Abstract
MicroRNAs (miRNAs) are endogenous non-coding small RNAs that downregulate target gene expression by imperfect base-pairing with the 3′ untranslated regions (3′UTRs) of target gene mRNAs. MiRNAs play important roles in regulating cancer cell proliferation, stemness maintenance, tumorigenesis, cancer metastasis, and cancer therapeutic resistance. While studies have shown that dysregulation of miRNA-205-5p (miR-205) expression is controversial in different types of human cancers, it is generally observed that miR-205-5p expression level is downregulated in breast cancer and that miR-205-5p exhibits a tumor suppressive function in breast cancer. This review focuses on the role of miR-205-5p dysregulation in different subtypes of breast cancer, with discussions on the effects of miR-205-5p on breast cancer cell proliferation, epithelial–mesenchymal transition (EMT), metastasis, stemness and therapy-resistance, as well as genetic and epigenetic mechanisms that regulate miR-205-5p expression in breast cancer. In addition, the potential diagnostic and therapeutic value of miR-205-5p in breast cancer is also discussed. A comprehensive list of validated miR-205-5p direct targets is presented. It is concluded that miR-205-5p is an important tumor suppressive miRNA capable of inhibiting the growth and metastasis of human breast cancer, especially triple negative breast cancer. MiR-205-5p might be both a potential diagnostic biomarker and a therapeutic target for metastatic breast cancer.
1. Introduction
Breast cancer is posing a tremendous threat to women’s health globally, and is the most prevalent female malignancy in the world [1]. Traditionally, breast cancer has been classified as in situ (ductal and lobular) or as an invasive disease based upon morphological markers [2]. Advancements in technology has allowed breast cancer to be further classified into different subtypes based upon molecular markers defined by immunohistochemistry (IHC): Estrogen (ER)/progesterone (PR), human epidermal growth factor receptor 2 (HER2), or triple negative breast cancer [2]. More recently, proteomics and gene-expression profiling of breast cancer has allowed for subtyping of breast cancer based upon its molecular profile. The most commonly used classification system stratifies breast cancer into four subtypes: Luminal A, luminal B, HER2 positive (+), and basal-like breast cancer [3]. Because the treatment of breast cancer has evolved from surgery to include systemic therapy determined by different molecular mechanisms and clinical phases, breast cancer therapy has achieved tremendous progress in patient survival [4]. However, despite this advancement, breast cancer remains the leading cause of cancer death in women, with 535,000 deaths in 2016 in 195 countries or territories across the world [5]. Approximately 90% of breast cancer-related death is ascribed to breast cancer metastasis [6]. Metastatic breast cancer is an incurable disease and has an unfavorable prognosis, with an average 5-year survival rate of around 25% [7]. Our limited understanding of the mechanisms underlying breast cancer metastasis constrains the current efficacy of therapy. This is especially true for triple negative breast cancer (TNBC), which is defined as estrogen receptor (ER), progesterone receptor (PR), and HER2 negative. TNBC usually displays higher recurrence, more aggressive metastasis, and worse clinical outcome compared with other breast cancer subtypes. Although TNBC shares many clinical characteristics, TNBC tumors display a higher level of molecular heterogeneity compared to other subtypes. Therefore, understanding the underlying mechanisms of TNBC is imperative, and more novel therapeutic strategies based on molecular mechanisms of TNBC need to be developed to improve patient prognosis [8].
MicroRNAs are endogenous small (19–25 bases) non-coding single-stranded RNAs nucleotides, which repress their target genes by typically pairing to the 3′UTR of mRNAs. In animals, mature microRNAs are derived from long primary transcripts (pri-miRNAs). Pri-miRNA is transcribed by RNA polymerase II from short open reading frames of DNA which do not encode for any proteins [9]. After being transcribed in the nucleus, pri-miRNA is cleaved by nuclear RNAse III Drosha, resulting in stem–loop intermediates termed precursor miRNA (pre-miRNA) [10,11]. After this cleavage event, pre-miRNA is exported out of the nucleus and subsequently truncated by another RNase in the cytoplasm, cytoplasmic RNase III Dicer, forming a 22 nt double-stranded RNA [12,13,14]. Afterwards, the strand with a less stable 5′ end is degraded, while the remaining strand is referred to as a mature miRNA [15,16]. The primary function of miRNAs is to downregulate expression of their target genes. They accomplish this by varying mechanisms, which include RNA degradation, induced decapping, induced deadenylation, altered cap protein binding, reduced ribosome occupancy, and sequestration of mRNA [17]. Usually, the seed sequence of miRNAs paired by base with 3′UTRs of mRNAs. The seed sequence usually consists of 2–8 bases and starts at the second and ends at the eighth nucleotide region counted from the 5′ end of miRNAs [18,19]. The seed sequence plays a significant role in recognition of target mRNAs and provides an important basis for miRNA target prediction.
MiRNAs are involved in the epigenetic regulation of cancer development. The functions of different miRNAs vary in different cancers, but can generally be classified as oncogenic or tumor suppressive. It has been reported that miRNAs participate in cancer initiation, tumorigenesis, proliferation, metastasis, epithelial mesenchymal transition (EMT), stemness maintenance, and therapeutic resistance by downregulating target oncogenes or tumor suppressive genes. Therefore, these dysregulated miRNAs in cancers are found as biomarkers for diagnosis and potential targets for cancer treatment [20].
In humans, miR-205 is located on chromosome 1q32.2 and is composed of a highly conserved structure (Figure 1). More specifically, miR-205 sits between the second and the third exon of LOC642587. MiR-205 is normally expressed in the breast, prostate, and thymus of humans and regulates development of these organs [21]. However, in cancer, miR-205-5p expression is context- and cancer-specific. For example, miR-205-5p is suppressed or silenced in breast, prostate, melanoma, and renal cell carcinoma, but is overexpressed in non-small cell lung carcinoma, bladder cancer, ovarian cancer, endometrioid adenocarcinoma, head and neck cancer, and esophageal adenocarcinoma [22,23,24,25,26,27,28,29].
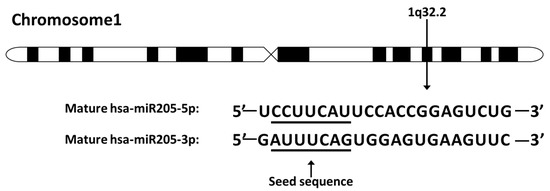
Figure 1.
The miR-205 location on chromosome and sequence. MiR-205 locates on human chromosome 1q32.2. The seed sequences of miR-205-5p and miR-205-3p are underlined. The miR-205 discussed in this review refers to miR-205-5p.
Since microRNAs have multiple targets, thus elucidating their underlying effects on cancers is complex. In support of this, miR-205-5p has been shown to play multiple roles in different cancers (Figure 2). For example, miR-205-5p is involved in embryogenesis, especially in epithelium morphogenesis [30], because it promotes epithelium differentiation in endoderm and ectoderm [31], but miR-205-5p is also involved in epithelial maintenance [32]. Furthermore, in gastric, breast, head, and neck cancer cell lines, epithelial identities, including morphology, increased E-cadherin expression, decreased Vimentin and N-cadherin expression, are restored by re-expressing miR-205-5p [33,34]. However, upregulation of miR-205-5p in cervical, lung, and renal cancer cell lines under hypoxic conditions promotes EMT by targeting ASPP2, one of the downstream apoptosis-stimulating proteins of p53 [35]. Interestingly, miR-205-5p was shown to suppress metastasis by downregulating LPR1, a migration factor, in lung cancer, which indicates a context-dependent function of miR-205-5p, even in the same type of cancer [36]. In addition to converging data with cell differentiation and migration among cancer types, miR-205-5p is also identified as both a positive and negative regulator of proliferation. In prostate cancer, miR-205-5p was shown to arrest cell growth by repressing the mitogen-activated protein kinase (MAPK) and androgen receptor (AR) pathways [37,38]. Whereas an inverse effect was reported in lung cancer as miR-205-5p inhibited PTEN signaling to enhance proliferation [39]. This review aims to summarize the expression and function of miR-205-5p in breast cancer subtypes and highlights the differential roles and targets of miR-205-5p in breast cancer initiation and progression.
