Centromeric Non-Coding RNAs: Conservation and Diversity in Function
Abstract
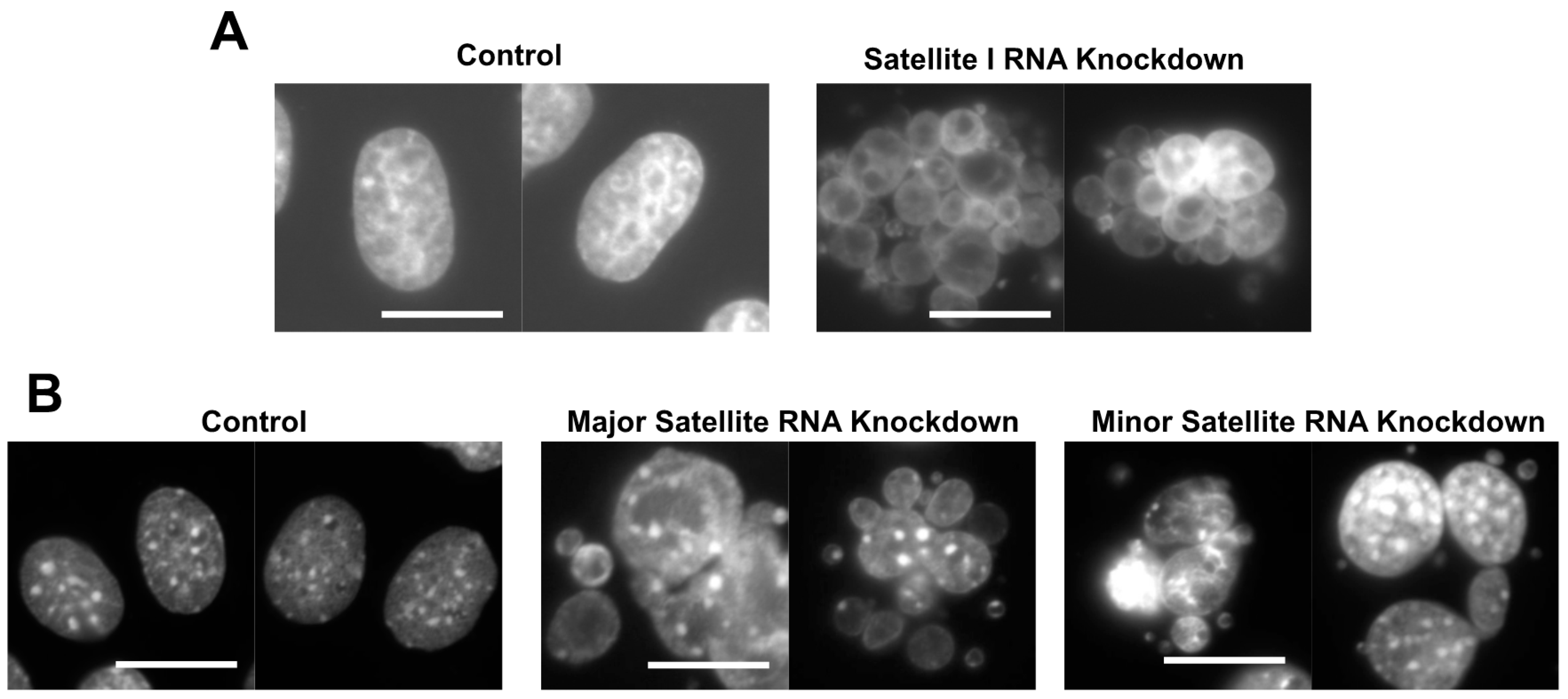
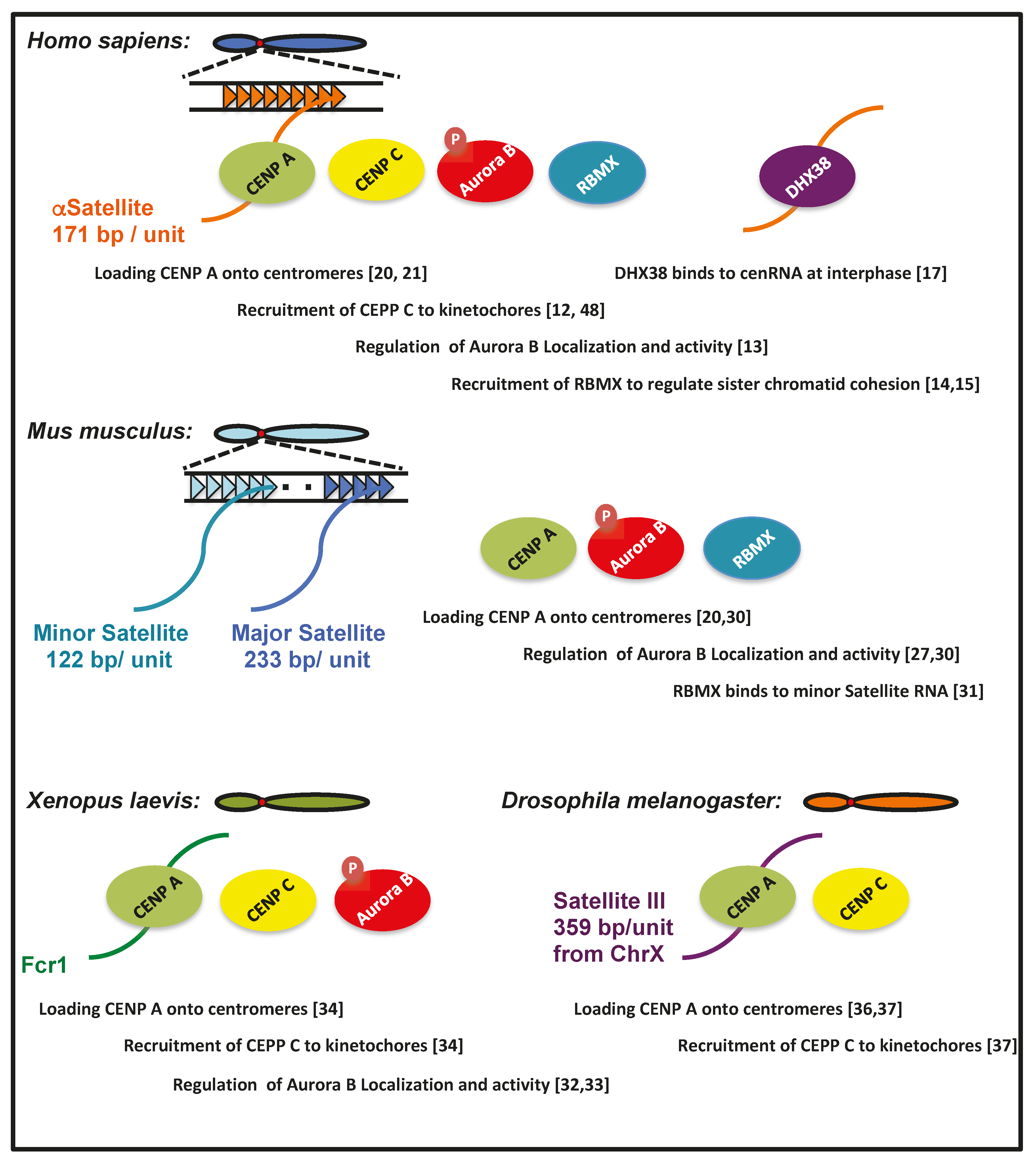
1. Roles of Human cenRNAs in Centromere Function
2. cenRNAs in Mice
3. Roles of cenRNA in Xenopus
4. cenRNA in Drosophila
5. cenRNAs in Plants
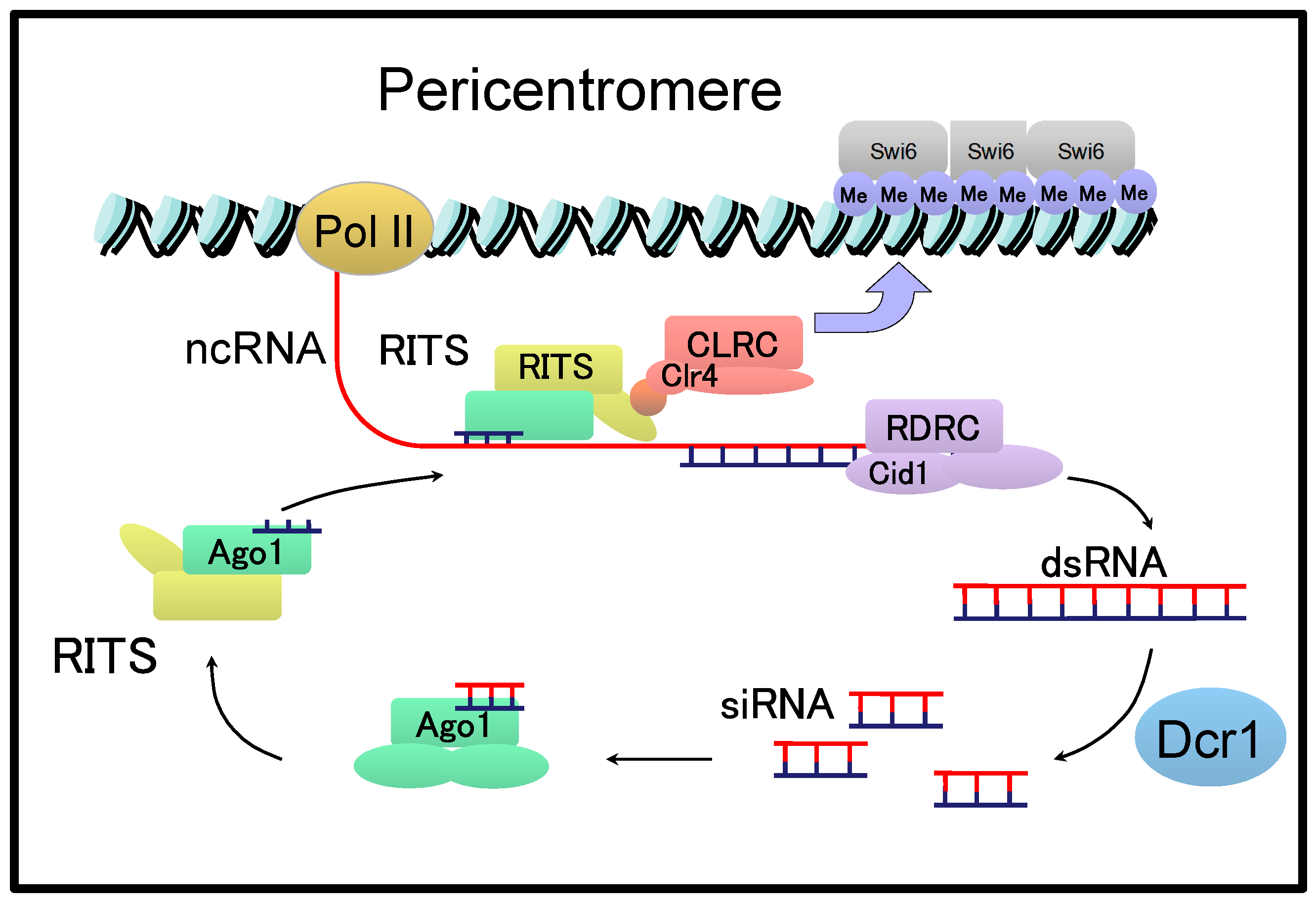
6. cenRNAs in Yeast
7. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- McKinley, K.L.; Cheeseman, I.M. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 2016, 17, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Rosic, S.; Erhardt, S. No longer a nuisance: Long non-coding RNAs join CENP-A in epigenetic centromere regulation. Cell. Mol. Life Sci. 2016, 73, 1387–1398. [Google Scholar] [CrossRef] [PubMed]
- Caceres-Gutierrez, R.; Herrera, L.A. Centromeric non-coding transcription: Opening the black box of chromosomal instability? Curr. Genomics 2017, 18, 227–235. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Perea-Resa, C.; Blower, M.D. Centromere biology: Transcription goes on stage. Mol. Cell. Biol. 2018, 38, e00263-18. [Google Scholar] [CrossRef]
- Talbert, P.B.; Henikoff, S. Transcribing centromeres: Noncoding RNAs and kinetochore assembly. Trends Genet. 2018, 34, 587–599. [Google Scholar] [CrossRef]
- Black, B.E.; Cleveland, D.W. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell 2011, 144, 471–479. [Google Scholar] [CrossRef]
- Fukagawa, T.; Earnshaw, W.C. The centromere: Chromatin foundation for the kinetochore machinery. Dev. Cell 2014, 30, 496–508. [Google Scholar] [CrossRef]
- Cheeseman, I.M.; Desai, A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008, 9, 33–46. [Google Scholar] [CrossRef]
- Ruchaud, S.; Carmena, M.; Earnshaw, W.C. Chromosomal passengers: Conducting cell division. Nat. Rev. Mol. Cell Biol. 2007, 8, 798–812. [Google Scholar] [CrossRef]
