Increased Extracellular Matrix Protein Production in Chronic Diabetic Complications: Implications of Non-Coding RNAs
Abstract
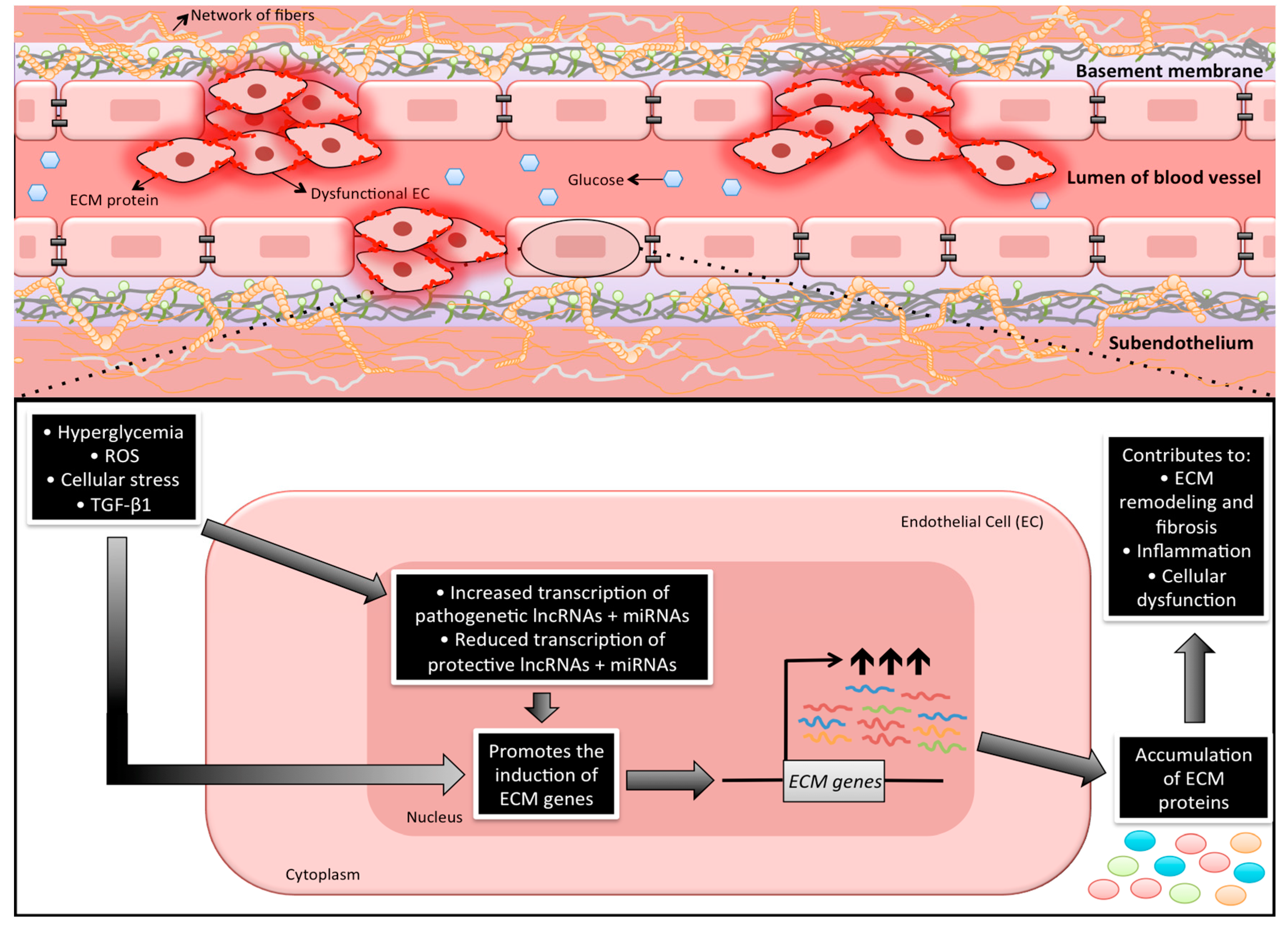
1. Introduction
2. ECM—The Footprint of Chronic Diabetic Complications
3. Cellular Phenotypic Changes Causing Increased ECM Protein Productions in Chronic Diabetic Complications
4. Hyperglycemia and Gene Transcription
4.1. Transcription Factors and Co-Activators
4.1.1. NF-κB and AP-1
4.1.2. p300 and Histone Acetylation
4.2. Non-Coding RNAs
4.2.1. Non-Coding RNAs
4.2.2. Long Non-Coding RNAs (lncRNAs)
5. Role of miRNAs in Chronic Diabetic Complications in Various Organs
5.1. miRNAs in Diabetic Nephropathy (DN)
5.2. miRNAs in Diabetic Cardiomyopathy (DCM)
5.3. miRNAs in Diabetic Retinopathy (DR)
6. Role of LncRNAs in Chronic Diabetic Complications in Various Organs
6.1. LncRNAs in Diabetic Nephropathy (DN)
6.2. LncRNAs in Diabetic Cardiomyopathy (DCM) and Other Cardiovascular Complications
6.3. lncRNAs in Diabetic Retinopathy (DR)
7. Relationship of lncRNAs, miRNAs, and Other Epigenetic Mechanisms on Causing Increased ECM Protein Production in Diabetes
7.1. Histone Methylation: PRC2 and lncRNAs
8. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cho, N.H.; Shaw, J.E.; Karuranga, S.; Huang, Y.; da Rocha Fernandes, J.D.; Ohlrogge, A.W.; Malanda, B. IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Pract. 2018, 138, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Gregg, E.W. Complications of diabetes in elderly people. BMJ 2002, 325, 916–917. [Google Scholar] [CrossRef]
- Chow, E.A.; Foster, H.; Gonzalez, V.; McIver, L. The Disparate Impact of Diabetes on Racial/Ethnic Minority Populations. Clin. Diabetes 2012, 30, 130–133. [Google Scholar] [CrossRef]
- Beckman, J.A.; Creager, M.A. Vascular complications of diabetes. Circ. Res. 2016, 118, 1771–1785. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.J.; McNamara, P.M.; Gordon, T.; Kannel, W.B. Morbidity and mortality in diabetics in the Framingham population. Sixteen year follow-up study. Diabetes Care 1974, 23, 105–111. [Google Scholar] [CrossRef]
- Biswas, S.; Thomas, A.A.; Chakrabarti, S. LncRNAs: Proverbial Genomic “Junk” or Key Epigenetic Regulators During Cardiac Fibrosis in Diabetes? Front. Cardiovasc. Med. 2018, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Brownlee, M. Biochemistry and molecular cell biology of diabetic complications. Nature 2001, 414, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Fioretto, P.; Mauer, M. Histopathology of Diabetic Nephropathy. Semin. Nephrol. 2007, 27, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Bae, E.; Amin, S.; Kim, D. Extracellular matrix, gap junctions, and retinal vascular homeostasis in diabetic retinopathy. Exp. Eye Res. 2015, 133, 58–68. [Google Scholar] [CrossRef]
- Asbun, J.; Villarreal, F.J. The pathogenesis of myocardial fibrosis in the setting of diabetic cardiomyopathy. J. Am. Coll. Cardiol. 2006, 47, 693–700. [Google Scholar] [CrossRef]
- Khan, Z.A.; Chakrabarti, S. Chronic Diabetic Complications: Endothelial Cells at the Frontline. Front. Cardiovasc. Drug Discov. 2010, 1, 121–137. [Google Scholar] [CrossRef]
- Kumagai, A.K. Glucose transport in brain and retina: Implications in the management and complications of diabetes. Diabetes Metab. Res. Rev. 1999, 15, 261–273. [Google Scholar] [CrossRef]