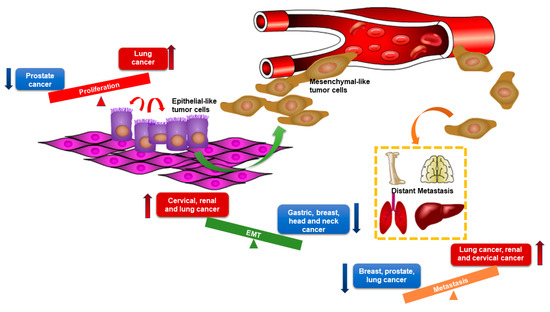
Figure 2.
The distinct roles of miR-205-5p in different types of cancers. Red arrows represent the facilitating effects of miR-205-5p and blue arrows represent the suppressive effects of miR-205-5p. The graph shows opposite roles of miR-205-5p in tumor proliferation, epithelial–mesenchymal transition (EMT), and metastasis among different types of cancers. MiR-205-5p exerts promoting effects in cancers listed in red boxes and exerts suppressive effects in cancers listed in blue boxes.
2. MiR-205-5p Expression in Normal Breast Tissues and Its Dysregulation in Different Subtypes of Breast Cancer Tissues
Originally, miR-205-5p expression was observed exclusively within myoepithelial cells in lobules and ducts of normal breast tissue [32]. However, compared to normal breast tissue, expression of miR-205-5p is decreased in breast cancer [40]. It is interesting to note that, although miR-205-5p expression is lost in breast cancer, the relative levels of miR-205-5p loss varies between subtypes. The level at which miR-205-5p is lost correlates with a variety of mechanisms for their biological behavior (Figure 3). MiR-205-5p is upregulated in ER/PR+ breast cancer compared with HER2+ breast cancer, and shows a significant correlation with ER/PR status [41]. Among all subtypes of breast cancer, triple negative breast cancers (TNBCs) express the least miR-205-5p [32,40,42]. Additionally, metastatic breast cancers express lower levels of miR-205-5p than non-metastatic breast cancers [32,43]. In support of this, our previous study found that lower expression of miR-205-5p was associated with worse recurrence-free survival and distant metastasis-free survival [44]. Low levels of miR-205-5p in serum has also been reported as a diagnostic biomarker for breast cancer patients, as well as an unfavorable clinical prognostic factor [45].
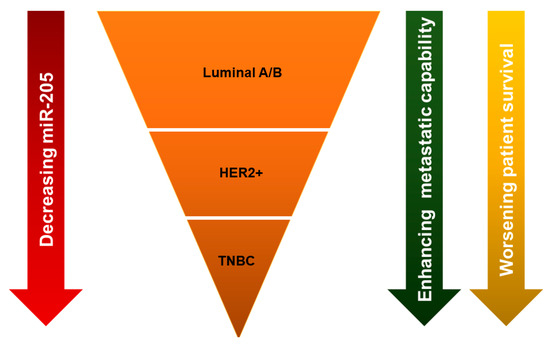
Figure 3.
Differential expression levels of miR-205 among breast cancer subtypes. The expression of miR-205-5p is lower in HER2+ than luminal A/B, and triple negative breast cancer (TNBC) has the lowest miR-205-5p level compared with the other subtypes. Decreasing miR-205-5p expression level is associated with enhanced metastatic capability and worsening of patient survival.
3. Mechanisms of MiR-205-5p Expression Regulation
Since the alteration of miR-205-5p expression is involved in breast cancer development, it is necessary to understand the regulatory mechanisms of miR-205-5p. MiR-205-5p is an intragenic miRNA, which is located within the intron of its host gene, as mentioned previously. One mechanism of miR-205-5p regulation is methylation of CpG islands within its promoter. In breast epithelial cells, overexpression of ERBB2 can drive miR-205-5p promotor methylation via Ras/Raf/MEK/ERK pathway-mediated DNMT upregulation [46]. In TNBC cells, Piovan et al. found that p53 binds a responsive element of the miR-205-5p host gene to increase miR-205-5p expression [47]. It was concluded that the high frequency of TP53 mutations in basal-like breast cancer correlated with suppressive expression of miR-205-5p [47]. Hairy and enhancer of split-1 (HES1) response elements were also found within the miR-205-5p promoter, and exert a negative effect on miR-205-5p expression in breast cancer [48].
4. Breast Cancer Subtype-Specific Roles of MiR-205-5p Dysregulation and Underlying Mechanisms
4.1. MiR-205-5p Dysregulation and Function in Hormonal Receptor Positive (Luminal A and B Subtypes) Breast Cancer
Hormonal receptor positive breast cancer constitutes approximately 80% of breast cancer [49]. Targeting the ER/PR-driven pathway has achieved great success in patients with hormonal receptor positive breast cancer. Tamoxifen, as a first-line endocrine therapy for ER/PR+ breast cancers, binds to the estrogen receptor to competitively block estrogen-induced target gene expression, which leads to suppression of cancer cell proliferation [50]. However, up to 30% of ER+ breast cancer patients are initially resistant to tamoxifen, and approximately 40% of patients with ER+ breast cancer that initially responded to tamoxifen eventually develop resistance [51]. A previous study demonstrated that miR-205-5p is involved in tamoxifen resistance of hormonal receptor positive breast cancer [52]. The molecular mechanisms of tamoxifen resistance can be summarized as follows: (1) Increased bidirectional ER/growth factor (GF) receptor cross-talk; (2) activated ER signaling downstream kinases, such as ERK, MAPK, and AKT; (3) absence of HDAC recruitment to ER corepressors; (4) acetylation of EGFR promotes receptor tyrosine phosphorylation and activation; and (5) corepressor of ER complexes inactivated and coactivator complexes activated [51]. Zhang et al. found that long noncoding RNA-ROR (lncRNA-ROR) was involved in the molecular mechanism of tamoxifen resistance, and found that lncRNA-ROR induced resistance by silencing miR-205-5p [52]. They generated a tamoxifen-resistant MCF7/TR5 cell line by chronic low-dose tamoxifen treatment [52], and found that the level of lncRNA-ROR was raised and miR-205-5p was decreased in these cells [52]. This negative correlation between lncRNA-ROR and miR-205-5p was further analyzed by Renilla luciferase activity, which indicated that lncRNA-ROR acted as a sponge and downregulated miR-205-5p expression [52]. Although they did not fully uncover how miR-205-5p is involved in tamoxifen resistance, it could be speculated that miR-205-5p might play a role in activating a downstream ER-dependent kinase.
Besides its function in endocrine therapy resistance, miR-205-5p has also been found to participate in cell proliferation of ER/PR+ breast cancer. In normal tissue, mammary duct cells are organized asymmetrically to an apical pole toward surrounding tissue and a basal pole that interfaces with the stroma and vasculature, and are mediated by tight junctions [53]. Dysfunction of the asymmetrical growth of the mammary duct is one of the mechanisms of breast cancer progress. Angiomotin (AMOT) is an adaptor protein that regulates tight junctions, and thus the spatial distribution of apical polarity proteins which controls apical asymmetry. Studies have shown that AMOT activates the ERK1/2 pathway to drive cell proliferation in ER+ breast cancer [54], and that miR-205-5p inhibits cell growth by direct targeting of AMOT in MCF-7 breast cancer cells [55]. This suggests that miR-205-5p function is critical for regulating breast cancer growth.