- Carmena, M.; Wheelock, M.; Funabiki, H.; Earnshaw, W.C. The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012, 13, 789–803. [Google Scholar] [CrossRef]
- Scott, K.C. Transcription and ncrnas: At the cent(rome)re of kinetochore assembly and maintenance. Chromosome Res. 2013, 21, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Chan, F.L.; Marshall, O.J.; Saffery, R.; Kim, B.W.; Earle, E.; Choo, K.H.; Wong, L.H. Active transcription and essential role of RNA polymerase ii at the centromere during mitosis. Proc. Natl. Acad. Sci. USA 2012, 109, 1979–1984. [Google Scholar] [CrossRef]
- Ideue, T.; Cho, Y.; Nishimura, K.; Tani, T. Involvement of satellite I noncoding RNA in regulation of chromosome segregation. Genes Cells 2014, 19, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Ideue, T.; Nagayama, M.; Araki, N.; Tani, T. RBMX is a component of the centromere noncoding RNP complex involved in cohesion regulation. Genes Cells 2018, 23, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, S.; Takata, H.; Morimoto, A.; Hayashihara, K.; Higashi, T.; Akatsuchi, K.; Mizusawa, E.; Yamakawa, M.; Ashida, M.; Matsunaga, T.M.; et al. RBMX: A regulator for maintenance and centromeric protection of sister chromatid cohesion. Cell Rep. 2012, 1, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Cordin, O.; Beggs, J.D. RNA helicases in splicing. RNA Biol. 2013, 10, 83–95. [Google Scholar] [CrossRef]
- Nishimura, K.; Cho, Y.; Tokunaga, K.; Nakao, M.; Tani, T.; Ideue, T. DEAH box RNA helicase DHX38 associates with satellite I noncoding rna involved in chromosome segregation. Genes Cells 2019, 24, 585–590. [Google Scholar] [CrossRef]
