; Chen, Y.-Q. A distinct set of long non-coding RNAs in childhood MLL-rearranged acute lymphoblastic leukemia: Biology and epigenetic target. Hum. Mol. Genet. 2014, 23, 3278–3288. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Camino, A.; Martin-Guerrero, I.; de Andoin, N.G.; Sastre, A.; Bañeres, A.C.; Astigarraga, I.; Navajas, A.; Garcia-Orad, A. Confirmation of involvement of new variants at CDKN2A/B in pediatric acute lymphoblastic leukemia susceptibility in the Spanish population. PLoS ONE 2017, 12, e0177421. [Google Scholar] [CrossRef] [PubMed]
- De la Rosa, R.; Villegas-Ruíz, V.; Caballero-Palacios, M.C.; Pérez-López, E.I.; Murata, C.; Zapata-Tarres, M.; Cárdenas-Cardos, R.; Paredes-Aguilera, R.; Rivera-Luna, R.; Juárez-Méndez, S. Expression of ZNF695 transcript variants in childhood B-cell acute lymphoblastic leukemia. Genes 2019, 10, 716. [Google Scholar] [CrossRef]
- Chen, S.; Chen, J.-Z.; Zhang, J.-Q.; Chen, H.-X.; Qiu, F.-N.; Yan, M.-L.; Tian, Y.-F.; Peng, C.-H.; Shen, B.-Y.; Chen, Y.-L.; et al. Silencing of long noncoding RNA LINC00958 prevents tumor initiation of pancreatic cancer by acting as a sponge of microRNA-330-5p to down-regulate PAX8. Cancer Lett. 2019, 446, 49–61. [Google Scholar] [CrossRef]
- Rodríguez-Malavé, N.I.; Fernando, T.R.; Patel, P.C.; Contreras, J.R.; Palanichamy, J.K.; Tran, T.M.; Anguiano, J.; Davoren, M.J.; Alberti, M.O.; Pioli, K.T.; et al. BALR-6 regulates cell growth and cell survival in B-lymphoblastic leukemia. Mol. Cancer 2015, 14, 214. [Google Scholar] [CrossRef]
- Wang, W.; Wu, F.; Ma, P.; Gan, S.; Li, X.; Chen, L.; Sun, L.; Sun, H.; Jiang, Z.; Guo, F. LncRNA CRNDE Promotes the Progression of B-cell Precursor Acute Lymphoblastic Leukemia by Targeting the miR-345-5p/CREB Axi. Mol. Cells 2020, 43, 718–727. [Google Scholar]
- Wang, Q.; Du, X.; Yang, M.; Xiao, S.; Cao, J.; Song, J.; Wang, L. LncRNA ZEB1-AS1 contributes to STAT3 activation by associating with IL-11 in B-lymphoblastic leukemia. Biotechnol. Lett. 2017, 39, 1801–1810. [Google Scholar] [CrossRef]
- Durinck, K.; Wallaert, A.; Van de Walle, I.; Van Loocke, W.; Volders, P.J.; Vanhauwaert, S.; Geerdens, E.; Benoit, Y.; Van Roy, N.; Poppe, B.; et al. The Notch driven long non-coding RNA repertoire in T-cell acute lymphoblastic leukemia. Haematologica 2014, 99, 1808. [Google Scholar] [CrossRef]
- Li, G.; Gao, L.; Zhao, J.; Liu, D.; Li, H.; Hu, M. LncRNA ANRIL/miR-7-5p/TCF4 axis contributes to the progression of T cell acute lymphoblastic leukemia. Cancer Cell Int. 2020, 20, 335. [Google Scholar] [CrossRef]
- Ngoc, P.C.T.; Tan, S.H.; Tan, T.K.; Chan, M.M.; Li, Z.; Yeoh, A.E.J.; Tenen, D.G.; Sanda, T. Identification of novel lncRNAs regulated by the TAL1 complex in T-cell acute lymphoblastic leukemia. Leukemia 2018, 32, 2138–2151. [Google Scholar] [CrossRef]
- Yousefi, H.; Purrahman, D.; Jamshidi, M.; Lak, E.; Keikhaei, B.; Mahmoudian-Sani, M.-R. Long non-coding RNA signatures and related signaling pathway in T-cell acute lymphoblastic leukemia. Clin. Transl. Oncol. 2022, 24, 2081–2089. [Google Scholar] [CrossRef]