4.2. MiR-205-5p Dysregulation and Function in Her2-Enriched (HER2+) Breast Cancer
HER2-enriched (HER2+) breast cancer is a distinct subtype characterized as high expression of HER2-regulated genes and low expression of hormonal receptors [56,57,58]. HER2+ is driven by the overexpression of ERBB2 (HER2), an oncogene coding for a tyrosine kinase receptor belonging to the human epidermal growth factor receptor (EGFR) family [59,60,61]. The other three members of the EGFR family are HER1 (EGFR), HER3 (ERBB3), and HER4 (ERBB4). After being bound by ligand, the receptors dimerize to either homo- or heterodimers to activate several intracellular signaling pathways, such as the Ras/MAPK and PI3K/Akt, which ultimately promote proliferation, survival, and motility [62]. The HER2/HER3 heterodimer in particular plays a significant role in breast cancer proliferation, and HER3 was found to be frequently co-expressed with HER2 [63]. Furthermore, even in the absence of ligand binding HER2, activation of the PI3K/Akt survival pathway strongly depends on HER3 phosphorylation [64]. Recent studies have shown that miR-205-5p directly targets HER3. This results in the inhibition of proliferation in SKBr3, MCF7, and MDA-MB-231 breast cancer cell lines [40,42].
Until the development of trastuzumab, a HER2-specific recombinant humanized monoclonal antibody, the diagnosis of HER2+ breast cancer had a poor prognosis [60,65,66]. Although trastuzumab has achieved great success in the targeted therapy of HER2+ breast cancer, the recent emerging trastuzumab-acquired resistance of cancer cells provides yet another barrier to overcome [67]. Work looking into the development of resistance has elucidated complex answers: Some classified HER2+ tumors express low HER2 expression, partial masking of the HER2 epitope, and/or poor HER2–T-DM1 complex internalization, among others [68]. In addition to acquired resistance, cancer stem cells (CSCs) also play an important role in trastuzumab resistance [69]. CSCs characteristically have an inherent drug resistance, and therefore are likely the cause of tumor recurrence. Overexpression of miR-205-5p in breast cancer stem cells contributed to the development of trastuzumab resistance by lowering ERB2 and EGFR expression [69]. Furthermore, De Cola et al. indicated that miR-205-5p was significantly upregulated in HER2+ patient-derived breast cancer stem cells (BCSCs) compared with the same cells grown in differentiating spheroid conditions, sphere-derived adherent cells (SDACs) [69]. Knocking-down the expression of miR-205-5p in BCSCs upregulated HER2 and EGFR and sensitivity to Lapatinib. Mechanistically, this group determined that p63 is a direct target of miR-205-5p, and there is a feedback loop between p63 and miR-205-5p, which determines some of the features of BCSCs [69].
4.3. MiR-205-5p Dysregulation and Function in Triple Negative Breast Cancer
Triple negative breast cancer (TNBC) is a group of breast cancer subtypes characterized by traits of aggressive tumor proliferation, early distant metastasis, and enhanced cancer relapse. Previous studies have identified miR-205-5p as a critical regulator of these three characteristics by modulating different signaling pathways.
Dysregulation of the cell cycle is one of the hallmarks of proliferative dysfunction. In addition to identifying putative response elements in the promoter of miR-205-5p, Piovan and colleagues also analyzed the effects of miR-205-5p on cell cycle progression [47]. They found that growth inhibition of MDA-MB-231 and BT-549 overexpressed with miR-205-5p was not due to changes in apoptosis, but rather that a reduction in E2F1 expression, a direct target of miR-205-5p, impaired G1/S phase transition, and promoted cell senescence [47]. Additionally, suppressing miR-205-5p contributed to tumor growth, likely by driving E2F expression and accelerating the G1/S transition [47]. In contrast, a separate study demonstrated that in normal mammary myoepithelial and stem cells, miR-205-5p enhanced cell growth by regulating the cell cycle through phosphatase and tensin homolog (PTEN), a well-studied tumor suppressor gene [70]. Increasing miR-205-5p resulted in reduced PTEN expression, which was found to promote cell G1/S transition with more cells in S phase and fewer cells in G0 phase [71].
The cancer microenvironment is critical to progression, because it not only directly interacts with cancer cells, but also extensively affects the biological behavior of the cell, including tumor proliferation. LAMC1 is a member of the laminin super family, a component of the extracellular matrix (ECM) and a significant ECM regulator [72]. In MDA-MB-231 TNBC cells, LAMC1 was found to be downregulated by miR-205-5p and was confirmed as a direct target [47]. Both repressing LAMC1 by shRNA and overexpression of miR-205-5p inhibited clone formation ability in MDA-MB-231 cells [47]. Although LAMC1 has multiple effects on biological activities, including cell adhesion, proliferation, migration, and differentiation, this study at least demonstrated that miR-205-5p inhibits breast cancer growth partly by targeting LAMC1 [47,72].
Alterations in EGFR signaling pathways drive tumor initiation and progression. As with HER2+ breast cancer, miR-205-5p can regulate TNBC growth by modulating members of the EGFR signaling pathway. Typically, tumors are unable to grow beyond a volume of 1–2 mm3 due to the lack of nutrients perfusing the whole tumor [73]. Therefore, cancer survival and growth relies upon angiogenesis, or the creation of new blood vessels. The high-mobility group box (HMGB) family is a family of non-histone DNA-binding proteins that participate in the formation of new blood vessels by binding to receptors, such as advanced glycation end products (RAGE), to improve angiogenic cytokine release [73]. In breast cancer patients, one member of this family (HMGB3) was found to be overexpressed, and its expression correlated with worse prognosis [49]. Elgamal et al. discovered that overexpression of miR-205-5p in MDA-MB-231 and BT549 TNBC cell lines had similar proliferation rates to those that suppressed HMGB by RNAi [49]. They also demonstrated that HMGB3 is the direct target of miR-205-5p by a dual-luciferase assay, thus proving that miR-205-5p impeded breast cancer growth by targeting HMGB3 [49].
In a different study, 16 miRNA expression profiles were scanned across 32 TNBC samples and their corresponding non-tumor adjacent tissues [74]. In these samples, the miR-205-5p levels were significantly suppressed in the lymph node metastatic group [74]. Compared with their non-tumor adjacent tissues, expression of miR-205-5p was found be decreased more than 2-fold in tumor tissues [74]. These data suggest that miR-205-5p may play a significant role in TNBC metastasis.
Epithelial–mesenchymal transition (EMT) has been demonstrated as a major contributor to cancer metastasis and invasion [75,76]. Polarized epithelial cells undergo a biochemical transformation into cells that display a more mesenchymal-like phenotype and morphology with enhanced migratory capacity, invasiveness, resistance to apoptosis, and productivity of ECM [77]. Through EMT, cancer cells acquire the critical features needed to initiate aggressive invasion, which advances to subsequent metastatic cascade [78]. On the molecular level, EMT is concordant with the upregulation of factors implicated in cytoskeletal changes and the mesenchymal phenotype, including ZEB1, ZEB2, SNAIL, SLUG, TWIST, SMAD 2/3, Vimentin and N-Cadherin, and repressed markers implicated in the maintenance of an epithelial phenotype, including E-Cadherin, Occludin, Claudin, and Laminin [79]. One of the first studies on miR-205-5p observed that expression of miR-205-5p and the miR-200 family in breast cancer was upregulated in epithelial-like phenotype compared with mesenchymal-like phenotype. They also negatively correlated this change in miR-205-5p expression with the expression level of ZEB1 and SIP1, and positively correlated with the expression level of E-cadherin. Then, they demonstrated that miR-205-5p and the miR-200 family targeted ZEB1 and SIP1 to regulate EMT/MET by restoring expression of E-cadherin in the kidney epithelial cell line [34]. Lee et al. further confirmed that miR-205-5p directly targeted ZEB1 and ZEB2 in MDA-MB-231 TNBC cells by a luciferase reporter assay [80]. They also found that polycomb group protein Mel-18, a well-known regulator of chromatin modifications [81], upregulates miR-205-5p expression by methylation of a CpG island within its promoter [80]. More mechanistically, they also found that Mel-18 prevented DNMT recruitment to the promoter of miR-205-5p to reduce methylation effects and rescue miR-205-5p expression [80]. The miR-205-5p re-expression inhibited the invasive and migratory phenotype of MDA-MB-231 and MCF-7 cells driven by shRNA knockdown of Mel-18 (shMel-18) [80]. Furthermore, in vivo NOD/SCID mice models were also established by MCF-7 cells treated with a control shRNA (shcon) or shMel-18. They found more aggressive and mesenchymal-like tumors in mice injected with knockdown of Mel-18, and an upregulation of mesenchymal markers and downregulation of epithelial markers [80]. In addition to being involved in angiogenesis and cell proliferation, HMGB3 is another target of miR-205-5p that is responsible for EMT in breast cancer [82]. In a transwell invasion assay, miR-205-5p was shown to suppress MDA-MB-231 and BT549 invasion [82]. These data suggest that miR-205-5p is an important regulator of EMT, and those phenotypic processes that are associated with EMT, such as cell migration.