- Mutazono, M.; Morita, M.; Tsukahara, C.; Chinen, M.; Nishioka, S.; Yumikake, T.; Dohke, K.; Sakamoto, M.; Ideue, T.; Nakayama, J.I.; et al. The intron in centromeric noncoding RNA facilitates rnai-mediated formation of heterochromatin. PLoS Genet. 2017, 13, e1006606. [Google Scholar] [CrossRef]
- Izuta, H.; Ikeno, M.; Suzuki, N.; Tomonaga, T.; Nozaki, N.; Obuse, C.; Kisu, Y.; Goshima, N.; Nomura, F.; Nomura, N.; et al. Comprehensive analysis of the ICEN (interphase centromere complex) components enriched in the CENP-A chromatin of human cells. Genes Cells 2006, 11, 673–684. [Google Scholar] [CrossRef]
- Quenet, D.; Dalal, Y. A long non-coding RNA is required for targeting centromeric protein a to the human centromere. Elife 2014, 3, e03254. [Google Scholar] [CrossRef]
- McNulty, S.M.; Sullivan, L.L.; Sullivan, B.A. Human centromeres produce chromosome-specific and array-specific alpha satellite transcripts that are complexed with CENP-A and CENP-C. Dev. Cell 2017, 42, 226–240. [Google Scholar] [CrossRef] [PubMed]
- Goulding, S.E.; Earnshaw, W.C. Shugoshin: A centromeric guardian senses tension. BioEssays 2005, 27, 588–591. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Qu, Q.; Warrington, R.; Rice, A.; Cheng, N.; Yu, H. Mitotic transcription installs Sgo1 at centromeres to coordinate chromosome segregation. Mol. Cell 2015, 59, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Fukagawa, T.; Nogami, M.; Yoshikawa, M.; Ikeno, M.; Okazaki, T.; Takami, Y.; Nakayama, T.; Oshimura, M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004, 6, 784–791. [Google Scholar] [CrossRef]
- Ichida, K.; Suzuki, K.; Fukui, T.; Takayama, Y.; Kakizawa, N.; Watanabe, F.; Ishikawa, H.; Muto, Y.; Kato, T.; Saito, M.; et al. Overexpression of satellite alpha transcripts leads to chromosomal instability via segregation errors at specific chromosomes. Int. J. Oncol. 2018, 52, 1685–1693. [Google Scholar] [CrossRef]