- Tan, S.H.; Leong, W.Z.; Ngoc, P.C.T.; Tan, T.K.; Bertulfo, F.C.; Lim, M.C.; An, O.; Li, Z.; Yeoh, A.E.J.; Fullwood, M.J.; et al. The enhancer RNA ARIEL activates the oncogenic transcriptional program in T-cell acute lymphoblastic leukemia. Blood 2019, 134, 239–251. [Google Scholar] [CrossRef]
- Drillis, G.; Goulielmaki, M.; Spandidos, D.A.; Aggelaki, S.; Zoumpourlis, V. Non-coding RNAs (miRNAs and lncRNAs) and their roles in lymphogenesis in all types of lymphomas and lymphoid malignancies (Review). Oncol. Lett. 2021, 21, 393. [Google Scholar] [CrossRef]
- Qian, C.-S.; Li, L.-J.; Huang, H.-W.; Yang, H.-F.; Wu, D.-P. MYC-regulated lncRNA NEAT1 promotes B cell proliferation and lymphomagenesis via the miR-34b-5p-GLI1 pathway in diffuse large B-cell lymphoma. Cancer Cell Int. 2020, 20, 87. [Google Scholar] [CrossRef]
- Benetatos, L.; Benetatou, A.; Vartholomatos, G. Long non-coding RNAs and MYC association in hematological malignancies. Ann. Hematol. 2020, 99, 2231–2242. [Google Scholar] [CrossRef]
- Yazdi, N.; Houshmand, M.; Atashi, A.; Kazemi, A.; Najmedini, A.A.; Zarif, M.N. Long noncoding RNA PVT1: Potential oncogene in the development of acute lymphoblastic leukemia. Turk. J. Biol. 2018, 42, 405–413. [Google Scholar] [CrossRef]
- Verboom, K.; Van Loocke, W.; Volders, P.-J.; Decaesteker, B.; Cobos, F.A.; Bornschein, S.; De Bock, C.E.; Atak, Z.K.; Clappier, E.; Aerts, S.; et al. A comprehensive inventory of TLX1 controlled long non-coding RNAs in T-cell acute lymphoblastic leukemia through polyA+ and total RNA sequencing. Haematologica 2018, 103, e585–e589. [Google Scholar] [CrossRef]
- Trimarchi, T.; Bilal, E.; Ntziachristos, P.; Fabbri, G.; Dalla-Favera, R.; Tsirigos, A.; Aifantis, I. Genome-wide Mapping and Characterization of Notch-Regulated Long Noncoding RNAs in Acute Leukemia. Cell 2014, 158, 593–606. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, P.; Lin, R.; Rong, L.; Xue, Y.; Fang, Y. LncRNA NALT interaction with NOTCH1 promoted cell proliferation in pediatric T cell acute lymphoblastic leukemia. Sci. Rep. 2015, 5, 13749. [Google Scholar] [CrossRef]
- Luo, Y.Y.; Wang, Z.H.; Yu, Q.; Yuan, L.L.; Peng, H.L.; Xu, Y.X. LncRNA-NEAT1 promotes proliferation of T-ALL cells via miR-146b-5p/NOTCH1 signaling pathway. Pathol. Res. Pract. 2020, 216, 153212. [Google Scholar] [CrossRef]
- Singh, N.; Padi, S.K.R.; Bearss, J.J.; Pandey, R.; Okumura, K.; Beltran, H.; Song, J.H.; Kraft, A.S.; Olive, V. PIM protein kinases regulate the level of the long noncoding RNA H19 to control stem cell gene transcription and modulate tumor growth. Mol. Oncol. 2020, 14, 974–990. [Google Scholar] [CrossRef]
- Chen, L.; Shi, Y.; Li, J.; Yang, X.; Li, R.; Zhou, X.; Zhu, L. LncRNA CDKN2B-AS1 contributes to tumorigenesis and chemoresistance in pediatric T-cell acute lymphoblastic leukemia through miR-335-3p/TRAF5 axis. Anti-Cancer Drugs 2020. [Google Scholar] [CrossRef]
- Li, X.; Song, F.; Sun, H. Long non-coding RNA AWPPH interacts with ROCK2 and regulates the proliferation and apoptosis of cancer cells in pediatric T-cell acute lymphoblastic leukemia. Oncol. Lett. 2020, 20. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, H.G.; Lu, C. A novel long non-coding RNA T-ALL-R-LncR1 knockdown and Par-4 cooperate to induce cellular apoptosis in T-cell acute lymphoblastic leukemia cells. Leuk. Lymphoma 2014, 55, 1373–1382. [Google Scholar] [CrossRef]