Integrins are a family of heterodimeric transmembrane receptors that bind extracellular matrix proteins, which are extensively involved in tumor cell adhesion, migration, invasion, and metastasis [83,84]. Integrin α5 (ITGA5) is one of the members of integrins and is found to play an essential role in breast cancer metastasis and growth [85,86]. According to our recent study, we found that ITGA5 expression was increased upon suppression of miR-205-5p in TNBC [44]. Overexpressing miR-205-5p in MDA-231-LM2 and SUM-159 TNBC cell lines drastically inhibited their migration and invasion capabilities [44]. Through rescue experiments, we found that re-expression of ITGA5 restores those metastatic characteristics. Further probing revealed that miR-205-5p directly targeted, and thus downregulated, ITGA5 and suppressed TNBC metastasis through the Src/Vav2/Rac1 signaling pathway [44].
Angiogenesis is not only necessary for tumor survival, but also plays an essential role in the dissemination and establishment of tumor metastases. In order to escape the primary tumor, tumor cells hijack endothelial cells to form new blood vessels. These neovessels allow for tumor cells to enter into the bloodstream. Therefore, the more highly vascularized the tumor is, the greater the chance that a cancer cell can metastasize [87]. Not surprisingly, miR-205-5p is also involved in angiogenesis. Wu et al. confirmed miR-205-5p paired to the 3′UTR of VEGF-A by a luciferase reporter assay and hypothesized that the observed reduced lung metastasis of MDA-MB-231 cell line transfected with miR-205-5p in vivo was partly due to repressed VEGF-A expression [40]. Cancer-associated fibroblasts (CAFs), a critical tumor stroma participant for tumor angiogenesis, were also found to be regulated by miR-205-5p in breast cancer. MiR-205-5p targets YAP1 (Yes-associated protein) in normal fibroblasts (NFs), preventing CAFs transformation from NFs, and suppresses angiogenesis, invasion, and metastasis of breast cancer cells in vivo [88].
Cancer stem cells mediate cancer relapse, metastasis, and therapy resistance. The stemness of cancer cells encompasses the capacity of asymmetrical self-renewal to one stem cell and one progenitor cell, as well as symmetrical division into two identical stem cells which can retain the original capacity for self-renewal [89]. MicroRNAs are a significant regulator of CSCs by targeting some critical members of pathways that regulate CSC phenotypes or EMT, including Bmi1, Suz12, Sox2, Klf4, and ZEB1/2 [34,90,91,92]. Sempere et al. reported that the low expression level of miR-205-5p was associated with increased relapse rate in patients with triple negative breast cancer that were enriched with breast cancer stem cells, implicating miR-205-5p in cell stemness [32]. In regard to EMT, the loss of E-cadherin during EMT discharges β-catenin from the plasma membrane and activates canonical Wnt signaling by β-catenin translocation to the nucleus, which is an essential pathway for cancer stem cells [93,94,95]. Loss of epithelial polarity also induces the Hippo pathway to promote mammosphere formation, an assay that can define stemness in vitro [96,97,98]. Because, disturbed epithelial polarity contributes to self-renewing symmetric divisions of CSCs that drives those CSC subgroups surging among cancer cell pools [99]. To further uncover the relation between miR-205-5p and self-renewing symmetric division, Chao et al. found that ZEB1, a direct target of miR-205-5p, and suppression of miR-205-5p results in enhanced ZEB1 expression. Enhanced ZEB1 expression drove self-renewing symmetric division and maintenance of stemness of CSCs by redistribution of NUMB, a protein primarily participating in controlling the polarity of stem cells [48]. Meanwhile, overexpression of miR-205-5p in MDA-MB-231 and BT-549 TNBC cell lines inhibited NOTCH2 expression, another direct target of miR-205-5p, which resulted in a loss of stem cell identity [48]. Intriguingly, Notch2 and its ligand, Jagged-1, could also upregulate HES1 expression to inhibit miR-205-5p, and constituted a negative feedback loop of Notch2/miR-205-5p/ZEB1 signaling in breast cancer cells [48].
The CSC-like property suppressive effects by miR-205-5p were also observed in our previous study [44]. A well-established serum-free suspension culture mammary sphere formation assay was performed to assess the CSC-like property of SUM-159 TNBC cells [44]. We found that the counts of spheres formed were reduced by miR-205-5p expression and increased by ITGA5 re-expression. These data support our in vivo data of nude mouse orthotopic mammary xenograft models, showing that miR-205-5p acts as a CSC inhibitor [44].
5. Potential Diagnostic and Therapeutic Values of MiR-205-5p in Breast Cancer
5.1. MiR-205-5p Abnormal Expression as a Potential Diagnostic Marker for Breast Cancer
Different types of cancers have varying microRNA molecular profiles. Therefore, characterizing specific microRNA levels for a particular cancer is helpful not only for confirming the diagnosis, but also for treatment decision, staging, and prognosis. Therefore, understanding the relationship between microRNAs and the cancer and subtype will contribute to a much more accurate diagnostic, subtype, grading, response, and prognosis prediction tool. In regards to miR-205-5p, Berber found that a 5-fold decrease in miR-205-5p expression levels compared to normal breast tissue was accurate to predict breast cancer lymph node metastasis [74]. This paper was able to obtain a sensitivity of 68.8% and specificity of 81.3% in a sample size of 32 patients with TNBC who underwent radical mastectomy and axillary dissection [74]. This study demonstrates the usefulness of miRNAs, particularly miR-205-5p, in patient prognosis prediction, and revealed that miR-205-5p expression levels might be one of the candidate indicators for TNBC metastasis [74].
It is well-known that intracellular miRNAs are excreted by cells into the blood stream after being packaged into exosomes or microvesicles [100]. This suggests that circulating miRNAs serve as ideal biomarkers for earlier detection of cancer [101]. Shaker and colleagues collected blood samples from 100 breast cancer patients and 30 healthy females, and identified the expression levels of miRNAs [102]. In this case, miR-205-5p was identified as a critical differential circulating miRNA between these groups. The sensitivity of miR-205-5p for detection of breast cancer was 98.8% and the specificity was 100% by using the optimal cutoff value [102]. Zhang et al. also performed a case control study on 58 breast cancer patients and 93 healthy controls [72]. The diagnostic proficiency of serum miR-205-5p for breast cancer was achieved at a sensitivity of 86.2% and a specificity of 82.8% [72]. Together, these studies demonstrate that miRNAs, and more specifically miR-205-5p, are potent diagnostic and prognostic prediction tools.
5.2. The Role of MiR-205-5p in Breast Cancer Treatment: A Potential Therapeutic Agent and Regulation of Drug Response/Resistance
As microRNAs are shown to be tremendous tumor suppressive effects, the prospects of anti-microRNAs and microRNAs themselves as therapeutics are promising. Since miR-205-5p shows differential effects between breast cancer subtypes, miR-205-5p therapy likely has the most patient benefit in TNBC. This is because many studies have shown that miR-205-5p is highly downregulated in TNBC, and re-expression of miR-205-5p in TNBC consistently inhibits tumor growth, stemness, and metastasis. However, in order to use miRNAs in the clinic, many barriers revolving around preventing microRNA degradation in vivo, inefficient systemic and targeted delivery, and reduced uptake by cells remains to be solved [103].
