Factors Regulating microRNA Expression and Function in Multiple Myeloma
Abstract
1. Introduction
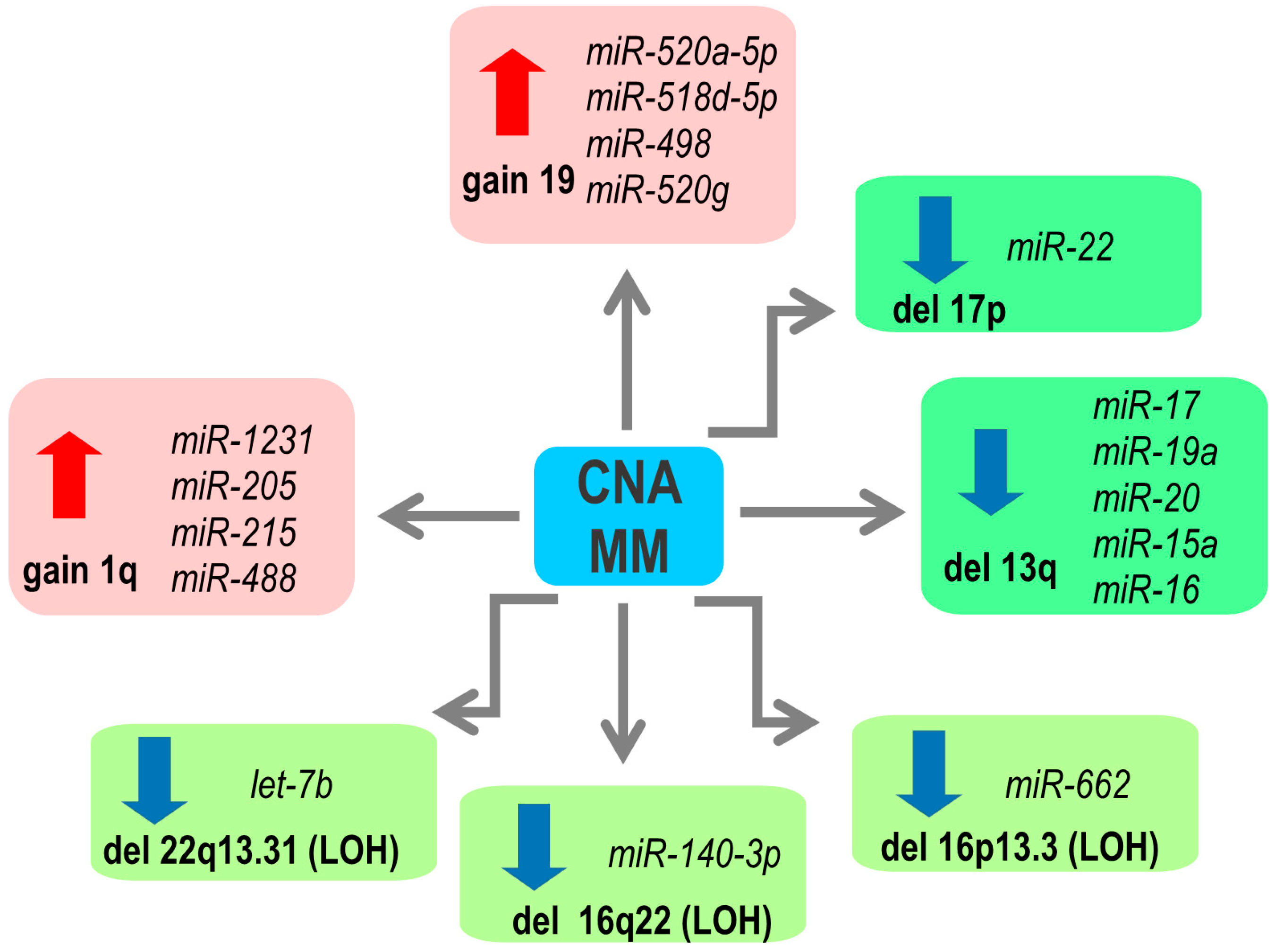
2. Copy Number Abnormalities Affect Expression of miRNAs
3. microRNA Expression and Epigenetics
4. Transcriptional Regulation of miRNA Expression
5. Impaired miRNA Processing may Deregulate miRNA Expression
6. Impact of miRNA–mRNA Associations
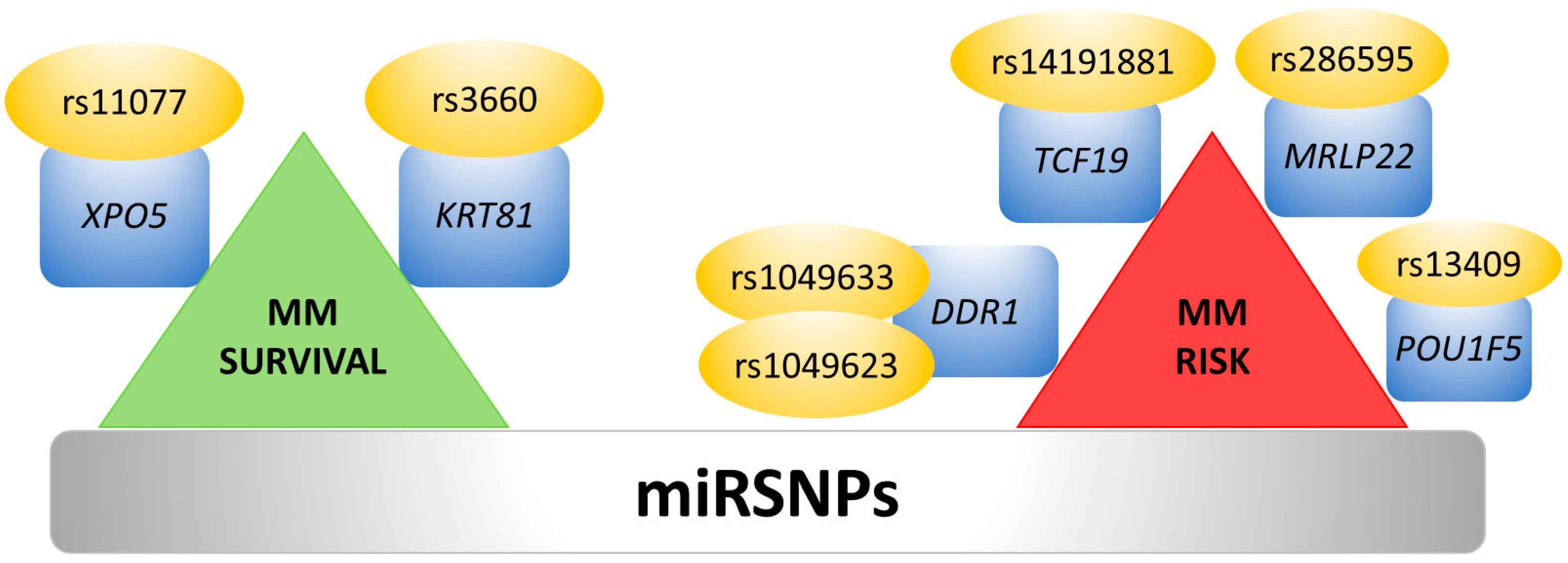
7. Single Nucleotide Polymorphisms and 3′UTR Polyadenylation Affect miRNA Binding Sites
8. Regulation of miRNA Expression by miRNA Sponges
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kumar, S.K.; Therneau, T.M.; Gertz, M.A.; Lacy, M.Q.; Dispenzieri, A.; Rajkumar, S.V.; Fonseca, R.; Witzig, T.E.; Lust, J.A.; Larson, D.R.; et al. Clinical course of patients with relapsed multiple myeloma. Mayo Clin. Proc. 2004, 79, 867–874. [Google Scholar] [CrossRef]
- Avet-Loiseau, H.; Li, C.; Magrangeas, F.; Gouraud, W.; Charbonnel, C.; Harousseau, J.-L.; Attal, M.; Marit, G.; Mathiot, C.; Facon, T.; et al. Prognostic significance of copy-number alterations in multiple myeloma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 4585–4590. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, R.; Harrington, D.; Oken, M.M.; Dewald, G.W.; Bailey, R.J.; Van Wier, S.A.; Henderson, K.J.; Blood, E.A.; Rajkumar, S.V.; Kay, N.E.; et al. Biological and prognostic significance of interphase fluorescence in situ hybridization detection of chromosome 13 abnormalities (delta13) in multiple myeloma: An eastern cooperative oncology group study. Cancer Res. 2002, 62, 715–720. [Google Scholar]
- Gutiérrez, N.C.; Castellanos, M.V.; Martín, M.L.; Mateos, M.V.; Hernández, J.M.; Fernández, M.; Carrera, D.; Rosiñol, L.; Ribera, J.M.; Ojanguren, J.M.; et al. Prognostic and biological implications of genetic abnormalities in multiple myeloma undergoing autologous stem cell transplantation: t(4;14) is the most relevant adverse prognostic factor, whereas RB deletion as a unique abnormality is not associated with adverse prognosis. Leukemia 2007, 21, 143–150. [Google Scholar] [PubMed]
- Morgan, G.J.; Walker, B.A.; Davies, F.E. The genetic architecture of multiple myeloma. Nat. Rev. Cancer 2012, 12, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Baskerville, S.; Bartel, D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 2005, 11, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Altuvia, Y.; Landgraf, P.; Lithwick, G.; Elefant, N.; Pfeffer, S.; Aravin, A.; Brownstein, M.J.; Tuschl, T.; Margalit, H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005, 33, 2697–2706. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]
- Ling, H.; Fabbri, M.; Calin, G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013, 12, 847–865. [Google Scholar] [CrossRef]
- Ambros, V. MicroRNAs and developmental timing. Curr. Opin. Genet. Dev. 2011, 21, 511–517. [Google Scholar] [CrossRef]
- Ambros, V.; Chen, X. The regulation of genes and genomes by small RNAs. Dev. Camb. Engl. 2007, 134, 1635–1641. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Gutiérrez, N.C.; Sarasquete, M.E.; Misiewicz-Krzeminska, I.; Delgado, M.; De Las Rivas, J.; Ticona, F.V.; Fermiñán, E.; Martín-Jiménez, P.; Chillón, C.; Risueño, A.; et al. Deregulation of microRNA expression in the different genetic subtypes of multiple myeloma and correlation with gene expression profiling. Leukemia 2010, 24, 629–637. [Google Scholar] [CrossRef]
- Pichiorri, F.; Suh, S.-S.; Ladetto, M.; Kuehl, M.; Palumbo, T.; Drandi, D.; Taccioli, C.; Zanesi, N.; Alder, H.; Hagan, J.P.; et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 12885–12890. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, M.; Biasiolo, M.; Agnelli, L.; Todoerti, K.; Mosca, L.; Fabris, S.; Sales, G.; Deliliers, G.L.; Bicciato, S.; Lombardi, L.; et al. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood 2009, 114, e20–e26. [Google Scholar] [CrossRef]
- Misiewicz-Krzeminska, I.; Sarasquete, M.E.; Quwaider, D.; Krzeminski, P.; Ticona, F.V.; Paíno, T.; Delgado, M.; Aires, A.; Ocio, E.M.; García-Sanz, R.; et al. Restoration of microRNA-214 expression reduces growth of myeloma cells through positive regulation of P53 and inhibition of DNA replication. Haematologica 2013, 98, 640–648. [Google Scholar] [CrossRef]
- Quwaider, D.; Corchete, L.A.; Misiewicz-Krzeminska, I.; Sarasquete, M.E.; Pérez, J.J.; Krzeminski, P.; Puig, N.; Mateos, M.V.; García-Sanz, R.; Herrero, A.B.; et al. DEPTOR maintains plasma cell differentiation and favorably affects prognosis in multiple myeloma. J. Hematol. Oncol. 2017, 10, 92. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, B.; Li, W.; Wang, L.; Yan, Z.; Li, H.; Yao, Y.; Yao, R.; Xu, K.; Li, Z. MiR-15a/16 regulates the growth of myeloma cells, angiogenesis and antitumor immunity by inhibiting Bcl-2, VEGF-A and IL-17 expression in multiple myeloma. Leuk. Res. 2016, 49, 73–79. [Google Scholar] [CrossRef]