- Feng, B.; Ruiz, M.A.; Chakrabarti, S. Oxidative-stress-induced epigenetic changes in chronic diabetic complications. Can. J. Physiol. Pharmacol. 2013, 91, 213–220. [Google Scholar] [CrossRef]
- Teti, A. Regulation of cellular functions by extracellular matrix. J. Am. Soc. Nephrol. 1992, 2 (Suppl. 10), S83–S87. [Google Scholar]
- Boudreau, N.J.; Jones, P.L. Extracellular matrix and integrin signalling: The shape of things to come. Biochem. J. 1999, 339, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Davis, G.E.; Senger, D.R. Endothelial extracellular matrix: Biosynthesis, remodeling, and functions during vascular morphogenesis and neovessel stabilization. Circ. Res. 2005, 97, 1093–1107. [Google Scholar] [CrossRef]
- Addison, C.L.; Nor, J.E.; Zhao, H.; Linn, S.A.; Polverini, P.J.; Delaney, C.E. The response of VEGF-stimulated endothelial cells to angiostatic molecules is substrate-dependent. BMC Cell Biol. 2005, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Lochter, A.; Bissell, M.J. Involvement of extracellular matrix constituents in breast cancer. Semin. Cancer Biol. 1995, 6, 165–173. [Google Scholar] [CrossRef]
- Midwood, K.S.; Orend, G. The role of tenascin-C in tissue injury and tumorigenesis. J. Cell Commun. Signal. 2009, 3, 287–310. [Google Scholar] [CrossRef]
- Noel, A.; Kebers, F.; Maquoi, E.; Foidart, J.M. Cell-cell and cell-matrix interactions during breast cancer progression. Curr. Top. Pathol. 1999, 93, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, A.G.; Iruela-Arispe, M.L. Extracellular matrix, inflammation, and the angiogenic response. Cardiovasc. Res. 2010, 86, 226–235. [Google Scholar] [CrossRef]
- Bishop, P.N. The role of extracellular matrix in retinal vascular development and preretinal neovascularization. Exp. Eye Res. 2015, 133, 30–36. [Google Scholar] [CrossRef]
- Jiang, B.; Liou, G.I.; Behzadian, M.A.; Caldwell, R.B. Astrocytes modulate retinal vasculogenesis: Effects on fibronectin expression. J. Cell Sci. 1994, 107, 2499–2508. [Google Scholar]
- Nishikawa, T.; Giardino, I.; Edelstein, D.; Brownlee, M. Changes in diabetic retinal matrix protein mRNA levels in a common transgenic mouse strain. Curr. Eye Res. 2003, 21, 581–587. [Google Scholar] [CrossRef]
- Silver, M.D.; Huckell, V.F.; Lorber, M. Basement membranes of small cardiac vessels in patients with diabetes and myxoedema: Preliminary observations. Pathology 1977, 9, 213–220. [Google Scholar] [CrossRef]
- Tsilibary, E.C. Microvascular basement membranes in diabetes mellitus. J. Pathol. 2003, 200, 537–546. [Google Scholar] [CrossRef]
- Pankov, R.; Yamada, K.M. Fibronectin at a glance. J. Cell Sci. 2002, 115 Pt 20, 3861–3863. [Google Scholar] [CrossRef]
- Khan, Z.A.; Cukiernik, M.; Gonder, J.R.; Chakrabarti, S. Oncofetal fibronectin in diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2004, 45, 287–295. [Google Scholar] [CrossRef]
- Khan, Z.A.; Chan, B.M.; Uniyal, S.; Barbin, Y.P.; Farhangkhoee, H.; Chen, S.; Chakrabarti, S. EDB fibronectin and angiogenesis—A novel mechanistic pathway. Angiogenesis 2005, 8, 183–196. [Google Scholar] [CrossRef]
- Grant, M.B.; Caballero, S.; Bush, D.M.; Spoerri, P.E. Fibronectin fragments modulate human retinal capillary cell proliferation and migration. Diabetes 1998, 47, 1335–1340. [Google Scholar] [CrossRef]
- Wilson, S.H.; Ljubimov, A.V.; Morla, A.O.; Caballero, S.; Shaw, L.C.; Spoerri, P.E.; Tarnuzzer, R.W.; Grant, M.B. Fibronectin fragments promote human retinal endothelial cell adhesion and proliferation and ERK activation through alpha5beta1 integrin and PI 3-kinase. Investig. Ophthalmol. Vis. Sci. 2003, 44, 1704–1715. [Google Scholar] [CrossRef]
- Yoon, Y.S.; Uchida, S.; Masuo, O.; Cejna, M.; Park, J.S.; Gwon, H.C.; Kirchmair, R.; Bahlman, F.; Walter, D.; Curry, C.; et al. Progressive attenuation of myocardial vascular endothelial growth factor expression is a seminal event in diabetic cardiomyopathy: Restoration of microvascular homeostasis and recovery of cardiac function in diabetic cardiomyopathy after replenishment of local vascular endothelial growth factor. Circulation 2005, 111, 2073–2085. [Google Scholar] [CrossRef]
- Chou, E.; Suzuma, I.; Way, K.J.; Opland, D.; Clermont, A.C.; Naruse, K.; Suzuma, K.; Bowling, N.L.; Vlahos, C.J.; Aiello, L.P.; et al. Decreased cardiac expression of vascular endothelial growth factor and its receptors in insulin-resistant and diabetic states: A possible explanation for impaired collateral formation in cardiac tissue. Circulation 2002, 105, 373–379. [Google Scholar] [CrossRef]
- Soldatos, G.; Cooper, M.E.; Jandeleit-Dahm, K.A.M. Advanced-glycation end products in insulin-resistant states. Curr. Hypertens. Rep. 2005, 7, 96–102. [Google Scholar] [CrossRef]
- Westermeier, F.; Riquelme, J.A.; Pavez, M.; Garrido, V.; Díaz, A.; Verdejo, H.E.; Castro, P.F.; García, L.; Lavandero, S. New molecular insights of insulin in diabetic cardiomyopathy. Front. Physiol. 2016, 7. [Google Scholar] [CrossRef]
- Law, B.; Fowlkes, V.; Goldsmith, J.G.; Carver, W.; Goldsmith, E.C. Diabetes-induced alterations in the extracellular matrix and their impact on myocardial function. Microsc. Microanal. 2012, 18, 22–34. [Google Scholar] [CrossRef]
- Fowlkes, V.; Clark, J.; Fix, C.; Law, B.A.; Morales, M.O.; Qiao, X.; Ako-Asare, K.; Goldsmith, J.G.; Carver, W.; Murray, D.B.; et al. Type II diabetes promotes a myofibroblast phenotype in cardiac fibroblasts. Life Sci. 2013, 92, 669–676. [Google Scholar] [CrossRef]
- Meran, S.; Steadman, R. Fibroblasts and myofibroblasts in renal fibrosis. Int. J. Exp. Pathol. 2011, 92, 158–167. [Google Scholar] [CrossRef]
- Li, J.; Qu, X.; Bertram, J.F. Endothelial-myofibroblast transition contributes to the early development of diabetic renal interstitial fibrosis in streptozotocin-induced diabetic mice. Am. J. Pathol. 2009, 175, 1380–1388. [Google Scholar] [CrossRef]
- Cao, Y.; Feng, B.; Chen, S.; Chu, Y.; Chakrabarti, S. Mechanisms of endothelial to mesenchymal transition in the retina in diabetes. Investig. Ophthalmol. Vis. Sci. 2014, 55, 7321–7331. [Google Scholar] [CrossRef]
- Schwartz, M.A.; Vestweber, D.; Simons, M. A unifying concept in vascular health and disease. Science 2018, 360, 270–271. [Google Scholar] [CrossRef] [PubMed]
- King, G.L.; Loeken, M.R. Hyperglycemia-induced oxidative stress in diabetic complications. Histochem. Cell Biol. 2004, 122, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Baynes, J.W.; Thorpe, S.R. Role of oxidative stress in diabetic complications: A new perspective on an old paradigm. Diabetes Care 1999, 48, 1–9. [Google Scholar] [CrossRef]
- Srivastava, S.K.; Ansari, N.H.; Liu, S.; Izban, A.; Das, B.; Szabo, G.; Bhatnagar, A. The effect of oxidants on biomembranes and cellular metabolism. Mol. Cell Biochem. 1989, 91, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, T.; Edelstein, D.; Du, X.L.; Yamagishi, S.I.; Matsumura, T.; Kaneda, Y.; Yorek, M.A.; Beebe, D.; Oates, P.J.; Hammes, H.P.; et al. Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature 2000, 404, 787–790. [Google Scholar] [CrossRef] [PubMed]
- Kiernan, R.; Brès, V.; Ng, R.W.M.; Coudart, M.; Messaoudi, S.E.; Sardet, C.; Jin, D.; Emiliani, S.; Benkirane, M. Post-activation Turn-off of NF-κB-dependent Transcription Is Regulated by Acetylation of p65. J. Biol. Chem. 2003, 278, 2758–2766. [Google Scholar] [CrossRef]
- Khan, Z.A.; Farhangkhoee, H.; Chakrabarti, S. Towards newer molecular targets for chronic diabetic complications. Curr. Vasc. Pharmacol. 2006, 4, 45–57. [Google Scholar] [CrossRef]
- Chen, S.; Khan, Z.A.; Cukiernik, M.; Chakrabarti, S. Differential activation of NF-κ B and AP-1 in increased fibronectin synthesis in target organs of diabetic complications. Am. J. Physiol. Endocrinol. Metab. 2003, 284, 1089–1097. [Google Scholar] [CrossRef]
- Chen, S.; Feng, B.; George, B.; Chakrabarti, R.; Chen, M.; Chakrabarti, S. Transcriptional coactivator p300 regulates glucose-induced gene expression in endothelial cells. Am. J. Physiol. Endocrinol. Metab. 2009, 298, 127–137. [Google Scholar] [CrossRef]
- Khan, Z.A.; Chakrabarti, S. Growth factors in proliferative diabetic retinopathy. Exp. Diabesity Res. 2003, 4, 287–301. [Google Scholar] [CrossRef]
- Kaur, H.; Chen, S.; Xin, X.; Chiu, J.; Khan, Z.A.; Chakrabarti, S. Diabetes-induced extracellular matrix protein expression is mediated by transcription coactivator p300. Diabetes 2006, 55, 3104–3111. [Google Scholar] [CrossRef] [PubMed]
- Quehenberger, P.; Bierhaus, A.; Fasching, P.; Muellner, C.; Klevesath, M.; Hong, M.; Stier, G.; Sattler, M.; Schleicher, E.; Speiser, W.; et al. Endothelin 1 transcription is controlled by nuclear factor-κB in AGE-stimulated cultured endothelial cells. Diabetes Care 2000, 49, 1561–1570. [Google Scholar] [CrossRef]
- Sanchez, A.P.; Sharma, K. Transcription factors in the pathogenesis of diabetic nephropathy. Expert Rev. Mol. Med. 2009, 11. [Google Scholar] [CrossRef]
- Rahman, I.; Marwick, J.; Kirkham, P. Redox modulation of chromatin remodeling: Impact on histone acetylation and deacetylation, NF-κB and pro-inflammatory gene expression. Biochem. Pharmacol. 2004, 68, 1255–1267. [Google Scholar] [CrossRef]
- Feng, B.; Chen, S.; McArthur, K.; Wu, Y.; Sen, S.; Ding, Q.; Feldman, R.D.; Chakrabarti, S. miR-146a-Mediated extracellular matrix protein production in chronic diabetes complications. Diabetes 2011, 60, 2975–2984. [Google Scholar] [CrossRef]
- Xie, X.; Peng, J.; Chang, X.; Huang, K.; Huang, J.; Wang, S.; Shen, X.; Liu, P.; Huang, H. Activation of RhoA/ROCK regulates NF-κB signaling pathway in experimental diabetic nephropathy. Mol. Cell. Endocrinol. 2013, 369, 86–97. [Google Scholar] [CrossRef]
- Shaulian, E.; Karin, M. AP-1 in cell proliferation and survival. Oncogene 2001, 20, 2390–2400. [Google Scholar] [CrossRef]
- Bakiri, L.; Lallemand, D.; Bossy-Wetzel, E.; Yaniv, M. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: A role in the control of cyclin D1 expression. EMBO J. 2000, 2, 2056–2068. [Google Scholar] [CrossRef]
- Chinenov, Y.; Kerppola, T.K. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 2001, 20, 2438–2452. [Google Scholar] [CrossRef]
- Fujioka, S.; Niu, J.; Schmidt, C.; Sclabas, G.M.; Peng, B.; Uwagawa, T.; Li, Z.; Evans, D.B.; Abbruzzese, J.L.; Chiao, P.J. NF-κB and AP-1 connection: Mechanism of NF-κB-dependent regulation of AP-1 activity. Mol. Cell. Biol. 2004, 24, 7806–7819. [Google Scholar] [CrossRef]
- Ramana, K.V.; Friedrich, B.; Srivastava, S.; Bhatnagar, A.; Srivastava, S.K. Activation of nulcear factor-κB by hyperglycemia in vascular smooth muscle cells is regulated by aldose reductase. Diabetes 2004, 53, 2910–2920. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.S.; Cho, M.H.; Lee, G.T.; Park, J.S.; Ahn, C.W.; Cha, B.S.; Lim, S.K.; Kim, K.R.; Ha, H.J.; Lee, H.C. The activation of NF-κB and AP-1 in peripheral blood mononuclear cells isolated from patients with diabetic nephropathy. Diabetes Res. Clin. Pract. 2008, 81, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Saberi, M.; Zmuda, E.; Wang, Y.; Altarejos, J.; Zhang, X.; Dentin, R.; Hedrick, S.; Bandyopadhyay, G.; Hai, T.; et al. Adipocyte CREB Promotes Insulin Resistance in Obesity. Cell Metab. 2009, 9, 277–286. [Google Scholar] [CrossRef]
- Nilsson, J.; Nilsson, L.M.; Chen, Y.W.; Molkentin, J.D.; Erlinge, D.; Gomez, M.F. High glucose activates nuclear factor of activated T cells in native vascular smooth muscle. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Solomon, S.S.; Borromeo, D.M.; Martinez-Hernandez, A.; Raghow, R. Insulin deprivation leads to deficiency of Sp1 transcription factor in H-411E hepatoma cells and in streptozotocin-induced diabetic ketoacidosis in the rat. Endocrinology 2001, 142, 1635–1642. [Google Scholar] [CrossRef]
- Cetkovic-Cvrlje, M.; Uckun, F.M. Effect of targeted disruption of signal transducer and activator of transcription (Stat)4 and Stat6 genes on the autoimmune diabetes development induced by multiple low doses of streptozotocin. Clin. Immunol. 2005, 114, 299–306. [Google Scholar] [CrossRef]
- Ponugoti, B.; Dong, G.; Graves, D.T. Role of Forkhead Transcription Factors in Diabetes-Induced Oxidative Stress. Exp. Diabetes Res. 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Yu, X.; Pan, F.Y.; Gao, X.; Xue, B.; Li, C.J. An early response transcription factor, Egr-1, enhances insulin resistance in type 2 diabetes with chronic hyperinsulinism. J. Biol. Chem. 2011, 286, 14508–14515. [Google Scholar] [CrossRef] [PubMed]
- Motterle, A.; Gattesco, S.; Caille, D.; Meda, P.; Regazzi, R. Involvement of long non-coding RNAs in beta cell failure at the onset of type 1 diabetes in NOD mice. Diabetologia 2015, 58, 1827–1835. [Google Scholar] [CrossRef]
- Yo, K.; Rünger, T.M. The long non-coding RNA FLJ46906 binds to the transcription factors NF-κB and AP-1 and regulates expression of aging-associated genes. Aging (Albany NY) 2018, 10, 2037–2050. [Google Scholar] [CrossRef]
- McKinsey, T.A.; Olson, E.N. Toward transcriptional therapies for the failing heart: Chemical screens to modulate genes. J. Clin. Investig. 2005, 115, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Yanazume, T.; Morimoto, T.; Wada, H.; Kawamura, T.; Hasegawa, K. Biological role of p300 in cardiac myocytes. Mol. Cell Biochem. 2003, 248, 115–119. [Google Scholar] [CrossRef] [PubMed]
- McKinsey, T.A.; Olson, E.N. Cardiac histone acetylation--therapeutic opportunities abound. Trends Genet. 2004, 20, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Goodman, R.H.; Smolik, S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000, 14, 1553–1577. [Google Scholar] [CrossRef]
- Chen, L.F.; Greene, W.C. Regulation of distinct biological activities of the NF-κB transcription factor complex by acetylation. J. Mol. Med. 2003, 81, 549–557. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. The CBP co-activator is a histone acetyltransferase. Nature 1996, 384, 641–643. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Nakagawa, H.; Tajima, A.; Yoshida, K.; Inque, I. ANRIL is implicated in the regulation of nucleus and potential transcriptional target of E2F1. Oncol. Rep. 2010, 24, 701–707. [Google Scholar] [CrossRef]
- Matouk, I.J.; DeGroot, N.; Mezan, S.; Ayesh, S.; Abu-lail, R.; Hochberg, A.; Galun, E. The H19 Non-Coding RNA Is Essential for Human Tumor Growth. PLoS ONE 2007, 845, e845. [Google Scholar] [CrossRef]
- Lottin, S.; Vercoutter-Edouart, A.S.; Adriaenssens, E.; Czeszak, X.; Lemoine, J.; Roudbaraki, M.; Coll, J.; Hondermarck, H.; Dugimont, T.; Curgy, J.J. Thioredoxin post-transcriptional regulation by H19 provides a new function to mRNA-like non-coding RNA. Oncogene 2002, 21, 1625–1631. [Google Scholar] [CrossRef]
- McArthur, K.; Feng, B.; Wu, Y.X.; Chen, S.; Chakrabarti, S. microRNA 200b regulates VEGF mediated alterations in diabetic retinopathy. Diabetes 2011, 60, 1314–1321. [Google Scholar] [CrossRef]
- Thomas, A.A.; Feng, B.; Chakrabarti, S. ANRIL: A regulator of VEGF in diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2017, 58, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Valencia-Sanchez, M.A.; Liu, J.; Hannon, G.J.; Parker, R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006, 20, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef]
- Chung, F.W.; Tellam, R.L. MicroRNA-26a targets the histone methyltransferase enhancer of zeste homolog 2 during myogenesis. J. Biol. Chem. 2008, 283, 9836–9843. [Google Scholar] [CrossRef]
- Williams, A.H.; Valdez, G.; Moresi, V.; Qi, X.; McAnally, J.; Elliott, J.L.; Bassel-Duby, R.; Sanes, J.R.; Olson, E.N. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 2009, 326, 1549–1554. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, Z.; Watabe, K.; Zhang, X.; Bai, C.; Xu, M.; Wu, F.; Mo, Y. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013, 20, 1558–1568. [Google Scholar] [CrossRef]
- Leucci, E.; Patella, F.; Waage, J.; Holmstrøm, K.; Lindow, M.; Porse, B.; Kauppinen, S.; Lund, A.H. MicroRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef]
- Fan, M.; Li, X.; Jiang, W.; Huang, Y.; Li, J.; Wang, Z. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp. Ther. Med. 2013, 5, 1143–1146. [Google Scholar] [CrossRef]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Ann. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, F.; Zhou, L.Y.; Long, B.; Yuan, S.M.; Wang, Y.; Liu, C.Y.; Sun, T.; Zhang, X.J.; Li, P.F. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 2014, 114, 1377–1388. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Huo, X.S.; Yuan, S.X.; Zhang, L.; Zhou, W.P.; Wang, F.; Sun, S.H. Repression of the Long Noncoding RNA-LET by Histone Deacetylase 3 Contributes to Hypoxia-Mediated Metastasis. Mol. Cell 2013, 49, 1083–1096. [Google Scholar] [CrossRef]
- Brown, C.J.; Ballabio, A.; Rupert, J.L.; Lafreniere, R.G.; Grompe, M.; Tonlorenzi, R.; Willard, H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 1991, 349, 38–44. [Google Scholar] [CrossRef]
- Lee, J.T. Epigenetic regulation by long noncoding RNAs. Science 2012, 338, 1435–1439. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; Lin, L.; Soh, B.S.; Stanton, L.W. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013, 29, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zheng, C.; Liu, J.; Lan, J.; Li, W.; Zhang, Y. CpG island methylation patterns and expressions of H19 gene in cloned fetus of goat. Sheng Wu Gong Cheng Xue Bao 2010, 26, 582–587. [Google Scholar] [PubMed]
- Bochenek, G.; Häsler, R.; Mokhtari, N.E.; König, I.N.; Loos, B.G.; Jepsen, S.; Rosenstiel, P.; Schreiber, S.; Schaefer, A. The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum. Mol. Genet. 2013, 22, 4516–4527. [Google Scholar] [CrossRef]
- Bernard, D.; Prasanth, K.V.; Tripathi, V.; Colasse, S.; Nakamura, T.; Xuan, Z.; Zhang, M.Q.; Sedel, F.; Jourdren, L.; Coulpier, F.; et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010, 29, 3082–3093. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Diederichs, S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012, 9, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.H.; Kim, K.M.; McClusky, W.G.; Abdelmohsen, K.; Gorospe, M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip. Rev. RNA 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15 INK4B tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Fraser, P. No-nonsense functions for long noncoding RNAs. Cell 2011, 145, 178–181. [Google Scholar] [CrossRef]
- Romero-Barrios, N.; Legascue, M.F.; Benhamed, M.; Ariel, F.; Crespi, M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018, 46, 2169–2184. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Zhang, J.; Wang, M.; Lanting, L.; Yuan, H.; Rossi, J.J.; Natarajan, R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc. Natl. Acad. Sci. USA 2007, 104, 3432–3437. [Google Scholar] [CrossRef]
- Deshpande, S.D.; Putta, S.; Wang, M.; Lai, J.Y.; Bitzer, M.; Nelson, R.G.; Lanting, L.L.; Kato, M.; Natarajan, R. Transforming growth factor-β induced cross talk between p53 and a MicroRNA in the pathogenesis of diabetic nephropathy. Diabetes 2013, 62, 3151–3162. [Google Scholar] [CrossRef]
- Kato, M.; Arce, L.; Wang, M.; Putta, S.; Lanting, L.; Natarajan, R. A microRNA circuit mediates transforming growth factor-B1 autoregulation in renal glomerular mesangial cells. Kidney Int. 2011, 80, 358–368. [Google Scholar] [CrossRef]
- Krupa, A.; Jenkins, R.; Luo, D.D.; Lewis, A.; Phillips, A.; Fraser, D. Loss of MicroRNA-192 Promotes Fibrogenesis in Diabetic Nephropathy. J. Am. Soc. Nephrol. 2010, 21, 438–447. [Google Scholar] [CrossRef]