MicroRNAs are extensively involved in therapeutic resistance [20,104,105,106]. The luminal A subtype of breast cancer is less sensitive to chemotherapy than other subtypes. Instead, treatment of luminal subtypes of breast cancer consists of endocrine therapy, which is highly effective. Although endocrine therapy is effective, chemotherapy is still required to be administered to those with early stage, or with high risk and advanced disease states. In addition to their involvement in drug resistance, miRNAs can also sensitize cells to chemotherapy, resulting in enhanced response and better patient prognosis. For example, circulating miR-205-5p was identified as a potential predictor of resistance to chemotherapy of epirubicin plus paclitaxel in luminal A subtype [107]. Additionally, miR-205-5p was also found to sensitize MDA-MB-231 TNBC and MCF-7 cells to docetaxel, both in vitro and in vivo [108]. In another study, Hu et al. analyzed 30 breast cancer tissues from patients and found that miR-205-5p had a positive correlation with neoadjuvant chemotherapy response rate [109]. MCF-7/A02 and CALDOX are chemoresistant breast cancer cell lines derived from the chemosensitive cell lines MCF-7 and Cal51 cells, respectively. MiR-205-5p was found to be downregulated in MCF-7/A02 and CALDOX compared with their parental cell lines [109]. Re-expression of miR-205-5p re-sensitized MCF-7/A02 and CALDOX response to doxorubicin and docetaxol [109]. It was further revealed that over-expression of miR-205-5p blocked the PI3K/AKT pathway by targeting VEGF-A and FGF2, which resulted in increased apoptosis upon chemotherapy treatment [109]. MiR-205-5p was also suggested to be an important negative mediator of radiotherapeutic resistance [110]. Ionizing radiation (IR) enhanced hypermethylation of CpG islands of the host gene of miR-205-5p and suppressed miR-205-5p expression [110]. A miR-205-5p mimic attenuated the IR induced Bcl-w increase, malignancy, and lung metastasis in H460 and MDA-MB-231 cells [110].
6. Summary and Perspectives
The studies on miR-205-5p have shed light on the complicated molecular mechanisms of breast cancer initiation and development. In general, miR-205-5p plays a tumor suppressive role in breast cancer. It inhibits tumor growth, metastasis, EMT, cancer stem cell maintenance, and drug resistance (Figure 4). The miR-205-5p expression is reduced in breast cancer compared with normal breast tissue. The expression of miR-205-5p in breast cancer varies among subtypes, with hormonal receptor positive (ER+) expressing the highest levels, HER2-enriched expressing lower levels, and triple negative breast cancer expressing the lowest levels of miR-205-5p. In hormonal receptor positive breast cancer, miR-205-5p not only suppresses tumor growth, but also participates in tamoxifen sensitivity maintenance. In HER2-enriched breast cancer, miR-205-5p plays a controversial role, which on the one hand downregulates HER3 to inhibit HER2/HER3 dimerization and effects on tumor growth, and on the other hand, downregulates HER2 expression, thus inducing lapatinib resistance. In TNBC, miR-205-5p is more extensively involved in tumor growth, metastasis, EMT, and stemness maintenance. The known direct targets of miR-205-5p in breast cancer are summarized in Table 1.
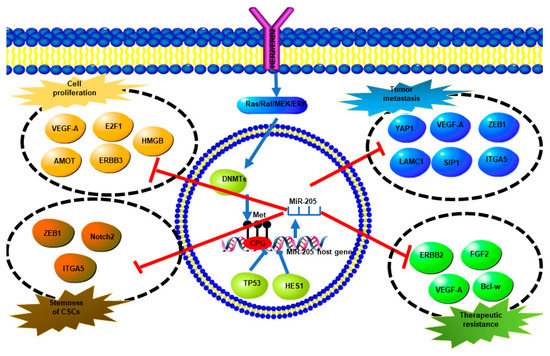
Figure 4.
A summary of miR-205-5p expression regulation and direct targets of miR-205-5p and their biological effects. Overexpression of ERBB2 promotes methylation of the miR-205-5p promoter via the Ras/Raf/MEK/ERK pathway which upregulates DNMTs, which finally results in miR-205-5p downregulation. TP53 and HES also inhibit miR-205-5p expression. MiR-205-5p targets different genes directly to regulate cell proliferation, tumor metastasis, stemness, and therapeutic resistance.

Table 1.
A summary of the validated direct targets of miR-205-5p in breast cancer.
Since more and more targets for miR-205-5p in breast cancer are being discovered, the mechanisms of how miR-205-5p regulate breast cancer development are expanding and are better understood. The roles of those target genes in breast cancer have been further discussed as well. Furthermore, the significance of miR-205-5p as a potential therapeutic target has been highlighted, as well as its target genes. However, there are still many questions that need to be answered in the future. Both miR-205-5p and the miR-200 family function similarly in breast cancer and even share some target genes, such as ZEB1 and SIP1 [34,74,115]. This opens up the possibility that these miRNAs share more commonalities, such as their regulating mechanism, as well as if they interact with each other, and even if these miRNAs have synergistic effects. The combination therapy of the two is also an understudied topic in the field. MiR-205-5p also interacts with other non-coding RNAs, such as lincRNA-ROR and lncRNA-PNUTS, which has opened a new horizon in studying miR-205-5p [52,116,117]. In addition, while studies clearly show that miR-205-5p displays strong tumor suppressive effects and has the potential to be a therapeutic agent for treating triple negative breast cancer, the challenge is how to safely and efficiently deliver miR-205-5p to tumor tissues.
Funding
This work was supported by a Research Scholar Grant (RGS-15-026-01-CSM) from the American Cancer Society, and a Postdoctoral Fellowship (PF-18-236-01-CCG) from the American Cancer Society–Michigan Cancer Research Fund.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ferlay, J.; Shin, H.R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 2010, 127, 2893–2917. [Google Scholar] [CrossRef] [PubMed]
- Vuong, D.; Simpson, P.T.; Green, B.; Cummings, M.C.; Lakhani, S.R. Molecular classification of breast cancer. Virchows Arch. 2014, 465, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Bluth, M.H.; Ali-Fehmi, R. Breast Carcinoma: Updates in Molecular Profiling 2018. Clin. Lab. Med. 2018, 38, 401–420. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Linden, H.M.; Anderson, B.O.; Li, C.I. Trends in 5-year survival rates among breast cancer patients by hormone receptor status and stage. Breast Cancer Res. Treat. 2014, 147, 609–616. [Google Scholar] [CrossRef]
- Fitzmaurice, C.; Akinyemiju, T.F.; Al Lami, F.H.; Alam, T.; Alizadeh-Navaei, R.; Allen, C.; Alsharif, U.; Alvis-Guzman, N.; Amini, E.; Anderson, B.O.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2018, 4, 1553–1568. [Google Scholar]
- Spano, D.; Heck, C.; De Antonellis, P.; Christofori, G.; Zollo, M. Molecular networks that regulate cancer metastasis. Semin. Cancer Biol. 2012, 22, 234–249. [Google Scholar] [CrossRef]
- Rabbani, S.A.; Mazar, A.P. Evaluating distant metastases in breast cancer: From biology to outcomes. Cancer Metastasis Rev. 2007, 26, 663–674. [Google Scholar] [CrossRef]