- Lehnertz, B.; Ueda, Y.; Derijck, A.A.; Braunschweig, U.; Perez-Burgos, L.; Kubicek, S.; Chen, T.; Li, E.; Jenuwein, T.; Peters, A.H. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003, 13, 1192–1200. [Google Scholar] [CrossRef]
- Bouzinba-Segard, H.; Guais, A.; Francastel, C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc. Natl. Acad. Sci. USA 2006, 103, 8709–8714. [Google Scholar] [CrossRef]
- Probst, A.V.; Okamoto, I.; Casanova, M.; El Marjou, F.; Le Baccon, P.; Almouzni, G. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev. Cell 2010, 19, 625–638. [Google Scholar] [CrossRef]
- Lu, J.; Gilbert, D.M. Proliferation-dependent and cell cycle regulated transcription of mouse pericentric heterochromatin. J. Cell Biol. 2007, 179, 411–421. [Google Scholar] [CrossRef]
- Ferri, F.; Bouzinba-Segard, H.; Velasco, G.; Hube, F.; Francastel, C. Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 2009, 37, 5071–5080. [Google Scholar] [CrossRef]
- Maison, C.; Bailly, D.; Roche, D.; Montes de Oca, R.; Probst, A.V.; Vassias, I.; Dingli, F.; Lombard, B.; Loew, D.; Quivy, J.P.; et al. Sumoylation promotes de novo targeting of HP1 alpha to pericentric heterochromatin. Nat. Genet. 2011, 43, 220–227. [Google Scholar] [CrossRef]
- Jambhekar, A.; Emerman, A.B.; Schweidenback, C.T.; Blower, M.D. RNA stimulates Aurora B kinase activity during mitosis. PLoS ONE 2014, 9, e100748. [Google Scholar] [CrossRef]
- Blower, M.D. Centromeric transcription regulates Aurora-B localization and activation. Cell Rep. 2016, 15, 1624–1633. [Google Scholar] [CrossRef] [PubMed]
- Grenfell, A.W.; Heald, R.; Strzelecka, M. Mitotic noncoding RNA processing promotes kinetochore and spindle assembly in Xenopus. J. Cell Biol. 2016, 214, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Okita, A.K. Transcriptional silencing of centromere repeats by heterochromatin safeguards chromosome integrity. Curr. Genet. 2019, 65, 1089–1098. [Google Scholar] [CrossRef]