- Amodio, N.; Stamato, M.A.; Gullà, A.M.; Morelli, E.; Romeo, E.; Raimondi, L.; Pitari, M.R.; Ferrandino, I.; Misso, G.; Caraglia, M.; et al. Therapeutic Targeting of miR-29b/HDAC4 Epigenetic Loop in Multiple Myeloma. Mol. Cancer Ther. 2016, 15, 1364–1375. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wu, D.; Wang, J.; Li, Y.; Chai, X.; Kang, Q. miR-320a regulates cell proliferation and apoptosis in multiple myeloma by targeting pre-B-cell leukemia transcription factor 3. Biochem. Biophys. Res. Commun. 2016, 473, 1315–1320. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Zhang, X.; Zhang, L.; Wang, Y.; Pan, H.; Xu, Z.; Pang, X. MicroRNA-497 suppresses cell proliferation and induces apoptosis through targeting PBX3 in human multiple myeloma. Am. J. Cancer Res. 2016, 6, 2880–2889. [Google Scholar] [PubMed]
- Yang, Y.; Li, F.; Saha, M.N.; Abdi, J.; Qiu, L.; Chang, H. miR-137 and miR-197 Induce Apoptosis and Suppress Tumorigenicity by Targeting MCL-1 in Multiple Myeloma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 2399–2411. [Google Scholar] [CrossRef]
- Saunders, M.A.; Liang, H.; Li, W.-H. Human polymorphism at microRNAs and microRNA target sites. Proc. Natl. Acad. Sci. USA 2007, 104, 3300–3305. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Rajewsky, N. Natural selection on human microRNA binding sites inferred from SNP data. Nat. Genet. 2006, 38, 1452–1456. [Google Scholar] [CrossRef] [PubMed]
- Hanamura, I.; Stewart, J.P.; Huang, Y.; Zhan, F.; Santra, M.; Sawyer, J.R.; Hollmig, K.; Zangarri, M.; Pineda-Roman, M.; van Rhee, F.; et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: Incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood 2006, 108, 1724–1732. [Google Scholar] [CrossRef]
- Pichiorri, F.; Suh, S.-S.; Rocci, A.; De Luca, L.; Taccioli, C.; Santhanam, R.; Zhou, W.; Benson, D.M.; Hofmainster, C.; Alder, H.; et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell 2010, 18, 367–381. [Google Scholar] [CrossRef]
- Zhang, Y.; Geng, L.; Talmon, G.; Wang, J. MicroRNA-520g confers drug resistance by regulating p21 expression in colorectal cancer. J. Biol. Chem. 2015, 290, 6215–6225. [Google Scholar] [CrossRef]
- Liu, R.; Liu, F.; Li, L.; Sun, M.; Chen, K. MiR-498 regulated FOXO3 expression and inhibited the proliferation of human ovarian cancer cells. Biomed. Pharmacother. 2015, 72, 52–57. [Google Scholar] [CrossRef]
- Roccaro, A.M.; Sacco, A.; Thompson, B.; Leleu, X.; Azab, A.K.; Azab, F.; Runnels, J.; Jia, X.; Ngo, H.T.; Melhem, M.R.; et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood 2009, 113, 6669–6680. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Zhang, R.; Qu, X.; Zhao, M.; Zhang, S.; Wu, H.; Jianyong, L.; Chen, L. MiR-15a, miR-16-1 and miR-17-92 cluster expression are linked to poor prognosis in multiple myeloma. Leuk. Res. 2012, 36, 1505–1509. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, M.; Agnelli, L.; Mosca, L.; Fabris, S.; Andronache, A.; Todoerti, K.; Ronchetti, D.; Deliliers, G.L.; Neri, A. Integrative high-resolution microarray analysis of human myeloma cell lines reveals deregulated miRNA expression associated with allelic imbalances and gene expression profiles. Genes. Chromosomes Cancer 2009, 48, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, Y.; Yang, T.; Liu, Y.; Li, A.; Fu, S.; Wu, M.; Pan, Z.; Zhou, W. microRNA-22, downregulated in hepatocellular carcinoma and correlated with prognosis, suppresses cell proliferation and tumourigenicity. Br. J. Cancer 2010, 103, 1215–1220. [Google Scholar] [CrossRef]
- Xia, S.-S.; Zhang, G.-J.; Liu, Z.-L.; Tian, H.-P.; He, Y.; Meng, C.-Y.; Li, L.-F.; Wang, Z.-W.; Zhou, T. MicroRNA-22 suppresses the growth, migration and invasion of colorectal cancer cells through a Sp1 negative feedback loop. Oncotarget 2017, 8, 36266–36278. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.-M.; Hu, L.-H.; Wang, Y.-Q.; Chen, P.; Huang, J.-G.; Lu, N.; He, J.-H.; Liao, C.-G. miR-22 is down-regulated in gastric cancer, and its overexpression inhibits cell migration and invasion via targeting transcription factor Sp1. Med. Oncol. 2013, 30, 542. [Google Scholar] [CrossRef] [PubMed]
- Ling, B.; Wang, G.-X.; Long, G.; Qiu, J.-H.; Hu, Z.-L. Tumor suppressor miR-22 suppresses lung cancer cell progression through post-transcriptional regulation of ErbB3. J. Cancer Res. Clin. Oncol. 2012, 138, 1355–1361. [Google Scholar] [CrossRef]
- Lee, H.; Han, S.; Kwon, C.S.; Lee, D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2016, 7, 100–113. [Google Scholar] [CrossRef]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef]
- Sampson, V.B.; Rong, N.H.; Han, J.; Yang, Q.; Aris, V.; Soteropoulos, P.; Petrelli, N.J.; Dunn, S.P.; Krueger, L.J. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007, 67, 9762–9770. [Google Scholar] [CrossRef]
- Takamizawa, J.; Konishi, H.; Yanagisawa, K.; Tomida, S.; Osada, H.; Endoh, H.; Harano, T.; Yatabe, Y.; Nagino, M.; Nimura, Y.; et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004, 64, 3753–3756. [Google Scholar] [CrossRef] [PubMed]
- Shell, S.; Park, S.-M.; Radjabi, A.R.; Schickel, R.; Kistner, E.O.; Jewell, D.A.; Feig, C.; Lengyel, E.; Peter, M.E. Let-7 expression defines two differentiation stages of cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 11400–11405. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Koch, A.; Joosten, S.C.; Feng, Z.; de Ruijter, T.C.; Draht, M.X.; Melotte, V.; Smits, K.M.; Veeck, J.; Herman, J.G.; Van Neste, L.; et al. Analysis of DNA methylation in cancer: Location revisited. Nat. Rev. Clin. Oncol. 2018, 15, 459–466. [Google Scholar] [CrossRef]
- Walker, B.A.; Wardell, C.P.; Chiecchio, L.; Smith, E.M.; Boyd, K.D.; Neri, A.; Davies, F.E.; Ross, F.M.; Morgan, G.J. Aberrant global methylation patterns affect the molecular pathogenesis and prognosis of multiple myeloma. Blood 2011, 117, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Heuck, C.J.; Mehta, J.; Bhagat, T.; Gundabolu, K.; Yu, Y.; Khan, S.; Chrysofakis, G.; Schinke, C.; Tariman, J.; Vickrey, E.; et al. Myeloma is characterized by stage-specific alterations in DNA methylation that occur early during myelomagenesis. J. Immunol. 2013, 190, 2966–2975. [Google Scholar] [CrossRef]
- Chim, C.S.; Wong, K.Y.; Qi, Y.; Loong, F.; Lam, W.L.; Wong, L.G.; Jin, D.Y.; Costello, J.F.; Liang, R. Epigenetic inactivation of the miR-34a in hematological malignancies. Carcinogenesis 2010, 31, 745–750. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, Y.E.; Zhang, Y.; Leleu, X.; Reagan, M.; Zhang, Y.; Mishima, Y.; Glavey, S.; Manier, S.; Sacco, A.; et al. Global epigenetic regulation of microRNAs in multiple myeloma. PLoS ONE 2014, 9, e110973. [Google Scholar] [CrossRef]
- Wong, K.-Y.; Liang, R.; So, C.-C.; Jin, D.-Y.; Costello, J.F.; Chim, C.-S. Epigenetic silencing of MIR203 in multiple myeloma. Br. J. Haematol. 2011, 154, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.Y.; Yim, R.L.H.; So, C.C.; Jin, D.-Y.; Liang, R.; Chim, C.S. Epigenetic inactivation of the MIR34B/C in multiple myeloma. Blood 2011, 118, 5901–5904. [Google Scholar] [CrossRef]
- Wong, K.-Y.; Yim, R.L.-H.; Kwong, Y.-L.; Leung, C.-Y.; Hui, P.-K.; Cheung, F.; Liang, R.; Jin, D.-Y.; Chim, C.-S. Epigenetic inactivation of the MIR129-2 in hematological malignancies. J. Hematol. Oncol. 2013, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wong, K.Y.; Chan, G.C.-F.; Chng, W.-J.; Chim, C.S. Epigenetic silencing of EVL/miR-342 in multiple myeloma. Transl. Res. J. Lab. Clin. Med. 2018, 192, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Tatekawa, S.; Chinen, Y.; Ri, M.; Narita, T.; Shimura, Y.; Matsumura-Kimoto, Y.; Tsukamoto, T.; Kobayashi, T.; Kawata, E.; Uoshima, N.; et al. Epigenetic repression of miR-375 is the dominant mechanism for constitutive activation of the PDPK1/RPS6KA3 signalling axis in multiple myeloma. Br. J. Haematol. 2017, 178, 534–546. [Google Scholar] [CrossRef] [PubMed]
- Moutinho, C.; Esteller, M. MicroRNAs and Epigenetics. Adv. Cancer Res. 2017, 135, 189–220. [Google Scholar] [PubMed]
- Zhao, S.; Wang, Y.; Liang, Y.; Zhao, M.; Long, H.; Ding, S.; Yin, H.; Lu, Q. MicroRNA-126 regulates DNA methylation in CD4+ T cells and contributes to systemic lupus erythematosus by targeting DNA methyltransferase 1. Arthritis Rheum. 2011, 63, 1376–1386. [Google Scholar] [CrossRef]
- Takata, A.; Otsuka, M.; Yoshikawa, T.; Kishikawa, T.; Hikiba, Y.; Obi, S.; Goto, T.; Kang, Y.J.; Maeda, S.; Yoshida, H.; et al. MicroRNA-140 acts as a liver tumor suppressor by controlling NF-κB activity by directly targeting DNA methyltransferase 1 (Dnmt1) expression. Hepatology 2013, 57, 162–170. [Google Scholar] [CrossRef]
- Yuan, L.; Chan, G.C.F.; Fung, K.L.; Chim, C.S. RANKL expression in myeloma cells is regulated by a network involving RANKL promoter methylation, DNMT1, microRNA and TNFα in the microenvironment. Biochim. Biophys. Acta 2014, 1843, 1834–1838. [Google Scholar] [CrossRef]
- Amodio, N.; D’Aquila, P.; Passarino, G.; Tassone, P.; Bellizzi, D. Epigenetic modifications in multiple myeloma: Recent advances on the role of DNA and histone methylation. Expert Opin. Ther. Targets 2017, 21, 91–101. [Google Scholar] [CrossRef]
- Amodio, N.; Leotta, M.; Bellizzi, D.; Di Martino, M.T.; D’Aquila, P.; Lionetti, M.; Fabiani, F.; Leone, E.; Gullà, A.M.; Passarino, G.; et al. DNA-demethylating and anti-tumor activity of synthetic miR-29b mimics in multiple myeloma. Oncotarget 2012, 3, 1246–1258. [Google Scholar] [CrossRef]
- Stamato, M.A.; Juli, G.; Romeo, E.; Ronchetti, D.; Arbitrio, M.; Caracciolo, D.; Neri, A.; Tagliaferri, P.; Tassone, P.; Amodio, N. Inhibition of EZH2 triggers the tumor suppressive miR-29b network in multiple myeloma. Oncotarget 2017, 8, 106527–106537. [Google Scholar] [CrossRef]
- Zhang, Y.-K.; Wang, H.; Leng, Y.; Li, Z.-L.; Yang, Y.-F.; Xiao, F.-J.; Li, Q.-F.; Chen, X.-Q.; Wang, L.-S. Overexpression of microRNA-29b induces apoptosis of multiple myeloma cells through down regulating Mcl-1. Biochem. Biophys. Res. Commun. 2011, 414, 233–239. [Google Scholar] [CrossRef]
- Jagannathan, S.; Vad, N.; Vallabhapurapu, S.; Vallabhapurapu, S.; Anderson, K.C.; Driscoll, J.J. MiR-29b replacement inhibits proteasomes and disrupts aggresome+autophagosome formation to enhance the antimyeloma benefit of bortezomib. Leukemia 2015, 29, 727–738. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, L.Q.; Wong, K.Y.; Li, Z.Y.; Chim, C.S. Infrequent DNA methylation of miR-9-1 and miR-9-3 in multiple myeloma. J. Clin. Pathol. 2015, 68, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Canella, A.; Cordero Nieves, H.; Sborov, D.W.; Cascione, L.; Radomska, H.S.; Smith, E.; Stiff, A.; Consiglio, J.; Caserta, E.; Rizzotto, L.; et al. HDAC inhibitor AR-42 decreases CD44 expression and sensitizes myeloma cells to lenalidomide. Oncotarget 2015, 6, 31134–31150. [Google Scholar] [CrossRef]
- Li, Z.; Wong, K.Y.; Chan, G.C.-F.; Chim, C.S. Epigenetic silencing of LPP/miR-28 in multiple myeloma. J. Clin. Pathol. 2018, 71, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; He, X.; Li, M.; Shi, F.; Wu, D.; Pan, M.; Guo, M.; Zhang, R.; Luo, S.; Gu, N.; et al. MiRNA-34a overexpression inhibits multiple myeloma cancer stem cell growth in mice by suppressing TGIF2. Am. J. Transl. Res. 2016, 8, 5433–5443. [Google Scholar] [PubMed]
- Wong, K.Y.; So, C.C.; Loong, F.; Chung, L.P.; Lam, W.W.L.; Liang, R.; Li, G.K.H.; Jin, D.-Y.; Chim, C.S. Epigenetic inactivation of the miR-124-1 in haematological malignancies. PLoS ONE 2011, 6, e19027. [Google Scholar] [CrossRef] [PubMed]
- Krzeminski, P.; Sarasquete, M.E.; Misiewicz-Krzeminska, I.; Corral, R.; Corchete, L.A.; Martín, A.A.; García-Sanz, R.; San Miguel, J.F.; Gutiérrez, N.C. Insights into epigenetic regulation of microRNA-155 expression in multiple myeloma. Biochim. Biophys. Acta 2015, 1849, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Bi, C.; Chung, T.-H.; Huang, G.; Zhou, J.; Yan, J.; Ahmann, G.J.; Fonseca, R.; Chng, W.J. Genome-wide pharmacologic unmasking identifies tumor suppressive microRNAs in multiple myeloma. Oncotarget 2015, 6, 26508–26518. [Google Scholar] [CrossRef] [PubMed]
- Schanen, B.C.; Li, X. Transcriptional regulation of mammalian miRNA genes. Genomics 2011, 97, 1–6. [Google Scholar] [CrossRef]
- Biasiolo, M.; Sales, G.; Lionetti, M.; Agnelli, L.; Todoerti, K.; Bisognin, A.; Coppe, A.; Romualdi, C.; Neri, A.; Bortoluzzi, S. Impact of host genes and strand selection on miRNA and miRNA* expression. PLoS ONE 2011, 6, e23854. [Google Scholar] [CrossRef]
- Ronchetti, D.; Lionetti, M.; Mosca, L.; Agnelli, L.; Andronache, A.; Fabris, S.; Deliliers, G.L.; Neri, A. An integrative genomic approach reveals coordinated expression of intronic miR-335, miR-342, and miR-561 with deregulated host genes in multiple myeloma. BMC Med. Genom. 2008, 1, 37. [Google Scholar] [CrossRef] [PubMed]
- Löffler, D.; Brocke-Heidrich, K.; Pfeifer, G.; Stocsits, C.; Hackermüller, J.; Kretzschmar, A.K.; Burger, R.; Gramatzki, M.; Blumert, C.; Bauer, K.; et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 2007, 110, 1330–1333. [Google Scholar] [CrossRef]
- Song Gao, J.; Zhang, Y.; Li, M.; Tucker, L.D.; Machan, J.T.; Quesenberry, P.; Rigoutsos, I.; Ramratnam, B. Atypical transcription of microRNA gene fragments. Nucleic Acids Res. 2010, 38, 2775–2787. [Google Scholar] [CrossRef] [PubMed]
- Amodio, N.; Di Martino, M.T.; Foresta, U.; Leone, E.; Lionetti, M.; Leotta, M.; Gullà, A.M.; Pitari, M.R.; Conforti, F.; Rossi, M.; et al. miR-29b sensitizes multiple myeloma cells to bortezomib-induced apoptosis through the activation of a feedback loop with the transcription factor Sp1. Cell Death Dis. 2012, 3, e436. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, L.; De Luca, A.; Morelli, E.; Giavaresi, G.; Tagliaferri, P.; Tassone, P.; Amodio, N. MicroRNAs: Novel Crossroads between Myeloma Cells and the Bone Marrow Microenvironment. BioMed Res. Int. 2016, 2016, 6504593. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Ray, R.M.; Johnson, L.R. STAT3-mediated transcription of Bcl-2, Mcl-1 and c-IAP2 prevents apoptosis in polyamine-depleted cells. Biochem. J. 2005, 392, 335–344. [Google Scholar] [CrossRef]
- Sepúlveda, P.; Encabo, A.; Carbonell-Uberos, F.; Miñana, M.D. BCL-2 expression is mainly regulated by JAK/STAT3 pathway in human CD34+ hematopoietic cells. Cell Death Differ. 2007, 14, 378–380. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Gong, W.; Liu, S.; Li, Q.; Guo, M.; Wang, J.; Wang, S.; Chen, N.; Wang, Y.; Liu, Q.; et al. Ibrutinib targets microRNA-21 in multiple myeloma cells by inhibiting NF-κB and STAT3. Tumour Biol. 2018, 40, 1010428317731369. [Google Scholar] [CrossRef]
- Raimondi, L.; Amodio, N.; Di Martino, M.T.; Altomare, E.; Leotta, M.; Caracciolo, D.; Gullà, A.; Neri, A.; Taverna, S.; D’Aquila, P.; et al. Targeting of multiple myeloma-related angiogenesis by miR-199a-5p mimics: In vitro and in vivo anti-tumor activity. Oncotarget 2014, 5, 3039–3054. [Google Scholar] [CrossRef]
- Borsi, E.; Perrone, G.; Terragna, C.; Martello, M.; Dico, A.F.; Solaini, G.; Baracca, A.; Sgarbi, G.; Pasquinelli, G.; Valente, S.; et al. Hypoxia inducible factor-1 alpha as a therapeutic target in multiple myeloma. Oncotarget 2014, 5, 1779–1792. [Google Scholar] [CrossRef] [PubMed]
- Holien, T.; Våtsveen, T.K.; Hella, H.; Waage, A.; Sundan, A. Addiction to c-MYC in multiple myeloma. Blood 2012, 120, 2450–2453. [Google Scholar] [CrossRef]
- Vigorito, E.; Perks, K.L.; Abreu-Goodger, C.; Bunting, S.; Xiang, Z.; Kohlhaas, S.; Das, P.P.; Miska, E.A.; Rodriguez, A.; Bradley, A.; et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 2007, 27, 847–859. [Google Scholar] [CrossRef]
- Danger, R.; Braza, F.; Giral, M.; Soulillou, J.-P.; Brouard, S. MicroRNAs, Major Players in B Cells Homeostasis and Function. Front. Immunol. 2014, 5, 98. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-C.; Yu, D.; Lee, Y.-S.; Wentzel, E.A.; Arking, D.E.; West, K.M.; Dang, C.V.; Thomas-Tikhonenko, A.; Mendell, J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008, 40, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, C.; Zhang, R.; Gao, X.; Qu, X.; Zhao, M.; Qiao, C.; Xu, J.; Li, J. miR-17-92 cluster microRNAs confers tumorigenicity in multiple myeloma. Cancer Lett. 2011, 309, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Novotny, G.W.; Sonne, S.B.; Nielsen, J.E.; Jonstrup, S.P.; Hansen, M.A.; Skakkebaek, N.E.; Rajpert-De Meyts, E.; Kjems, J.; Leffers, H. Translational repression of E2F1 mRNA in carcinoma in situ and normal testis correlates with expression of the miR-17-92 cluster. Cell Death Differ. 2007, 14, 879–882. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839–843. [Google Scholar] [CrossRef]
- Concepcion, C.P.; Bonetti, C.; Ventura, A. The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J. Sudbury Mass 2012, 18, 262–267. [Google Scholar] [CrossRef]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef]
- Ventura, A.; Young, A.G.; Winslow, M.M.; Lintault, L.; Meissner, A.; Erkeland, S.J.; Newman, J.; Bronson, R.T.; Crowley, D.; Stone, J.R.; et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008, 132, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Morelli, E.; Biamonte, L.; Federico, C.; Amodio, N.; Di Martino, M.T.; Gallo Cantafio, M.E.; Manzoni, M.; Scionti, F.; Samur, M.K.; Gullà, A.; et al. Therapeutic vulnerability of multiple myeloma to MIR17PTi, a first-in-class inhibitor of pri-miR-17-92. Blood 2018, 132, 1050–1063. [Google Scholar] [CrossRef] [PubMed]
- Fulciniti, M.; Amodio, N.; Bandi, R.L.; Cagnetta, A.; Samur, M.K.; Acharya, C.; Prabhala, R.; D’Aquila, P.; Bellizzi, D.; Passarino, G.; et al. miR-23b/SP1/c-myc forms a feed-forward loop supporting multiple myeloma cell growth. Blood Cancer J. 2016, 6, e380. [Google Scholar] [CrossRef] [PubMed]
- Saha, M.N.; Abdi, J.; Yang, Y.; Chang, H. MiRNA-29a as a tumor suppressor mediates PRIMA-1Met-induced anti-myeloma activity by targeting c-Myc. Oncotarget 2016, 7, 7149–7160. [Google Scholar] [CrossRef] [PubMed]
- Tsang, J.; Zhu, J.; van Oudenaarden, A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell 2007, 26, 753–767. [Google Scholar] [CrossRef] [PubMed]
- Mott, J.L.; Kurita, S.; Cazanave, S.C.; Bronk, S.F.; Werneburg, N.W.; Fernandez-Zapico, M.E. Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. J. Cell. Biochem. 2010, 110, 1155–1164. [Google Scholar] [CrossRef]
- Herrero, A.B.; Rojas, E.A.; Misiewicz-Krzeminska, I.; Krzeminski, P.; Gutiérrez, N.C. Molecular Mechanisms of p53 Deregulation in Cancer: An Overview in Multiple Myeloma. Int. J. Mol. Sci. 2016, 17, 2003. [Google Scholar] [CrossRef]
- Feng, Z.; Zhang, C.; Wu, R.; Hu, W. Tumor suppressor p53 meets microRNAs. J. Mol. Cell Biol. 2011, 3, 44–50. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Zhao, Y.; Feng, Z. MicroRNA Control of p53. J. Cell. Biochem. 2017, 118, 7–14. [Google Scholar] [CrossRef]
- Leotta, M.; Biamonte, L.; Raimondi, L.; Ronchetti, D.; Di Martino, M.T.; Botta, C.; Leone, E.; Pitari, M.R.; Neri, A.; Giordano, A.; et al. A p53-dependent tumor suppressor network is induced by selective miR-125a-5p inhibition in multiple myeloma cells. J. Cell. Physiol. 2014, 229, 2106–2116. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Calin, G.A.; Lopez-Berestein, G.; Sood, A.K. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov. 2016, 6, 235–246. [Google Scholar] [CrossRef]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.-H.; Nam, J.-W.; Heo, I.; Rhee, J.-K.; Sohn, S.Y.; Cho, Y.; Zhang, B.-T.; Kim, V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef]
- Zeng, Y.; Cullen, B.R. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004, 32, 4776–4785. [Google Scholar] [CrossRef]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Kuchenbauer, F.; Mah, S.M.; Heuser, M.; McPherson, A.; Rüschmann, J.; Rouhi, A.; Berg, T.; Bullinger, L.; Argiropoulos, B.; Morin, R.D.; et al. Comprehensive analysis of mammalian miRNA* species and their role in myeloid cells. Blood 2011, 118, 3350–3358. [Google Scholar] [CrossRef]
- Marco, A.; Macpherson, J.I.; Ronshaugen, M.; Griffiths-Jones, S. MicroRNAs from the same precursor have different targeting properties. Silence 2012, 3, 8. [Google Scholar] [CrossRef]
- Shi, Q.; He, Q.; Wei, J. MicroRNA-342 prohibits proliferation and invasion of melanoma cells by directly targeting Zinc-finger E-box binding homeobox 1. Oncol. Res. 2018, 26, 1447–1455. [Google Scholar] [CrossRef]
- Langenberger, D.; Bermudez-Santana, C.; Hertel, J.; Hoffmann, S.; Khaitovich, P.; Stadler, P.F. Evidence for human microRNA-offset RNAs in small RNA sequencing data. Bioinform. Oxf. Engl. 2009, 25, 2298–2301. [Google Scholar] [CrossRef]
- Bortoluzzi, S.; Biasiolo, M.; Bisognin, A. MicroRNA-offset RNAs (moRNAs): By-product spectators or functional players? Trends Mol. Med. 2011, 17, 473–474. [Google Scholar] [CrossRef]
- Nepal, C.; Coolen, M.; Hadzhiev, Y.; Cussigh, D.; Mydel, P.; Steen, V.M.; Carninci, P.; Andersen, J.B.; Bally-Cuif, L.; Müller, F.; et al. Transcriptional, post-transcriptional and chromatin-associated regulation of pri-miRNAs, pre-miRNAs and moRNAs. Nucleic Acids Res. 2016, 44, 3070–3081. [Google Scholar] [CrossRef] [PubMed]
- Umbach, J.L.; Strelow, L.I.; Wong, S.W.; Cullen, B.R. Analysis of rhesus rhadinovirus microRNAs expressed in virus-induced tumors from infected rhesus macaques. Virology 2010, 405, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Taft, R.J.; Simons, C.; Nahkuri, S.; Oey, H.; Korbie, D.J.; Mercer, T.R.; Holst, J.; Ritchie, W.; Wong, J.J.-L.; Rasko, J.E.J.; et al. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat. Struct. Mol. Biol. 2010, 17, 1030–1034. [Google Scholar] [CrossRef]
- Lu, G.; Middleton, R.E.; Sun, H.; Naniong, M.; Ott, C.J.; Mitsiades, C.S.; Wong, K.-K.; Bradner, J.E.; Kaelin, W.G. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 2014, 343, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Krönke, J.; Udeshi, N.D.; Narla, A.; Grauman, P.; Hurst, S.N.; McConkey, M.; Svinkina, T.; Heckl, D.; Comer, E.; Li, X.; et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 2014, 343, 301–305. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, L.; Barlogie, B.; Stephens, O.; Wu, X.; Williams, D.R.; Cartron, M.-A.; van Rhee, F.; Nair, B.; Waheed, S.; et al. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc. Natl. Acad. Sci. USA 2010, 107, 7904–7909. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Hou, Y.; Langlais, P.; Erickson, P.; Zhu, J.; Shi, C.-X.; Luo, M.; Zhu, Y.; Xu, Y.; Mandarino, L.J.; et al. Expression of the cereblon binding protein argonaute 2 plays an important role for multiple myeloma cell growth and survival. BMC Cancer 2016, 16, 297. [Google Scholar] [CrossRef]
- Asai, T.; Suzuki, Y.; Matsushita, S.; Yonezawa, S.; Yokota, J.; Katanasaka, Y.; Ishida, T.; Dewa, T.; Kiwada, H.; Nango, M.; et al. Disappearance of the angiogenic potential of endothelial cells caused by Argonaute2 knockdown. Biochem. Biophys. Res. Commun. 2008, 368, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Yu, W.; Qu, X.; Wang, R.; Xu, J.; Zhang, Q.; Xu, J.; Li, J.; Chen, L. Argonaute 2 promotes myeloma angiogenesis via microRNA dysregulation. J. Hematol. Oncol. 2014, 7, 40. [Google Scholar] [CrossRef] [PubMed]
- Sarasquete, M.E.; Gutiérrez, N.C.; Misiewicz-Krzeminska, I.; Paiva, B.; Chillón, M.C.; Alcoceba, M.; García-Sanz, R.; Hernández, J.M.; González, M.; San-Miguel, J.F. Upregulation of Dicer is more frequent in monoclonal gammopathies of undetermined significance than in multiple myeloma patients and is associated with longer survival in symptomatic myeloma patients. Haematologica 2011, 96, 468–471. [Google Scholar] [CrossRef]
- Tardito, D.; Mallei, A.; Popoli, M. Lost in translation. New unexplored avenues for neuropsychopharmacology: Epigenetics and microRNAs. Expert Opin. Investig. Drugs 2013, 22, 217–233. [Google Scholar] [CrossRef]
- Croce, C.M.; Calin, G.A. miRNAs, cancer, and stem cell division. Cell 2005, 122, 6–7. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Tuo, W.; Lian, H.; Liu, Q.; Zhu, X.-Q.; Gao, H. Strategies to identify microRNA targets: New advances. New Biotechnol. 2010, 27, 734–738. [Google Scholar] [CrossRef]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human MicroRNA targets. PLoS Biol. 2004, 2, e363. [Google Scholar] [CrossRef]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Kertesz, M.; Iovino, N.; Unnerstall, U.; Gaul, U.; Segal, E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007, 39, 1278–1284. [Google Scholar] [CrossRef]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.-S.; Tam, W.-L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar] [CrossRef] [PubMed]
- Jayaswal, V.; Lutherborrow, M.; Ma, D.D.F.; Yang, Y.H. Identification of microRNA-mRNA modules using microarray data. BMC Genom. 2011, 12, 138. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, R.; Reiss, D.J.; Shannon, P.; Facciotti, M.; Hood, L.; Baliga, N.S.; Thorsson, V. The Inferelator: An algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. Genome Biol. 2006, 7, R36. [Google Scholar] [CrossRef]
- Farah, M.J.; Wallace, M.A. Semantically-bounded anomia: Implications for the neural implementation of naming. Neuropsychologia 1992, 30, 609–621. [Google Scholar] [CrossRef]
- Bisognin, A.; Sales, G.; Coppe, A.; Bortoluzzi, S.; Romualdi, C. MAGIA2: From miRNA and genes expression data integrative analysis to microRNA-transcription factor mixed regulatory circuits (2012 update). Nucleic Acids Res. 2012, 40, W13–W21. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef]
- Oliveira, A.C.; Bovolenta, L.A.; Nachtigall, P.G.; Herkenhoff, M.E.; Lemke, N.; Pinhal, D. Combining Results from Distinct MicroRNA Target Prediction Tools Enhances the Performance of Analyses. Front. Genet. 2017, 8, 59. [Google Scholar] [CrossRef]
- Elton, T.S.; Yalowich, J.C. Experimental procedures to identify and validate specific mRNA targets of miRNAs. EXCLI J. 2015, 14, 758–790. [Google Scholar]
- Thomson, D.W.; Bracken, C.P.; Goodall, G.J. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011, 39, 6845–6853. [Google Scholar] [CrossRef]
- Kuhn, D.E.; Martin, M.M.; Feldman, D.S.; Terry, A.V.; Nuovo, G.J.; Elton, T.S. Experimental validation of miRNA targets. Methods San Diego Calif. 2008, 44, 47–54. [Google Scholar] [CrossRef]
- Biasiolo, M.; Forcato, M.; Possamai, L.; Ferrari, F.; Agnelli, L.; Lionetti, M.; Todoerti, K.; Neri, A.; Marchiori, M.; Bortoluzzi, S.; et al. Critical analysis of transcriptional and post-transcriptional regulatory networks in multiple myeloma. Pac. Symp. Biocomput. Pac. Symp. Biocomput. 2010, 397–408. [Google Scholar]
- Calura, E.; Bisognin, A.; Manzoni, M.; Todoerti, K.; Taiana, E.; Sales, G.; Morgan, G.J.; Tonon, G.; Amodio, N.; Tassone, P.; et al. Disentangling the microRNA regulatory milieu in multiple myeloma: Integrative genomics analysis outlines mixed miRNA-TF circuits and pathway-derived networks modulated in t(4;14) patients. Oncotarget 2016, 7, 2367–2378. [Google Scholar] [CrossRef] [PubMed]
- Cipolla, G.A.; Park, J.K.; de Oliveira, L.A.; Lobo-Alves, S.C.; de Almeida, R.C.; Farias, T.D.J.; de Lemos, D.S.; Malheiros, D.; Lavker, R.M.; Petzl-Erler, M.L. A 3′UTR polymorphism marks differential KLRG1 mRNA levels through disruption of a miR-584-5p binding site and associates with pemphigus foliaceus susceptibility. Biochim. Biophys. Acta 2016, 1859, 1306–1313. [Google Scholar] [CrossRef]
- Salzman, D.W.; Weidhaas, J.B. SNPing cancer in the bud: microRNA and microRNA-target site polymorphisms as diagnostic and prognostic biomarkers in cancer. Pharmacol. Ther. 2013, 137, 55–63. [Google Scholar] [CrossRef] [PubMed]
- de Larrea, C.F.; Navarro, A.; Tejero, R.; Tovar, N.; Díaz, T.; Cibeira, M.T.; Rosiñol, L.; Ferrer, G.; Rovira, M.; Rozman, M.; et al. Impact of MiRSNPs on survival and progression in patients with multiple myeloma undergoing autologous stem cell transplantation. Clin. Cancer Res. 2012, 18, 3697–3704. [Google Scholar] [CrossRef]
- Macauda, A.; Calvetti, D.; Maccari, G.; Hemminki, K.; Försti, A.; Goldschmidt, H.; Weinhold, N.; Houlston, R.; Andersen, V.; Vogel, U.; et al. Identification of miRSNPs associated with the risk of multiple myeloma. Int. J. Cancer 2017, 140, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C.; Bartel, D.P. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 2009, 138, 673–684. [Google Scholar] [CrossRef]
- Wu, X.; Bartel, D.P. Widespread Influence of 3′-End Structures on Mammalian mRNA Processing and Stability. Cell 2017, 169, 905–917.e11. [Google Scholar] [CrossRef] [PubMed]
- Misiewicz-Krzeminska, I.; Sarasquete, M.E.; Vicente-Dueñas, C.; Krzeminski, P.; Wiktorska, K.; Corchete, L.A.; Quwaider, D.; Rojas, E.A.; Corral, R.; Martín, A.A.; et al. Post-transcriptional Modifications Contribute to the Upregulation of Cyclin D2 in Multiple Myeloma. Clin. Cancer Res. 2016, 22, 207–217. [Google Scholar] [CrossRef]
- Bak, R.O.; Mikkelsen, J.G. miRNA sponges: Soaking up miRNAs for regulation of gene expression. Wiley Interdiscip. Rev. RNA 2014, 5, 317–333. [Google Scholar] [CrossRef] [PubMed]
- Ghini, F.; Rubolino, C.; Climent, M.; Simeone, I.; Marzi, M.J.; Nicassio, F. Endogenous transcripts control miRNA levels and activity in mammalian cells by target-directed miRNA degradation. Nat. Commun. 2018, 9, 3119. [Google Scholar] [CrossRef]
- Bonizzato, A.; Gaffo, E.; Te Kronnie, G.; Bortoluzzi, S. CircRNAs in hematopoiesis and hematological malignancies. Blood Cancer J. 2016, 6, e483. [Google Scholar] [CrossRef]
- Panda, A.C. Circular RNAs Act as miRNA Sponges. Adv. Exp. Med. Biol. 2018, 1087, 67–79. [Google Scholar] [PubMed]
- Lee, Y.-R.; Chen, M.; Pandolfi, P.P. The functions and regulation of the PTEN tumour suppressor: New modes and prospects. Nat. Rev. Mol. Cell Biol. 2018, 19, 547–562. [Google Scholar] [CrossRef] [PubMed]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Kats, L.; Salmena, L.; Weiss, D.; Tan, S.M.; Ala, U.; Karreth, F.; Poliseno, L.; Provero, P.; Di Cunto, F.; et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011, 147, 344–357. [Google Scholar] [CrossRef]
- Gao, H.; Song, X.; Kang, T.; Yan, B.; Feng, L.; Gao, L.; Ai, L.; Liu, X.; Yu, J.; Li, H. Long noncoding RNA CRNDE functions as a competing endogenous RNA to promote metastasis and oxaliplatin resistance by sponging miR-136 in colorectal cancer. OncoTargets Ther. 2017, 10, 205–216. [Google Scholar] [CrossRef]
- Han, P.; Li, J.-W.; Zhang, B.-M.; Lv, J.-C.; Li, Y.-M.; Gu, X.-Y.; Yu, Z.-W.; Jia, Y.-H.; Bai, X.-F.; Li, L.; et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol. Cancer 2017, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.-B.; He, X.; Huang, Y.-F.; Wu, Q.-N.; Zhou, Y.-C.; Hao, D.-J. Long Noncoding RNA CRNDE Promotes Multiple Myeloma Cell Growth by Suppressing miR-451. Oncol. Res. 2017, 25, 1207–1214. [Google Scholar] [CrossRef]
- Chen, L.; Hu, N.; Wang, C.; Zhao, H.; Gu, Y. Long non-coding RNA CCAT1 promotes multiple myeloma progression by acting as a molecular sponge of miR-181a-5p to modulate HOXA1 expression. Cell Cycle Georget. Tex 2018, 17, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Amodio, N.; Stamato, M.A.; Juli, G.; Morelli, E.; Fulciniti, M.; Manzoni, M.; Taiana, E.; Agnelli, L.; Cantafio, M.E.G.; Romeo, E.; et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia 2018, 32, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Lin, J.; Fang, H.; Fang, J.; Li, C.; Chen, W.; Liu, S.; Ondrejka, S.; Gong, Z.; Reu, F.; et al. Targeting the MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in multiple myeloma. Leukemia 2018, 32, 2250–2262. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Xiao, X.; Yang, S. LncRNA MALAT1 acts as an oncogene in multiple myeloma through sponging miR-509-5p to modulate FOXP1 expression. Oncotarget 2017, 8, 101984–101993. [Google Scholar] [CrossRef]
- Ebert, M.S.; Sharp, P.A. MicroRNA sponges: Progress and possibilities. RNA 2010, 16, 2043–2050. [Google Scholar] [CrossRef] [PubMed]
- Tay, F.C.; Lim, J.K.; Zhu, H.; Hin, L.C.; Wang, S. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv. Drug Deliv. Rev. 2015, 81, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Mignacca, L.; Saint-Germain, E.; Benoit, A.; Bourdeau, V.; Moro, A.; Ferbeyre, G. Sponges against miR-19 and miR-155 reactivate the p53-Socs1 axis in hematopoietic cancers. Cytokine 2016, 82, 80–86. [Google Scholar] [CrossRef]



| miRNA | Methylation Status in NPC, MM and HMCLs | Function in MM | Ref. |
|---|---|---|---|
| 9-3 and 9-1 | Unmethylated in MM, methylated in HMCLs | CD44 overexpression, (glycoprotein that has been associated with lenalidomide and dexamethasone resistance in myeloma) | [63,64] |
| 10b-5p | Methylated in MM Upregulated after 5-aza-CdR in HMCLs | Not tested | [48] |
| 28 | Unmethylated in healthy controls and at MM relapse, methylated in some HMCLs | Inverse correlation between CCND1 expression and LPP/miR-28 methylation | [65] |
| 34a | Methylated in primary MM, no difference between diagnosis and relapse, highly methylated in HMCLs | Inhibition of MM growth and reduction of bone lesions by targeting TGIF2 | [47,66] |
| 34b/c | Methylated in MM, increased methylation in relapsed MM, highly methylated in HMCLs | Inhibition of cellular proliferation and induction of apoptosis in MM cells | [50] |
| 124-1 | Methylated in primary MM, highly methylated in HMCLs | Repression of CDK6 | [67] |
| 129 | Methylation increases from MGUS to MM at diagnosis and at relapse/progression | Downregulation of SOX4 | [51] |
| 152 | Highly methylated in MM and in HMCLs Upregulated after 5-aza-CdR in HMCLs Targets DNMT1 | Decrease of DNMT1 and E2F2 Induction of apoptosis in MM cells | [48] |
| 155 | Highly methylated in HMCLs Expression induced by LPS only when unmethylated in HMCLs | High expression of miR-155 was associated with improved overall survival (OS) in MM. | [68] |
| 192, 194 and 215 | Methylated in MM cell lines | High expression of miR-194 was associated with improved OS in MM. Overexpression associated with inhibition of cellular proliferation and migration | [28] |
| 198, 135a*, 200c, 125a-3p, 188-5p, 483-5p, 663, 630 | Methylated in MM Upregulated after 5-aza-CdR in HMCLs | Not tested | [69] |
| 203 | Methylated in HMCLs | Downregulation of CREB1 protein and inhibition of proliferation of myeloma cells | [49] |
| 342-3p | Methylated in MGUS and increased methylation in relapsed MM Upregulated after 5-aza-CdR in HMCLs | Not tested | [52] |
| 375 | Methylated in MM and HMCLs | Downregulation of PDPK1 and IGF1R | [53] |
| Gene Locus | miRNA | Host Gene | Regulation Mechanism | Ref. |
|---|---|---|---|---|
| 7q32.2 | miR-335 | MEST | Not by CNA | [33,72] |
| 14q32.2 | miR-342-3p | EVL | By promoter DNA methylation of its host gene, not by CNA | [33,72] |
| 2q32.1-q32.2 | miR-561 | GULP1 | Not by CNA | [33,72] |
| 17q21.32 | miR-152 | COPZ2 | Unknown | [33] |
| 3q27.3-q28 | miR-28-5p | LPP | By promoter DNA methylation of the host gene | [65] |
| 17q23.1 | miR-21 | TMEM49 | By an upstream enhancer, overlapping with intronic region of the host gene | [73] |
| 21q21.3 | miR-155 | BIC | By promoter DNA methylation of the host gene | [68] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Misiewicz-Krzeminska, I.; Krzeminski, P.; Corchete, L.A.; Quwaider, D.; Rojas, E.A.; Herrero, A.B.; Gutiérrez, N.C. Factors Regulating microRNA Expression and Function in Multiple Myeloma. Non-Coding RNA 2019, 5, 9. https://doi.org/10.3390/ncrna5010009
Misiewicz-Krzeminska I, Krzeminski P, Corchete LA, Quwaider D, Rojas EA, Herrero AB, Gutiérrez NC. Factors Regulating microRNA Expression and Function in Multiple Myeloma. Non-Coding RNA. 2019; 5(1):9. https://doi.org/10.3390/ncrna5010009
Chicago/Turabian StyleMisiewicz-Krzeminska, Irena, Patryk Krzeminski, Luis A. Corchete, Dalia Quwaider, Elizabeta A. Rojas, Ana Belén Herrero, and Norma C. Gutiérrez. 2019. "Factors Regulating microRNA Expression and Function in Multiple Myeloma" Non-Coding RNA 5, no. 1: 9. https://doi.org/10.3390/ncrna5010009
APA StyleMisiewicz-Krzeminska, I., Krzeminski, P., Corchete, L. A., Quwaider, D., Rojas, E. A., Herrero, A. B., & Gutiérrez, N. C. (2019). Factors Regulating microRNA Expression and Function in Multiple Myeloma. Non-Coding RNA, 5(1), 9. https://doi.org/10.3390/ncrna5010009