- Denkert, C.; Liedtke, C.; Tutt, A.; von Minckwitz, G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet 2017, 389, 2430–2442. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Kim, V.N. MicroRNA precursors in motion: Exportin-5 mediates their nuclear export. Trends Cell Biol. 2004, 14, 156–159. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Grishok, A.; Pasquinelli, A.E.; Conte, D.; Li, N.; Parrish, S.; Ha, I.; Baillie, D.L.; Fire, A.; Ruvkun, G.; Mello, C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001, 106, 23–34. [Google Scholar] [CrossRef]
- Hutvagner, G.; McLachlan, J.; Pasquinelli, A.E.; Balint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Hutvagner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Mott, J.L.; Mohr, A.M. Overview of MicroRNA Biology. Semin. Liver Dis. 2015, 35, 3–11. [Google Scholar]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Ma, F.; Liu, X.; Li, D.; Wang, P.; Li, N.; Lu, L.; Cao, X. MicroRNA-466l upregulates IL-10 expression in TLR-triggered macrophages by antagonizing RNA-binding protein tristetraprolin-mediated IL-10 mRNA degradation. J. Immunol. 2010, 184, 6053–6059. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef]
- Vosgha, H.; Salajegheh, A.; Smith, R.A.; Lam, A.K. The important roles of miR-205 in normal physiology, cancers and as a potential therapeutic target. Curr. Cancer Drug Targets 2014, 14, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.A.; Majid, S.; de Semir, D.; Nosrati, M.; Bezrookove, V.; Kashani-Sabet, M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J. Biol. Chem. 2011, 286, 16606–16614. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Gottardo, F.; Liu, C.G.; Ferracin, M.; Calin, G.A.; Fassan, M.; Bassi, P.; Sevignani, C.; Byrne, D.; Negrini, M.; Pagano, F.; et al. Micro-RNA profiling in kidney and bladder cancers. Urol. Oncol. 2007, 25, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Leung, H.; Khan, K.M.; Miller, C.G.; Subbaramaiah, K.; Falcone, D.J.; Dannenberg, A.J. Tobacco smoke induces urokinase-type plasminogen activator and cell invasiveness: Evidence for an epidermal growth factor receptor dependent mechanism. Cancer Res. 2007, 67, 8966–8972. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Davidson, B.; Zhang, Z.; Kleinberg, L.; Li, M.; Florenes, V.A.; Wang, T.L.; Shih Ie, M. Gene expression signatures differentiate ovarian/peritoneal serous carcinoma from diffuse malignant peritoneal mesothelioma. Clin Cancer Res. 2006, 12, 5944–5950. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Lin, Z.; Zhuang, Z.; Liang, X. Expression profile of mammalian microRNAs in endometrioid adenocarcinoma. Eur. J. Cancer Prev. 2009, 18, 50–55. [Google Scholar] [CrossRef]
- Lebanony, D.; Benjamin, H.; Gilad, S.; Ezagouri, M.; Dov, A.; Ashkenazi, K.; Gefen, N.; Izraeli, S.; Rechavi, G.; Pass, H.; et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J. Clin. Oncol. 2009, 27, 2030–2037. [Google Scholar] [CrossRef]
- Kimura, S.; Naganuma, S.; Susuki, D.; Hirono, Y.; Yamaguchi, A.; Fujieda, S.; Sano, K.; Itoh, H. Expression of microRNAs in squamous cell carcinoma of human head and neck and the esophagus: miR-205 and miR-21 are specific markers for HNSCC and ESCC. Oncol. Rep. 2010, 23, 1625–1633. [Google Scholar]
- Shingara, J.; Keiger, K.; Shelton, J.; Laosinchai-Wolf, W.; Powers, P.; Conrad, R.; Brown, D.; Labourier, E. An optimized isolation and labeling platform for accurate microRNA expression profiling. RNA 2005, 11, 1461–1470. [Google Scholar] [CrossRef]
- Darnell, D.K.; Kaur, S.; Stanislaw, S.; Konieczka, J.H.; Yatskievych, T.A.; Antin, P.B. MicroRNA expression during chick embryo development. Dev. Dyn. 2006, 235, 3156–3165. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Christensen, M.; Silahtaroglu, A.; Bak, M.; Heath, C.V.; Schwartz, G.; Wells, W.; Kauppinen, S.; Cole, C.N. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007, 67, 11612–11620. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Li, M.; Zhang, L.; Bi, Y.; Wang, P.; Li, J.; Jiang, X. MicroRNA-205 suppresses the invasion and epithelial-mesenchymal transition of human gastric cancer cells. Mol. Med. Rep. 2016, 13, 4767–4773. [Google Scholar] [CrossRef][Green Version]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yu, M.; Zhao, K.; He, M.; Ge, W.; Sun, Y.; Wang, Y.; Sun, H.; Hu, Y. Upregulation of MiR-205 under hypoxia promotes epithelial-mesenchymal transition by targeting ASPP2. Cell Death Dis. 2016, 7, 2517. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Bu, G. MicroRNA-205 inhibits tumor cell migration through down-regulating the expression of the LDL receptor-related protein 1. Biochem. Biophys. Res. Commun. 2009, 388, 400–405. [Google Scholar] [CrossRef]
- Boll, K.; Reiche, K.; Kasack, K.; Morbt, N.; Kretzschmar, A.K.; Tomm, J.M.; Verhaegh, G.; Schalken, J.; von Bergen, M.; Horn, F.; et al. MiR-130a, miR-203 and miR-205 jointly repress key oncogenic pathways and are downregulated in prostate carcinoma. Oncogene 2013, 32, 277–285. [Google Scholar] [CrossRef]
- Gandellini, P.; Folini, M.; Longoni, N.; Pennati, M.; Binda, M.; Colecchia, M.; Salvioni, R.; Supino, R.; Moretti, R.; Limonta, P.; et al. miR-205 Exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 2009, 69, 2287–2295. [Google Scholar] [CrossRef]
- Bai, J.; Zhu, X.; Ma, J.; Wang, W. miR-205 regulates A549 cells proliferation by targeting PTEN. Int. J. Clin. Exp. Pathol. 2015, 8, 1175–1183. [Google Scholar]
- Wu, H.; Zhu, S.; Mo, Y.Y. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009, 19, 439–448. [Google Scholar] [CrossRef]
- Mattie, M.D.; Benz, C.C.; Bowers, J.; Sensinger, K.; Wong, L.; Scott, G.K.; Fedele, V.; Ginzinger, D.; Getts, R.; Haqq, C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol. Cancer 2006, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Casalini, P.; Piovan, C.; Di Leva, G.; Merlo, A.; Triulzi, T.; Menard, S.; Croce, C.M.; Tagliabue, E. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009, 69, 2195–2200. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Li, Y.; Tao, H.; Humphries, B.; Li, A.; Jiang, Y.; Yang, C.; Luo, R.; Wang, Z. Integrin alpha5 down-regulation by miR-205 suppresses triple negative breast cancer stemness and metastasis by inhibiting the Src/Vav2/Rac1 pathway. Cancer Lett. 2018, 433, 199–290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, B.; Zhao, H.; Chang, J. The expression and clinical significance of serum miR-205 for breast cancer and its role in detection of human cancers. Int. J. Clin. Exp. Med. 2015, 8, 3034–3043. [Google Scholar]
- Hasegawa, T.; Adachi, R.; Iwakata, H.; Takeno, T.; Sato, K.; Sakamaki, T. ErbB2 signaling epigenetically suppresses microRNA-205 transcription via Ras/Raf/MEK/ERK pathway in breast cancer. FEBS Open Bio 2017, 7, 1154–1165. [Google Scholar] [CrossRef]
- Piovan, C.; Palmieri, D.; Di Leva, G.; Braccioli, L.; Casalini, P.; Nuovo, G.; Tortoreto, M.; Sasso, M.; Plantamura, I.; Triulzi, T.; et al. Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol. Oncol. 2012, 6, 458–472. [Google Scholar] [CrossRef]
- Chao, C.H.; Chang, C.C.; Wu, M.J.; Ko, H.W.; Wang, D.; Hung, M.C.; Yang, J.Y.; Chang, C.J. MicroRNA-205 signaling regulates mammary stem cell fate and tumorigenesis. J. Clin. Investig. 2014, 124, 3093–3106. [Google Scholar] [CrossRef]
- Anderson, W.F.; Chatterjee, N.; Ershler, W.B.; Brawley, O.W. Estrogen receptor breast cancer phenotypes in the Surveillance, Epidemiology, and End Results database. Breast Cancer Res. Treat. 2002, 76, 27–36. [Google Scholar] [CrossRef]
- Lewis, J.S.; Jordan, V.C. Selective estrogen receptor modulators (SERMs): Mechanisms of anticarcinogenesis and drug resistance. Mutat. Res. 2005, 591, 247–263. [Google Scholar] [CrossRef]
- Badia, E.; Oliva, J.; Balaguer, P.; Cavailles, V. Tamoxifen resistance and epigenetic modifications in breast cancer cell lines. Curr. Med. Chem. 2007, 14, 3035–3045. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Liang, F.; Zhang, J.W.; Wang, F.; Wang, L.; Kang, X.G. Effects of long noncoding RNA-ROR on tamoxifen resistance of breast cancer cells by regulating microRNA-205. Cancer Chemother. Pharmacol. 2017, 79, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Itoh, M.; Bissell, M.J. The organization of tight junctions in epithelia: Implications for mammary gland biology and breast tumorigenesis. J. Mammary Gland. Biol. Neoplasia 2003, 8, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Ranahan, W.P.; Han, Z.; Smith-Kinnaman, W.; Nabinger, S.C.; Heller, B.; Herbert, B.S.; Chan, R.; Wells, C.D. The adaptor protein AMOT promotes the proliferation of mammary epithelial cells via the prolonged activation of the extracellular signal-regulated kinases. Cancer Res. 2011, 71, 2203–2211. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Fan, Q. MicroRNA-205 inhibits the proliferation and invasion of breast cancer by regulating AMOT expression. Oncol. Rep. 2015, 34, 2163–2170. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carey, L.A.; Dees, E.C.; Sawyer, L.; Gatti, L.; Moore, D.T.; Collichio, F.; Ollila, D.W.; Sartor, C.I.; Graham, M.L.; Perou, C.M. The triple negative paradox: Primary tumor chemosensitivity of breast cancer subtypes. Clin. Cancer Res. 2007, 13, 2329–2334. [Google Scholar] [CrossRef] [PubMed]
- Carey, L.A.; Perou, C.M.; Livasy, C.A.; Dressler, L.G.; Cowan, D.; Conway, K.; Karaca, G.; Troester, M.A.; Tse, C.K.; Edmiston, S.; et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 2006, 295, 2492–2502. [Google Scholar] [CrossRef]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Baselga, J. Why the epidermal growth factor receptor? The rationale for cancer therapy. Oncologist 2002, 7, 2–8. [Google Scholar] [CrossRef]
- Slamon, D.J.; Clark, G.M.; Wong, S.G.; Levin, W.J.; Ullrich, A.; McGuire, W.L. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987, 235, 177–182. [Google Scholar] [CrossRef]
- Cobleigh, M.A.; Vogel, C.L.; Tripathy, D.; Robert, N.J.; Scholl, S.; Fehrenbacher, L.; Wolter, J.M.; Paton, V.; Shak, S.; Lieberman, G.; et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J. Clin. Oncol. 1999, 17, 2639–2648. [Google Scholar] [CrossRef] [PubMed]
- Moasser, M.M. The oncogene HER2: Its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene 2007, 26, 6469–6487. [Google Scholar] [CrossRef] [PubMed]
- Naidu, R.; Yadav, M.; Nair, S.; Kutty, M.K. Expression of c-erbB3 protein in primary breast carcinomas. Br. J. Cancer 1998, 78, 1385–1390. [Google Scholar] [CrossRef] [PubMed]
- Holbro, T.; Beerli, R.R.; Maurer, F.; Koziczak, M.; Barbas, C.F., III; Hynes, N.E. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc. Natl. Acad. Sci. USA 2003, 100, 8933–8938. [Google Scholar] [CrossRef] [PubMed]
- Slamon, D.J.; Leyland-Jones, B.; Shak, S.; Fuchs, H.; Paton, V.; Bajamonde, A.; Fleming, T.; Eiermann, W.; Wolter, J.; Pegram, M.; et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 2001, 344, 783–792. [Google Scholar] [CrossRef]
- Piccart-Gebhart, M.J.; Procter, M.; Leyland-Jones, B.; Goldhirsch, A.; Untch, M.; Smith, I.; Gianni, L.; Baselga, J.; Bell, R.; Jackisch, C.; et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N. Engl. J. Med. 2005, 353, 1659–1672. [Google Scholar] [CrossRef] [PubMed]
- Nahta, R.; Yu, D.; Hung, M.C.; Hortobagyi, G.N.; Esteva, F.J. Mechanisms of disease: Understanding resistance to HER2-targeted therapy in human breast cancer. Nat. Clin. Pr. Oncol. 2006, 3, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Barok, M.; Joensuu, H.; Isola, J. Trastuzumab emtansine: Mechanisms of action and drug resistance. Breast Cancer Res. 2014, 16, 209. [Google Scholar] [CrossRef]
- De Cola, A.; Volpe, S.; Budani, M.C.; Ferracin, M.; Lattanzio, R.; Turdo, A.; D’Agostino, D.; Capone, E.; Stassi, G.; Todaro, M.; et al. miR-205-5p-mediated downregulation of ErbB/HER receptors in breast cancer stem cells results in targeted therapy resistance. Cell Death Dis. 2015, 6, e1823. [Google Scholar] [CrossRef]
- Greene, S.B.; Gunaratne, P.H.; Hammond, S.M.; Rosen, J.M. A putative role for microRNA-205 in mammary epithelial cell progenitors. J. Cell Sci. 2010, 123 Pt 4, 606–618. [Google Scholar] [CrossRef]
- Sun, H.; Lesche, R.; Li, D.M.; Liliental, J.; Zhang, H.; Gao, J.; Gavrilova, N.; Mueller, B.; Liu, X.; Wu, H. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 6199–6204. [Google Scholar] [CrossRef] [PubMed]
- Aumailley, M. The laminin family. Cell Adh. Migr. 2013, 7, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Avery-Kiejda, K.A.; Braye, S.G.; Forbes, J.F.; Scott, R.J. The expression of Dicer and Drosha in matched normal tissues, tumours and lymph node metastases in triple negative breast cancer. BMC Cancer 2014, 14, 253. [Google Scholar] [CrossRef] [PubMed]
- Berber, U.; Yilmaz, I.; Narli, G.; Haholu, A.; Kucukodaci, Z.; Demirel, D. miR-205 and miR-200c: Predictive Micro RNAs for Lymph Node Metastasis in Triple Negative Breast Cancer. J. Breast Cancer 2014, 17, 143–148. [Google Scholar] [CrossRef]
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef] [PubMed]
- Said, N.A.; Simpson, K.J.; Williams, E.D. Strategies and challenges for systematically mapping biologically significant molecular pathways regulating carcinoma epithelial-mesenchymal transition. Cells Tissues Organs 2013, 197, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- Thiery, J.P. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef]
- De Wever, O.; Pauwels, P.; De Craene, B.; Sabbah, M.; Emami, S.; Redeuilh, G.; Gespach, C.; Bracke, M.; Berx, G. Molecular and pathological signatures of epithelial-mesenchymal transitions at the cancer invasion front. Histochem. Cell Biol. 2008, 130, 481–494. [Google Scholar] [CrossRef]
- Lee, J.Y.; Park, M.K.; Park, J.H.; Lee, H.J.; Shin, D.H.; Kang, Y.; Lee, C.H.; Kong, G. Loss of the polycomb protein Mel-18 enhances the epithelial-mesenchymal transition by ZEB1 and ZEB2 expression through the downregulation of miR-205 in breast cancer. Oncogene 2014, 33, 1325–1335. [Google Scholar] [CrossRef]
- Valk-Lingbeek, M.E.; Bruggeman, S.W.; van Lohuizen, M. Stem cells and cancer; the polycomb connection. Cell 2004, 118, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Elgamal, O.A.; Park, J.K.; Gusev, Y.; Azevedo-Pouly, A.C.; Jiang, J.; Roopra, A.; Schmittgen, T.D. Tumor suppressive function of mir-205 in breast cancer is linked to HMGB3 regulation. PLoS ONE 2013, 8, e76402. [Google Scholar] [CrossRef] [PubMed]
- Hood, J.D.; Cheresh, D.A. Role of integrins in cell invasion and migration. Nat. Rev. Cancer 2002, 2, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Q.; Lu, J.T.; Tan, C.C.; Wang, Q.S.; Feng, Y.M. RUNX2 promotes breast cancer bone metastasis by increasing integrin alpha5-mediated colonization. Cancer Lett. 2016, 380, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Chen, X.; Wu, Y.; Feng, Z.; He, T.; Wang, L.; Liao, L.; Xu, J. Steroid receptor coactivator-1 upregulates integrin alpha(5) expression to promote breast cancer cell adhesion and migration. Cancer Res. 2011, 71, 1742–1751. [Google Scholar] [CrossRef] [PubMed]
- Bielenberg, D.R.; Zetter, B.R. The Contribution of Angiogenesis to the Process of Metastasis. Cancer J. 2015, 21, 267–273. [Google Scholar] [CrossRef]
- Du, Y.E.; Tu, G.; Yang, G.; Li, G.; Yang, D.; Lang, L.; Xi, L.; Sun, K.; Chen, Y.; Shu, K.; et al. MiR-205/YAP1 in Activated Fibroblasts of Breast Tumor Promotes VEGF-independent Angiogenesis through STAT3 Signaling. Theranostics 2017, 7, 3972–3988. [Google Scholar] [CrossRef]
- Clarke, M.F.; Dick, J.E.; Dirks, P.B.; Eaves, C.J.; Jamieson, C.H.; Jones, D.L.; Visvader, J.; Weissman, I.L.; Wahl, G.M. Cancer stem cells—Perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006, 66, 9339–9344. [Google Scholar] [CrossRef]
- Wellner, U.; Schubert, J.; Burk, U.C.; Schmalhofer, O.; Zhu, F.; Sonntag, A.; Waldvogel, B.; Vannier, C.; Darling, D.; zur Hausen, A.; et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009, 11, 1487–1495. [Google Scholar] [CrossRef]
- Shimono, Y.; Zabala, M.; Cho, R.W.; Lobo, N.; Dalerba, P.; Qian, D.; Diehn, M.; Liu, H.; Panula, S.P.; Chiao, E.; et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 2009, 138, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Iliopoulos, D.; Lindahl-Allen, M.; Polytarchou, C.; Hirsch, H.A.; Tsichlis, P.N.; Struhl, K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell 2010, 39, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Onder, T.T.; Gupta, P.B.; Mani, S.A.; Yang, J.; Lander, E.S.; Weinberg, R.A. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008, 68, 3645–3654. [Google Scholar] [CrossRef] [PubMed]
- Alexander, C.M.; Goel, S.; Fakhraldeen, S.A.; Kim, S. Wnt signaling in mammary glands: Plastic cell fates and combinatorial signaling. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H.; Nusse, R. Wnt/beta-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef]
- Yu, F.X.; Guan, K.L. The Hippo pathway: Regulators and regulations. Genes Dev. 2013, 27, 355–371. [Google Scholar] [CrossRef]
- Cordenonsi, M.; Zanconato, F.; Azzolin, L.; Forcato, M.; Rosato, A.; Frasson, C.; Inui, M.; Montagner, M.; Parenti, A.R.; Poletti, A.; et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 2011, 147, 759–772. [Google Scholar] [CrossRef]
- Chen, D.; Sun, Y.; Wei, Y.; Zhang, P.; Rezaeian, A.H.; Teruya-Feldstein, J.; Gupta, S.; Liang, H.; Lin, H.K.; Hung, M.C.; et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat. Med. 2012, 18, 1511–1517. [Google Scholar] [CrossRef]
- Cicalese, A.; Bonizzi, G.; Pasi, C.E.; Faretta, M.; Ronzoni, S.; Giulini, B.; Brisken, C.; Minucci, S.; Di Fiore, P.P.; Pelicci, P.G. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 2009, 138, 1083–1095. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, K.Y.; Liu, S.M.; Sen, S. Tumor-associated circulating microRNAs as biomarkers of cancer. Molecules 2014, 19, 1912–1938. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Shaker, O.; Maher, M.; Nassar, Y.; Morcos, G.; Gad, Z. Role of microRNAs -29b-2, -155, -197 and -205 as diagnostic biomarkers in serum of breast cancer females. Gene 2015, 560, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Simonson, B.; Das, S. MicroRNA Therapeutics: The Next Magic Bullet? Mini Rev. Med. Chem. 2015, 15, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Lin, T.S. The Role of MicroRNAs in Diagnosis, Prognosis, Metastasis and Resistant Cases in Breast Cancer. Curr. Pharm. Des. 2016, 23, 1845–1859. [Google Scholar]
- Zhou, K.; Liu, M.; Cao, Y. New Insight into microRNA Functions in Cancer: Oncogene-microRNA-Tumor Suppressor Gene Network. Front. Mol. Biosci. 2017, 4, 46. [Google Scholar] [CrossRef]
- Wang, J.; Yang, M.; Li, Y.; Han, B. The Role of MicroRNAs in the Chemoresistance of Breast Cancer. Drug Dev. Res. 2015, 76, 368–374. [Google Scholar] [CrossRef]
- Li, Q.; Liu, M.; Ma, F.; Luo, Y.; Cai, R.; Wang, L.; Xu, N.; Xu, B. Circulating miR-19a and miR-205 in serum may predict the sensitivity of luminal A subtype of breast cancer patients to neoadjuvant chemotherapy with epirubicin plus paclitaxel. PLoS ONE 2014, 9, e104870. [Google Scholar] [CrossRef]
- Cai, Y.; Yan, X.; Zhang, G.; Zhao, W.; Jiao, S. MicroRNA-205 increases the sensitivity of docetaxel in breast cancer. Oncol. Lett. 2016, 11, 1105–1109. [Google Scholar] [CrossRef]
- Hu, Y.; Qiu, Y.; Yague, E.; Ji, W.; Liu, J.; Zhang, J. miRNA-205 targets VEGFA and FGF2 and regulates resistance to chemotherapeutics in breast cancer. Cell Death Dis. 2016, 7, 2291. [Google Scholar] [CrossRef]
- Kim, E.S.; Choi, J.Y.; Hwang, S.J.; Bae, I.H. Hypermethylation of miR-205-5p by IR Governs Aggressiveness and Metastasis via Regulating Bcl-w and Src. Mol. Nucl. Acids 2019, 14, 450–464. [Google Scholar] [CrossRef]
- Wang, S.; Huang, J.; Lyu, H.; Lee, C.K.; Tan, J.; Wang, J.; Liu, B. Functional cooperation of miR-125a, miR-125b, and miR-205 in entinostat-induced downregulation of erbB2/erbB3 and apoptosis in breast cancer cells. Cell Death Dis. 2013, 4, e556. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Kang, F.B.; Wang, J.; Yang, C.; He, D.W. Downregulation of miR-205 contributes to epithelial-mesenchymal transition and invasion in triple-negative breast cancer by targeting HMGB1-RAGE signaling pathway. Anti-Cancer Drugs 2019, 30, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Moon, Y.; Choi, J.; Yoon, S.; Jung, K.H.; Cheon, J.; Kim, W.; Kim, D.; Lee, C.H.; Kim, S.W.; et al. The GTP binding activity of transglutaminase 2 promotes bone metastasis of breast cancer cells by downregulating microRNA-205. Am. J. Cancer Res. 2019, 9, 597–607. [Google Scholar] [PubMed]
- Adachi, R.; Horiuchi, S.; Sakurazawa, Y.; Hasegawa, T.; Sato, K.; Sakamaki, T. ErbB2 down-regulates microRNA-205 in breast cancer. Biochem. Biophys. Res. Commun. 2011, 411, 804–808. [Google Scholar] [CrossRef]
- Dong, Y.; Si, J.W.; Li, W.T.; Liang, L.; Zhao, J.; Zhou, M.; Li, D.; Li, T. miR-200a/miR-141 and miR-205 upregulation might be associated with hormone receptor status and prognosis in endometrial carcinomas. Int. J. Clin. Exp. Pathol. 2015, 8, 2864–2875. [Google Scholar]
- Grelet, S.; Link, L.A.; Howley, B.; Obellianne, C.; Palanisamy, V.; Gangaraju, V.K.; Diehl, J.A.; Howe, P.H. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 2017, 19, 1105–1115. [Google Scholar] [CrossRef]
- Hou, P.; Zhao, Y.; Li, Z.; Yao, R.; Ma, M.; Gao, Y.; Zhao, L.; Zhang, Y.; Huang, B.; Lu, J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014, 5, e1287. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).