- Wang, B.; Herman-Edelstein, M.; Koh, P.; Burns, W.; Jandeleit-Dahm, K.; Watson, A.; Saleem, M.; Goodall, G.J.; Twigg, S.M.; Cooper, M.E.; et al. E-cadherin expression is regulated by miR-192/215 by a mechanism that is independent of the profibrotic effects of transforming growth factor-β. Diabetes 2010, 59, 1794–1802. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, Y.; Minto, A.W.; Wang, J.; Shi, Q.; Li, X.; Quigg, R.J. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. 2008, 22, 4126–4135. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Jha, J.C.; Hagiwara, S.; McClelland, A.D.; Jandeleit-Dahm, K.; Thomas, M.C.; Cooper, M.E.; Kantharidis, P. Transforming growth factor-β1-mediated renal fibrosis is dependent on the regulation of transforming growth factor receptor 1 expression by let-7b. Kidney Int. 2014, 85, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Wang, Y.; Wang, W.; Chang, B.H.J.; Danesh, F.R. Identification of microRNA-93 as a novel regulator of vascular endothelial growth factor in hyperglycemic conditions. J. Biol. Chem. 2010, 285, 23457–23465. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Komers, R.; Carew, R.; Winbanks, C.E.; Xu, B.; Herman-Edelstein, M.; Koh, P.; Thomas, M.; Jandeleit-Dahm, K.; Gregorevic, P.; et al. Suppression of microRNA-29 Expression by TGF-b1Promotes Collagen Expression and Renal Fibrosis. J. Am. Soc. Nephrol. 2012, 23, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Chung, A.C.; Huang, X.R.; Meng, X.; Hui, D.S.C.; Yu, C.; Sung, J.J.Y.; Lan, H.Y. TGF-β/Smad3 Signaling Promotes Renal Fibrosis by Inhibiting miR-29. J. Am. Soc. Nephrol. 2011, 22, 1462–1474. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Wang, Y.; Danesh, F.R. MicroRNA-29c Is a Signature MicroRNA under High. Glucose Conditions That Targets Sprouty Homolog 1, and Its in Vivo Knockdown Prevents Progression of Diabetic Nephropathy. J. Biol. Chem. 2011, 286, 11837–11848. [Google Scholar] [CrossRef]
- Loeffler, I.; Liebisch, M.; Wolf, G. Collagen VIII influences epithelial phenotypic changes in experimental diabetic nephropathy. Am. J. Physiol. Physiol. 2012, 303, F733–F745. [Google Scholar] [CrossRef]
- Park, J.T.; Kato, M.; Yuan, H.; Castro, N.; Lanting, L.; Wang, M.; Natarajan, R. FOG2 protein down-regulation by transforming growth factor-β1-induced microRNA-200b/c leads to Akt kinase activation and glomerular mesangial hypertrophy related to diabetic nephropathy. J. Biol. Chem. 2013, 288, 22469–22480. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Zhang, Y.; Luo, Y.; Wang, Z.; Bi, S.; Song, D.; Dai, Y.; Wang, T.; Qiu, L.; Wen, L.; et al. Aldose reductase regulates miR-200a-3p/141-3p to coordinate Keap1-Nrf2, Tgfβ1/2, and Zeb1/2 signaling in renal mesangial cells and the renal cortex of diabetic mice. Free Radic. Biol. Med. 2014, 67, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, M.L.; Khosroheidari, M.; Eddy, E.; Kiefer, J. Role of MicroRNA 1207-5P and Its Host Gene, the Long Non-Coding RNA Pvt1, as Mediators of Extracellular Matrix Accumulation in the Kidney: Implications for Diabetic Nephropathy. PLoS ONE 2013, 8, e77468. [Google Scholar] [CrossRef]
- Dey, N.; Das, F.; Ghosh-Choudhury, N.; Mandal, C.C.; Parekh, D.J.; Block, K.; Kasinath, B.S.; Abboud, H.E.; Choudhury, G.G. microRNA-21 governs TORC1 activation in renal cancer cell proliferation and invasion. PLoS ONE 2012, 7, e37366. [Google Scholar] [CrossRef]
- Chen, S.; Feng, B.; Thomas, A.; Chakrabarti, S. miR-146a regulates glucose induced upregulation of inflammatory cytokines and extracellular matrix proteins in the retina and kidney in diabetes. PLoS ONE 2017, 12, e0173918. [Google Scholar] [CrossRef]
- Zhong, X.; Chung, A.C.; Chen, H.Y.; