Inverse-Folding Design of Yeast Telomerase RNA Increases Activity In Vitro
Abstract
:1. Introduction
2. Results
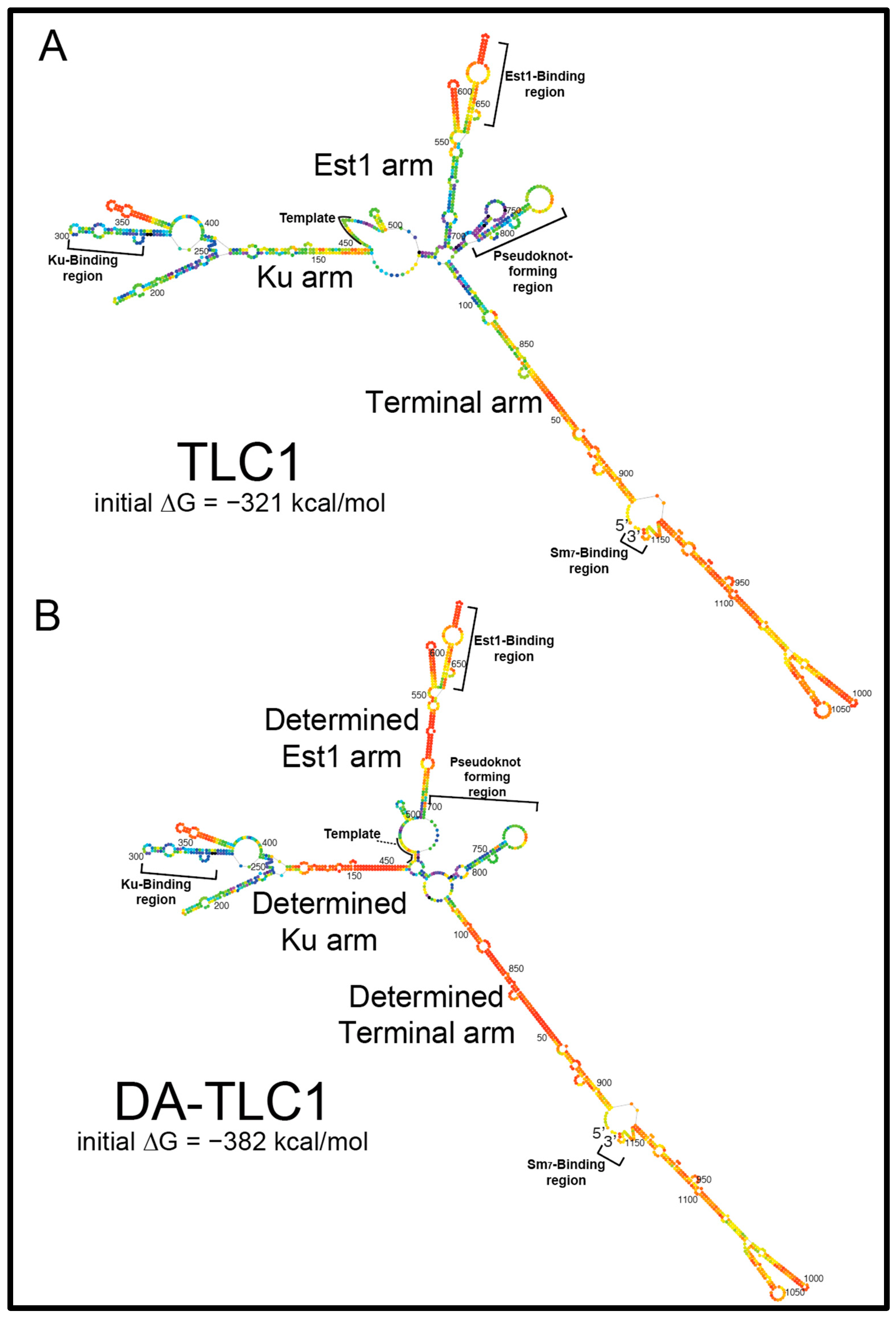
2.1. Inverse Design of a TLC1 RNA That Folds More Readily into Its Active Biological Structure: Determined-Arm TLC1 (DA-TLC1)
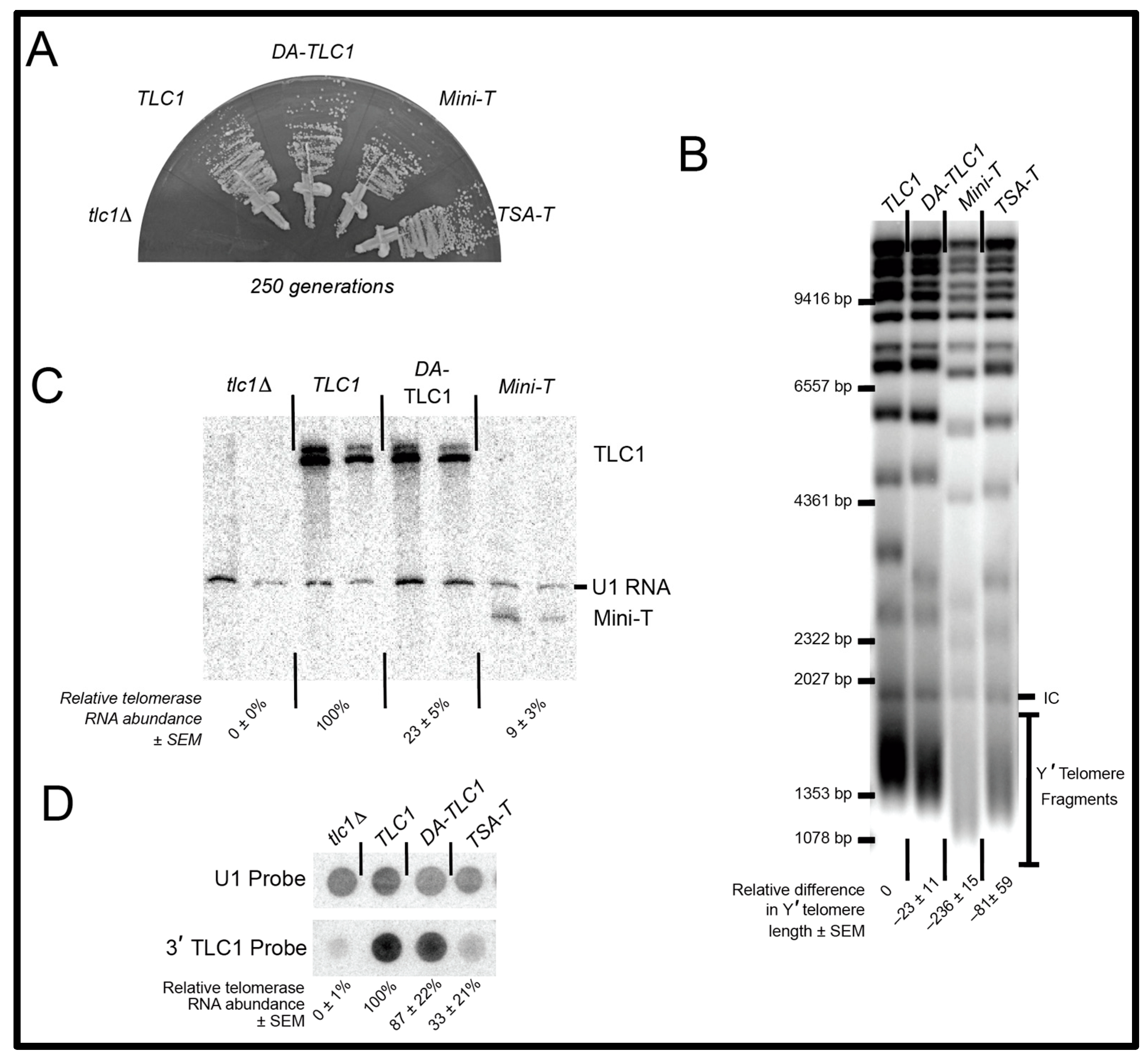
2.2. DA-TLC1 Retains Function In Vivo
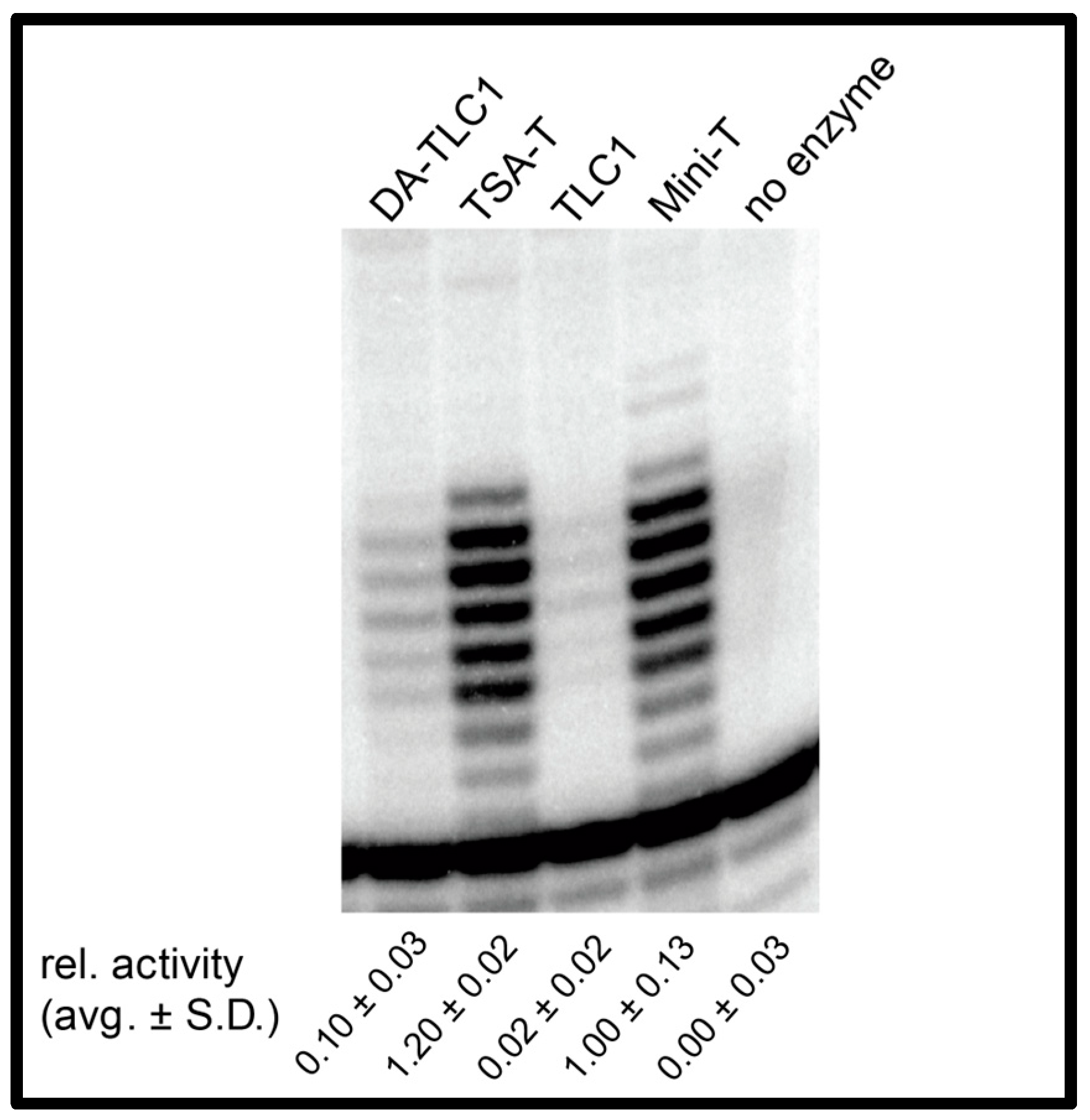
2.3. DA-TLC1 Has Increased Activity In Vitro Compared to Wild-Type TLC1
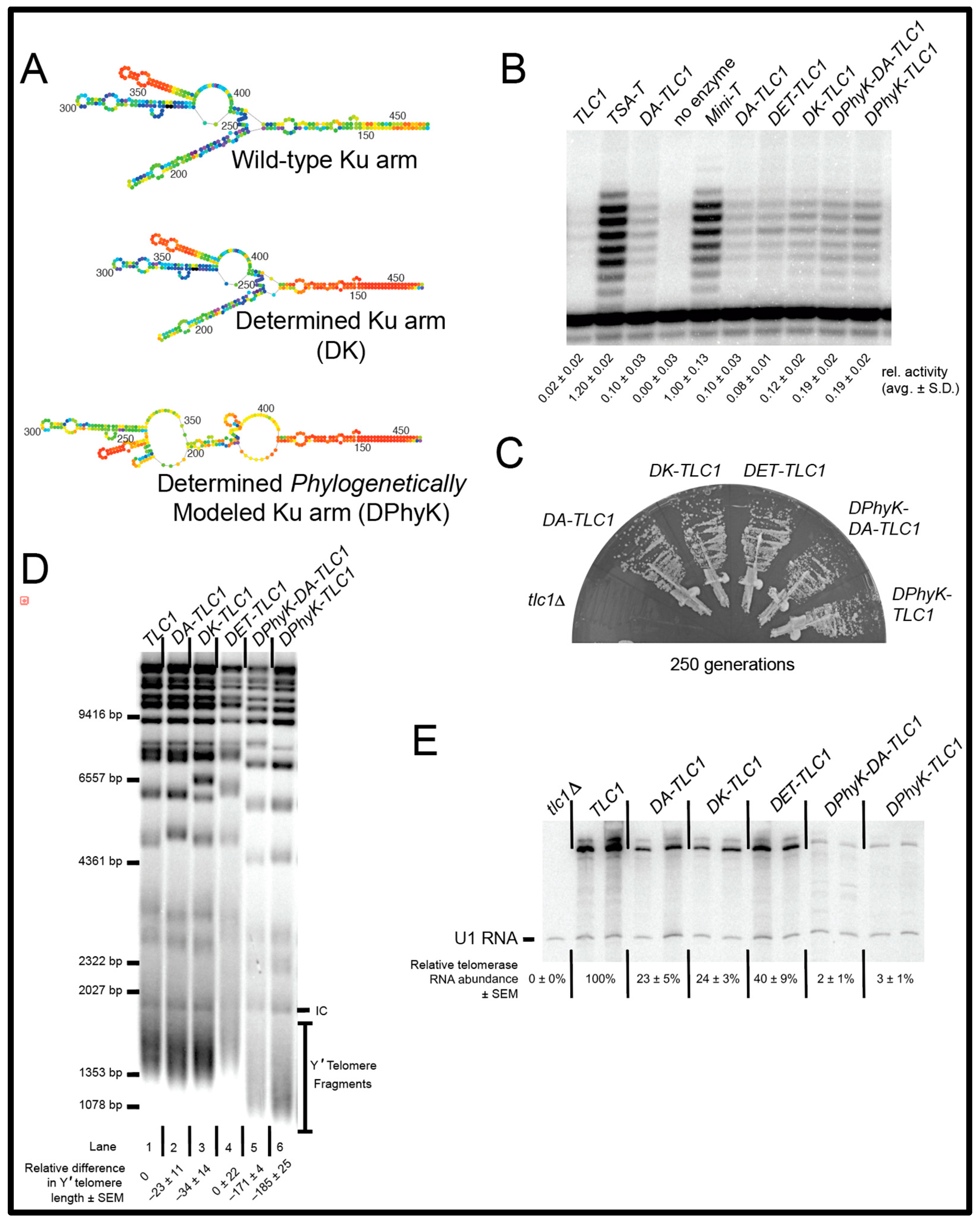
2.4. The Secondary Structure of the Ku Arm Affects Telomerase Function In Vivo and In Vitro
3. Discussion
4. Materials and Methods
4.1. Design of DA-TLC1
4.2. Experiments in Yeast
4.3. Nucleic Acid Blots
4.4. Reconstituted Telomerase Activity Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Watson, J.D. Origin of concatemeric T7 DNA. Nat. New Biol. 1972, 239, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Hayflick, L.; Moorhead, P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar] [CrossRef]
- Weinert, T.A.; Hartwell, L.H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 1988, 241, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Greider, C.W.; Blackburn, E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985, 43, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Weinrich, S.L.; Pruzan, R.; Ma, L.; Ouellette, M.; Tesmer, V.M.; Holt, S.E.; Bodnar, A.G.; Lichtsteiner, S.; Kim, N.W.; Trager, J.B.; et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 1997, 17, 498–502. [Google Scholar] [CrossRef]