- Bobkov, G.O.M.; Gilbert, N.; Heun, P. Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation. J. Cell Biol. 2018, 217, 1957–1972. [Google Scholar] [CrossRef]
- Rosic, S.; Kohler, F.; Erhardt, S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell Biol. 2014, 207, 335–349. [Google Scholar] [CrossRef]
- Topp, C.N.; Zhong, C.X.; Dawe, R.K. Centromere-encoded RNAs are integral components of the Maize kinetochore. Proc. Natl. Acad. Sci. USA 2004, 101, 15986–15991. [Google Scholar] [CrossRef]
- Du, Y.; Topp, C.N.; Dawe, R.K. DNA binding of centromere protein C (CenpC) is stabilized by single-stranded RNA. PLoS Genet. 2010, 6, e1000835. [Google Scholar] [CrossRef]
- May, B.P.; Lippman, Z.B.; Fang, Y.; Spector, D.L.; Martienssen, R.A. Differential regulation of strand-specific transcripts from Arabidopsis centromeric satellite repeats. PLoS Genet. 2005, 1, 79. [Google Scholar] [CrossRef]
- Ling, Y.H.; Yuen, K.W.Y. Point centromere activity requires an optimal level of centromeric noncoding RNA. Proc. Natl. Acad. Sci. USA 2019, 116, 6270–6279. [Google Scholar] [CrossRef] [PubMed]
- Allshire, R.C.; Ekwall, K. Epigenetic regulation of chromatin states in Schizosaccharomyces pombe. Cold Spring Harb. Perspect. Biol. 2015, 7, a018770. [Google Scholar] [CrossRef] [PubMed]
- White, S.A.; Allshire, R.C. RNAi-mediated chromatin silencing in fission yeast. Curr. Top. Microbiol. Immunol. 2008, 320, 157–183. [Google Scholar] [PubMed]
- Bayne, E.H.; Portoso, M.; Kagansky, A.; Kos-Braun, I.C.; Urano, T.; Ekwall, K.; Alves, F.; Rappsilber, J.; Allshire, R.C. Splicing factors facilitate RNAi-directed silencing in fission yeast. Science 2008, 322, 602–606. [Google Scholar] [CrossRef] [PubMed]
- Chinen, M.; Morita, M.; Fukumura, K.; Tani, T. Involvement of the spliceosomal U4 small nuclear RNA in heterochromatic gene silencing at fission yeast centromeres. J. Biol. Chem. 2010, 285, 5630–5638. [Google Scholar] [CrossRef]
- Kallgren, S.P.; Andrews, S.; Tadeo, X.; Hou, H.; Moresco, J.J.; Tu, P.G.; Yates, J.R., 3rd; Nagy, P.L.; Jia, S. The proper splicing of RNAi factors is critical for pericentric heterochromatin assembly in fission yeast. PLoS Genet. 2014, 10, e1004334. [Google Scholar] [CrossRef]
- Choi, E.S.; Stralfors, A.; Castillo, A.G.; Durand-Dubief, M.; Ekwall, K.; Allshire, R.C. Identification of noncoding transcripts from within CENP-A chromatin at fission yeast centromeres. J. Biol. Chem. 2011, 286, 23600–23607. [Google Scholar] [CrossRef]
- Wong, L.H.; Brettingham-Moore, K.H.; Chan, L.; Quach, J.M.; Anderson, M.A.; Northrop, E.L.; Hannan, R.; Saffery, R.; Shaw, M.L.; Williams, E.; et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007, 17, 1146–1160. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ideue, T.; Tani, T. Centromeric Non-Coding RNAs: Conservation and Diversity in Function. Non-Coding RNA 2020, 6, 4. https://doi.org/10.3390/ncrna6010004
Ideue T, Tani T. Centromeric Non-Coding RNAs: Conservation and Diversity in Function. Non-Coding RNA. 2020; 6(1):4. https://doi.org/10.3390/ncrna6010004
Chicago/Turabian StyleIdeue, Takashi, and Tokio Tani. 2020. "Centromeric Non-Coding RNAs: Conservation and Diversity in Function" Non-Coding RNA 6, no. 1: 4. https://doi.org/10.3390/ncrna6010004
APA StyleIdeue, T., & Tani, T. (2020). Centromeric Non-Coding RNAs: Conservation and Diversity in Function. Non-Coding RNA, 6(1), 4. https://doi.org/10.3390/ncrna6010004