- Yang, J.; Yang, Y. Long noncoding RNA endogenous bornavirus-like nucleoprotein acts as an oncogene by regulating microRNA-655-3p expression in T-cell acute lymphoblastic leukemia. Bioengineered 2022, 13, 6409–6419. [Google Scholar] [CrossRef]
- Yang, T.; Jin, X.; Lan, J.; Wang, W. Long non-coding RNA SNHG16 has Tumor suppressing effect in acute lymphoblastic leukemia by inverse interaction on hsa-miR-124-3p. IUBMB Life 2019, 71, 134–142. [Google Scholar] [CrossRef]
- Nashwa, E.K.; Aziz, M.A.A.; Hesham, M.; Matbouly, S.; Mostafa, S.A.; Bakkar, A.; Abouelnile, M.; Noufal, Y.; Mahran, N.A.; Abd Elkhalek, M.A.; et al. Upregulation of leukemia-induced non-coding activator RNA (LUNAR1) predicts poor outcome in pediatric T-acute lymphoblastic leukemia. Immunobiology 2021, 226, 152149. [Google Scholar]
- Asadi, M.; Gholampour, M.A.; Kompani, F.; Alizadeh, S. Expression of Long Non-Coding RNA H19 in Acute Lymphoblastic Leukemia. Cell J. 2023, 25, 1. [Google Scholar]
- Li, J.; Muhammad, J.; Xie, T.; Sun, J.; Lei, Y.; Wei, Z.; Pan, S.; Qin, H.; Shao, L.; Jiang, D.; et al. LINC00853 restrains T cell acute lymphoblastic leukemia invasion and infiltration by regulating CCR9/CCL25. Mol. Immunol. 2021, 140, 267–275. [Google Scholar] [CrossRef]
- Li, G.; Lei, X.; Zhang, Y.; Liu, Z.; Zhu, K. LncRNA PPM1A-AS Regulate Tumor Development Through Multiple Signal Pathways in T-Cell Acute Lymphoblastic Leukemia. Front. Oncol. 2021, 11, 4278. [Google Scholar] [CrossRef] [PubMed]
- Renou, L.; Boelle, P.-Y.; Deswarte, C.; Spicuglia, S.; Benyoucef, A.; Calvo, J.; Uzan, B.; Belhocine, M.; Cieslak, A.; Landman-Parker, J.; et al. Homeobox protein TLX3 activates miR-125b expression to promote T-cell acute lymphoblastic leukemia. Blood Adv. 2017, 1, 733–747. [Google Scholar] [CrossRef] [PubMed]
- El-Khazragy, N.; Esmaiel, M.A.; Mohamed, M.M.; Hassan, N.S. Upregulation of long noncoding RNA Lnc-IRF2-3 and Lnc-ZNF667-AS1 is associated with poor survival in B-chronic lymphocytic leukemia. Int. J. Lab. Hematol. 2020, 42, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.R.; Ruppert, A.S.; Fobare, S.; Chen, T.L.; Liu, C.; Lehman, A.; Blachly, J.S.; Zhang, X.; Lucas, D.M.; Grever, M.R.; et al. The long noncoding RNA, treRNA, decreases DNA damage and is associated with poor response to chemotherapy in chronic lymphocytic leukemia. Oncotarget 2017, 8, 25942–25954. [Google Scholar] [CrossRef]
- Sattari, A.; Siddiqui, H.; Moshiri, F.; Ngankeu, A.; Nakamura, T.; Kipps, T.J.; Croce, C.M. Upregulation of long noncoding RNA MIAT in aggressive form of chronic lymphocytic leukemias. Oncotarget 2016, 7, 54174–54182. [Google Scholar] [CrossRef]
- Jing, Z.; Gao, L.; Wang, H.; Chen, J.; Nie, B.; Hong, Q. Long non-coding RNA GAS5 regulates human B lymphocytic leukaemia tumourigenesis and metastasis by sponging miR-222. Cancer Biomark. 2019, 26, 385–392. [Google Scholar] [CrossRef]
- Elwafa, R.A.; Elrahman, A.A.; Ghallab, O. Long intergenic non-coding RNA-p21 is associated with poor prognosis in chronic lymphocytic leukemia. Clin. Transl. Oncol. 2020, 23, 92–99. [Google Scholar] [CrossRef]
- Wang, L.Q.; Wong, K.Y.; Li, Z.H.; Chim, C.S. Epigenetic silencing of tumor suppressor long non-coding RNA BM742401 in chronic lymphocytic leukemia. Oncotarget 2016, 7, 82400–82410. [Google Scholar] [CrossRef]
- Du, X.; Liu, H.; Yang, C.; Shi, X.; Cao, L.; Zhao, X.; Miao, Y.; Zhu, H.; Wang, L.; Xu, W.; et al. LncRNA landscape analysis identified LncRNA LEF-AS1 as an oncogene that upregulates LEF1 and promotes survival in chronic lymphocytic leukemia. Leuk. Res. 2021, 110, 106706. [Google Scholar] [CrossRef]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.d.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef]
- Gholami, A.; Farhadi, K.; Sayyadipour, F.; Soleimani, M.; Saba, F. Long noncoding RNAs (lncRNAs) in human lymphomas. Genes Dis. 2021, 9, 900–914. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zou, J.; Wan, X.; Sun, C.; Peng, F.; Chu, Z.; Hu, Y. The Role of Noncoding RNAs in B-Cell Lymphoma. Front. Oncol. 2020, 10, 577890. [Google Scholar] [CrossRef] [PubMed]
- Mu, G.; Liu, Q.; Wu, S.; Xia, Y.; Fang, Q. Long Noncoding RNA HAGLROS Promotes the Process of Mantle Cell Lymphoma by Regulating miR-100/ATG5 Axis and Involving in PI3K/AKT/mTOR Signal (Retraction of Vol 47, Pg 3649, 2019); Taylor & Francis Ltd.: Abingdon, OR, USA, 2020. [Google Scholar]
- Hu, G.; Gupta, S.K.; Troska, T.P.; Nair, A.; Gupta, M. Long non-coding RNA profile in mantle cell lymphoma identifies a functional lncRNA ROR1-AS1 associated with EZH2/PRC2 complex. Oncotarget 2017, 8, 80223. [Google Scholar] [CrossRef]
- Wen, S.; Zeng, M.; Li, Y.; Hu, X.; Li, S.; Liang, X.; Zhu, L.; Yang, S. Downregulation of MANCR inhibits cancer cell proliferation in mantle cell lymphoma possibly by interacting with RUNX2. Acta Biochim. Biophys. Sin. 2019, 51, 1142–1147. [Google Scholar] [CrossRef]
- Wang, X.; Sehgal, L.; Jain, N.; Khashab, T.; Mathur, R.; Samaniego, F. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J. Transl. Med. 2016, 14, 346. [Google Scholar] [CrossRef]
- Guo, C.; Gong, M.; Li, Z. Knockdown of lncRNA MCM3AP-AS1 Attenuates Chemoresistance of Burkitt Lymphoma to Doxorubicin Treatment via Targeting the miR-15a/EIF4E Axis. Cancer Manag. Res. 2020, 12, 5845–5855. [Google Scholar] [CrossRef] [PubMed]
- Doose, G.; Haake, A.; Bernhart, S.H.; Duggimpudi, S.; Wojciech, F.; Bergmann, A.K.; Borkhardt, A.; Burkhardt, B.; Claviez, A.; Dimitrova, L.; et al. MINCR is a MYC-induced lncRNA able to modulate MYC’s transcriptional network in Burkitt lymphoma cells. Proc. Natl. Acad. Sci. USA 2015, 112, E5261–E5270. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lv, Y.; Wang, J.; Zhu, X.; Chen, J.; Zhang, W.; Wang, C.; Jiang, L. LncRNA NORAD Mediates the Proliferation and Apoptosis of Diffuse Large-B-Cell Lymphoma via Regulation of miR-345-3p/TRAF6 Axis. Arch. Med. Res. 2022, 53, 271–279. [Google Scholar] [CrossRef]
- Sehgal, L.; Mathur, R.; Braun, F.K.; Wise, J.; Berkova, Z.; Neelapu, S.S.; Kwak, L.; Samaniego, F.J. FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia 2014, 28, 2376–2387. [Google Scholar] [CrossRef]
- Nobili, L.; Ronchetti, D.; Taiana, E.; Neri, A. Long non-coding RNAs in B-cell malignancies: A comprehensive overview. Oncotarget 2017, 8, 60605–60623. [Google Scholar] [CrossRef]
- Zhao, L.; Liu, Y.; Zhang, J.; Liu, Y.; Qi, Q. LncRNA SNHG14/miR-5590-3p/ZEB1 positive feedback loop promoted diffuse large B cell lymphoma progression and immune evasion through regulating PD-1/PD-L1 checkpoint. Cell Death Dis. 2019, 10, 731. [Google Scholar] [CrossRef]
- Tan, L.; Zhu, Y.; Fan, T.; Peng, C.; Wang, J.; Sun, L.; Chen, C. OsZIP7 functions in xylem loading in roots and inter-vascular transfer in nodes to deliver Zn/Cd to grain in rice. Biochem. Biophys. Res. Commun. 2019, 512, 112–118. [Google Scholar] [CrossRef]
- Meng, H.; Zhao, B.; Wang, Y. FOXM1-induced upregulation of lncRNA OR3A4 promotes the progression of diffuse large B-cell lymphoma via Wnt/β-catenin signaling pathway. Exp. Mol. Pathol. 2020, 115, 104451. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Tian, X. Retracted: Knockdown of long noncoding RNA HOTAIR inhibits cell growth of human lymphoma cells by upregulation of miR-148b. J. Cell. Biochem. 2019, 120, 12348–12359. [Google Scholar] [CrossRef]
- Peng, W.; Feng, J. Long noncoding RNA LUNAR1 associates with cell proliferation and predicts a poor prognosis in diffuse large B-cell lymphoma. Biomed. Pharmacother. 2016, 77, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Wu, J.; Feng, J. Long noncoding RNA HULC predicts poor clinical outcome and represents pro-oncogenic activity in diffuse large B-cell lymphoma. Biomed. Pharmacother. 2016, 79, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zhao, H.; Xu, W.; Bao, S.; Cheng, L.; Sun, J. Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma. Mol. Cancer 2017, 16, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Qian, W.; Ye, X. Long Noncoding RNAs in Diffuse Large B-Cell Lymphoma: Current Advances and Perspectives. OncoTargets Ther. 2020, 13, 4295–4303. [Google Scholar] [CrossRef]
- Baytak, E.; Gong, Q.; Akman, B.; Yuan, H.; Chan, W.C.; Küçük, C. Whole transcriptome analysis reveals dysregulated oncogenic lncRNAs in natural killer/T-cell lymphoma and establishes MIR155HG as a target of PRDM1. Tumor Biol. 2017, 39, 1010428317701648. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, S.H.; Yang, W.I.; Kim, S.J.; Yoon, S.O. Association of the long non-coding RNA MALAT1 with the polycomb repressive complex pathway in T and NK cell lymphoma. Oncotarget 2017, 8, 31305. [Google Scholar] [CrossRef]
- Mularoni, V.; Donati, B.; Tameni, A.; Manicardi, V.; Reggiani, F.; Sauta, E.; Zanelli, M.; Tigano, M.; Vitale, E.; Torricelli, F.; et al. Long non-coding RNA mitophagy and ALK–anaplastic lymphoma associated transcript: A novel regulator of mitophagy in T cell lymphoma. Haematologica 2020. [Google Scholar] [CrossRef] [PubMed]
- Fragliasso, V.; Verma, A.; Manzotti, G.; Tameni, A.; Bareja, R.; Heavican, T.B.; Iqbal, J.; Wang, R.; Fiore, D.; Mularoni, V.; et al. The novel lncRNA BlackMamba controls the neoplastic phenotype of ALK− anaplastic large cell lymphoma by regulating the DNA helicase HELLS. Leukemia 2020, 34, 2964–2980. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.-S.; Chung, I.-H.; Lin, Y.-H.; Lin, T.-K.; Chen, W.-J.; Lin, K.-H. The Long Non-Coding RNA MIR503HG Enhances Proliferation of Human ALK-Negative Anaplastic Large-Cell Lymphoma. Int. J. Mol. Sci. 2018, 19, 1463. [Google Scholar] [CrossRef] [PubMed]
- Gourvest, M.; Brousset, P.; Bousquet, M. Long Noncoding RNAs in Acute Myeloid Leukemia: Functional Characterization and Clinical Relevance. Cancers 2019, 11, 1638. [Google Scholar] [CrossRef]
- Sole, C.; Arnaiz, E.; Manterola, L.; Otaegui, D.; Lawrie, C.H. The circulating transcriptome as a source of cancer liquid biopsy biomarkers. In Seminars in Cancer Biology; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Pahle, J.; Walther, W. Vectors and strategies for nonviral cancer gene therapy. Expert Opin. Biol. Ther. 2016, 16, 443–461. [Google Scholar] [CrossRef]
- Sakuma, T.; Yamamoto, T. Acceleration of cancer science with genome editing and related technologies. Cancer Sci. 2018, 109, 3679–3685. [Google Scholar] [CrossRef]

| Alteration | Subtype | Target | Function | Ref. | |
|---|---|---|---|---|---|
| Oncogenic lncRNAs | |||||
| TCL6 | Upregulated | ETV6-RUNX1 | Unknown | Low expression associated with poor disease-free survival | [71] |
| LINC0098 | Upregulated | Pre B-ALL t(12:21) ETV6-RUNX1 and High hyperdiploid | miR-330-5p | miRNA sponge | [85] |
| BALR-2 | Upregulated | B-ALL t (12;21), t (1;19) and MLL-rearrangement | JUN and BIM | Increases cell growth and prednisolone resistance, associated with poor survival | [74] |
| RP11-137H2.4 | Upregulated | B-ALL | NRAS/BRAF/NF-κB MAPK pathways | Inhibits proliferation and migration | [80] |
| ENST00000443469 | Upregulated | B-ALL MLL-rearrangement | LAMP5 | Promotes cell proliferation and inhibits apoptosis | [82] |
| LINC00152 | Upregulated | B-ALL | Unknown | Associated with risk of early relapse | [81] |
| BALR-6 | Upregulated | B-ALL MLL-rearrangement | SP1-mediated transcription of CREB1 | Promotes cell proliferation and inhibits apoptosis | [86] |
| CRNDE | Upregulated | B-ALL | miR-345-5p | Promotes cell proliferation and inhibits apoptosis | [87] |
| ZEB1-AS1 | Upregulated | B-ALL | IL11/STAT3 pathway | Increases cell proliferation | [88] |
| Tumor suppressive lncRNAs | |||||
| IUR | Downregulated | B-ALL Ph+ | STAT5 and CD71 | Increases cell growth and survival | [75] |
| LINC00221 | Downregulated | B-ALL | miR 152-3P and ATP2A2 | Promotes apoptosis and inhibits tumor proliferation | [76] |
| CASC15 | Upregulated | B-ALL ETV6-RUNX1 | SOX4 | Promotes apoptosis and inhibits tumor proliferation | [74,78] |
| lnc-NKX2-3-1 | Upregulated | B-ALL ETV6-RUNX1 | Unknown | Unknown | [72] |
| lnc-TIMM21-5 | Upregulated | B-ALL ETV6-RUNX1 | Unknown | Unknown | [72] |
| lnc-ASTN1-1 | Upregulated | B-ALL ETV6-RUNX1 | Unknown | Unknown | [72] |
| lnc-RTN4R-1 | Upregulated | B-ALL ETV6-RUNX1 | Unknown | Unknown | [72] |
| Alteration | Target | Function | Ref. | |
|---|---|---|---|---|
| SNHG1 | Upregulated | miR 124-3P | Promotes cell proliferation and migration | [107] |
| PVT1 | Upregulated | miR-486-5p, NOP2 and Myc | Increases cell viability, deregulates cell cycle, and inhibits apoptosis | [97] |
| ARIEL | Upregulated | ARID5B | Increases cell growth and survival | [93] |
| LUNAR1 | Upregulated | IGF1R | Increases cell growth | [99,108] |
| NALT | Upregulated | Notch signaling | Induces cell proliferation | [100] |
| NEAT1 | Upregulated | miR-146b-5p | Promotes cell proliferation and growth | [101] |
| AWPPH | Upregulated | ROCK2 | Promotes cell proliferation and inhibits apoptosis | [104] |
| T-ALL-R-LncR1 | Upregulated | Par-4/THAP1 protein complex | Inhibits apoptosis | [105] |
| H19 | Upregulated | SOX2, OCT4 and NANOG | Maintains stemness, promotes cell proliferation, and inhibits apoptosis | [102,109] |
| LINC00853 | Downregulated | CCR9 expression | Inhibits cell proliferation and migration | [110] |
| EBLN3P | Upregulated | miR-655-3p | Increases proliferation, invasion, and migration | [106] |
| PPM1A-AS | Upregulated | Notch, PI3K-AKT, and JAK-STAT signaling | Regulates cell proliferation and apoptosis | [111] |
| LINC00478 | Upregulated | mir-125b | Increases cell growth and invasiveness | [112] |
| Alteration | Target | Function | Ref. | |
|---|---|---|---|---|
| Lnc-IRF2-3 | Upregulated | Unknown | Promotes cell proliferation and inhibits apoptosis | [113] |
| Lnc-ZNF667-AS1 | Upregulated | Unknown | Poor prognosis and survival | [113] |
| treRNA | Upregulated | ZAP70 | Poor prognosis and survival | [114] |
| MIAT | Upregulated | OCT4 | Inhibits apoptosis and increases aggressiveness | [115] |
| GAS5 | Downregulated | miR-222 | Promotes tumorigenesis and metastasis | [116] |
| LincRNA-P21 | Downregulated | p53 | Inhibits apoptosis | [117] |
| BM742401 | Downregulated | Cas9 and 8 | Promotes cell proliferation and inhibits apoptosis | [118] |
| DLEU2 | Downregulated | miR-15a and miR-16-1 | A tumor suppressor | [37] |
| LEF1-AS1 | Upregulated | LEF1 | Inhibits apoptosis | [119] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baghdadi, H.; Heidari, R.; Zavvar, M.; Ahmadi, N.; Shakouri Khomartash, M.; Vahidi, M.; Mohammadimehr, M.; Bashash, D.; Ghorbani, M. Long Non-Coding RNA Signatures in Lymphopoiesis and Lymphoid Malignancies. Non-Coding RNA 2023, 9, 44. https://doi.org/10.3390/ncrna9040044
Baghdadi H, Heidari R, Zavvar M, Ahmadi N, Shakouri Khomartash M, Vahidi M, Mohammadimehr M, Bashash D, Ghorbani M. Long Non-Coding RNA Signatures in Lymphopoiesis and Lymphoid Malignancies. Non-Coding RNA. 2023; 9(4):44. https://doi.org/10.3390/ncrna9040044
Chicago/Turabian StyleBaghdadi, Hamed, Reza Heidari, Mahdi Zavvar, Nazanin Ahmadi, Mehdi Shakouri Khomartash, Mahmoud Vahidi, Mojgan Mohammadimehr, Davood Bashash, and Mahdi Ghorbani. 2023. "Long Non-Coding RNA Signatures in Lymphopoiesis and Lymphoid Malignancies" Non-Coding RNA 9, no. 4: 44. https://doi.org/10.3390/ncrna9040044
APA StyleBaghdadi, H., Heidari, R., Zavvar, M., Ahmadi, N., Shakouri Khomartash, M., Vahidi, M., Mohammadimehr, M., Bashash, D., & Ghorbani, M. (2023). Long Non-Coding RNA Signatures in Lymphopoiesis and Lymphoid Malignancies. Non-Coding RNA, 9(4), 44. https://doi.org/10.3390/ncrna9040044