- Lingner, J.; Hughes, T.R.; Shevchenko, A.; Mann, M.; Lundblad, V.; Cech, T.R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 1997, 276, 561–567. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 1989, 337, 331–337. [Google Scholar] [CrossRef]
- Lendvay, T.S.; Morris, D.K.; Sah, J.; Balasubramanian, B.; Lundblad, V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 1996, 144, 1399–1412. [Google Scholar] [CrossRef]
- Dandjinou, A.T.; Levesque, N.; Larose, S.; Lucier, J.F.; Abou Elela, S.; Wellinger, R.J. A phylogenetically based secondary structure for the yeast telomerase RNA. Curr. Biol. 2004, 14, 1148–1158. [Google Scholar] [CrossRef]
- Zappulla, D.C.; Cech, T.R. Yeast telomerase RNA: A flexible scaffold for protein subunits. Proc. Natl. Acad. Sci. USA 2004, 101, 10024–10029. [Google Scholar] [CrossRef]
- Mefford, M.A.; Rafiq, Q.; Zappulla, D.C. RNA connectivity requirements between conserved elements in the core of the yeast telomerase RNP. EMBO J. 2013, 32, 2980–2993. [Google Scholar] [CrossRef]
- Niederer, R.O.; Zappulla, D.C. Refined secondary-structure models of the core of yeast and human telomerase RNAs directed by SHAPE. RNA 2015, 21, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Ly, H.; Hussain, A.; Abraham, M.; Pearl, S.; Tzfati, Y.; Parslow, T.G.; Blackburn, E.H. A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proc. Natl. Acad. Sci. USA 2004, 101, 14713–14718. [Google Scholar] [CrossRef]
- Zappulla, D.C.; Goodrich, K.; Cech, T.R. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. NSMB 2005, 12, 1072–1077. [Google Scholar] [CrossRef]
- Mefford, M.A.; Hass, E.P.; Zappulla, D.C. A 4-base pair core-enclosing helix in telomerase RNA is essential and binds to the TERT catalytic protein subunit. Mol. Cell. Biol. 2020, 40, e0023920. [Google Scholar] [CrossRef]
- Qiao, F.; Cech, T.R. Triple-helix structure in telomerase RNA contributes to catalysis. NSMB 2008, 15, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Zappulla, D.C.; Cech, T.R. RNA as a flexible scaffold for proteins: Yeast telomerase and beyond. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Zappulla, D.C. Yeast Telomerase RNA Flexibly Scaffolds Protein Subunits: Results and Repercussions. Molecules 2020, 25, 2750. [Google Scholar] [CrossRef]
- Zappulla, D.C.; Goodrich, K.J.; Arthur, J.R.; Gurski, L.A.; Denham, E.M.; Stellwagen, A.E.; Cech, T.R. Ku can contribute to telomere lengthening in yeast at multiple positions in the telomerase RNP. RNA 2011, 17, 298–311. [Google Scholar] [CrossRef]
- Hass, E.P.; Zappulla, D.C. Repositioning the Sm-Binding Site in Saccharomyces cerevisiae Telomerase RNA Reveals RNP Organizational Flexibility and Sm-Directed 3′-End Formation. Noncoding RNA 2020, 6, 9. [Google Scholar] [CrossRef]
- Peterson, S.E.; Stellwagen, A.E.; Diede, S.J.; Singer, M.S.; Haimberger, Z.W.; Johnson, C.O.; Tzoneva, M.; Gottschling, D.E. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 2001, 27, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Seto, A.G.; Livengood, A.J.; Tzfati, Y.; Blackburn, E.H.; Cech, T.R. A bulged stem tethers Est1p to telomerase RNA in budding yeasts. Genes. Dev. 2002, 16, 2800–2810. [Google Scholar] [CrossRef]
- Seto, A.G.; Zaug, A.J.; Sobel, S.G.; Wolin, S.L.; Cech, T.R. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 1999, 401, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Lebo, K.J.; Niederer, R.O.; Zappulla, D.C. A second essential function of the Est1-binding arm of yeast telomerase RNA. RNA 2015, 21, 862–876. [Google Scholar] [CrossRef]
- Cohn, M.; Blackburn, E.H. Telomerase in yeast. Science 1995, 269, 396–400. [Google Scholar] [CrossRef]
- Lebo, K.J.; Zappulla, D.C. Stiffened yeast telomerase RNA supports RNP function in vitro and in vivo. RNA 2012, 18, 1666–1678. [Google Scholar] [CrossRef]
- Lemieux, B.; Laterreur, N.; Perederina, A.; Noel, J.F.; Dubois, M.L.; Krasilnikov, A.S.; Wellinger, R.J. Active Yeast Telomerase Shares Subunits with Ribonucleoproteins RNase P and RNase MRP. Cell 2016, 165, 1171–1181. [Google Scholar] [CrossRef]
- Churkin, A.; Retwitzer, M.D.; Reinharz, V.; Ponty, Y.; Waldispuhl, J.; Barash, D. Design of RNAs: Comparing programs for inverse RNA folding. Brief. Bioinform. 2018, 19, 350–358. [Google Scholar] [CrossRef]
- Bonnet, E.; Rzazewski, P.; Sikora, F. Designing RNA Secondary Structures Is Hard. J. Comput. Biol. 2020, 27, 302–316. [Google Scholar] [CrossRef]
- Hofacker, I.L.; Fontana, W.; Stadler, P.F.; Bonhoeffer, L.S.; Tacker, M.; Schuster, P. Fast Folding and Comparison of RNA Secondary Structures. Monatshefte Chem. 1994, 125, 167–188. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Mathews, D.H.; Sabina, J.; Zuker, M.; Turner, D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999, 288, 911–940. [Google Scholar] [CrossRef]
- Zuker, M.; Jacobson, A.B. “Well-determined” regions in RNA secondary structure prediction: Analysis of small subunit ribosomal RNA. Nucleic Acids Res. 1995, 23, 2791–2798. [Google Scholar] [CrossRef]
- Zuker, M.; Jacobson, A.B. Using reliability information to annotate RNA secondary structures. RNA 1998, 4, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Kim, Y.; Cruickshank, C.; Theimer, C.A. Thermodynamic characterization of the Saccharomyces cerevisiae telomerase RNA pseudoknot domain in vitro. RNA 2012, 18, 973–991. [Google Scholar] [CrossRef]
- Le, S.; Moore, J.K.; Haber, J.E.; Greider, C.W. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 1999, 152, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Lundblad, V.; Blackburn, E.H. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell 1993, 73, 347–360. [Google Scholar] [CrossRef] [PubMed]
- McEachern, M.J.; Blackburn, E.H. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes. Dev. 1996, 10, 1822–1834. [Google Scholar] [CrossRef]
- Singer, M.S.; Gottschling, D.E. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science 1994, 266, 404–409. [Google Scholar] [CrossRef]
- Lundblad, V.; Szostak, J.W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 1989, 57, 633–643. [Google Scholar] [CrossRef]
- Marin-Gonzalez, A.; Aicart-Ramos, C.; Marin-Baquero, M.; Martín-González, A.; Suomalainen, M.; Kannan, A.; Vilhena, J.G.; Greber, U.F.; Moreno-Herrero, F.; Pérez, R. Double-stranded RNA bending by AU-tract sequences. Nucleic Acids Res. 2020, 22, 12917–12928. [Google Scholar] [CrossRef] [PubMed]
- Stellwagen, A.E.; Haimberger, Z.W.; Veatch, J.R.; Gottschling, D.E. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes. Dev. 2003, 17, 2384–2395. [Google Scholar] [CrossRef] [PubMed]
- Kohrer, K.; Domdey, H. Preparation of high molecular weight RNA. Methods Enzymol. 1991, 194, 398–405. [Google Scholar] [CrossRef]
- Friedman, K.L.; Cech, T.R. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes. Dev. 1999, 13, 2863–2874. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lebo, K.J.; Zappulla, D.C. Inverse-Folding Design of Yeast Telomerase RNA Increases Activity In Vitro. Non-Coding RNA 2023, 9, 51. https://doi.org/10.3390/ncrna9050051
Lebo KJ, Zappulla DC. Inverse-Folding Design of Yeast Telomerase RNA Increases Activity In Vitro. Non-Coding RNA. 2023; 9(5):51. https://doi.org/10.3390/ncrna9050051
Chicago/Turabian StyleLebo, Kevin J., and David C. Zappulla. 2023. "Inverse-Folding Design of Yeast Telomerase RNA Increases Activity In Vitro" Non-Coding RNA 9, no. 5: 51. https://doi.org/10.3390/ncrna9050051
APA StyleLebo, K. J., & Zappulla, D. C. (2023). Inverse-Folding Design of Yeast Telomerase RNA Increases Activity In Vitro. Non-Coding RNA, 9(5), 51. https://doi.org/10.3390/ncrna9050051






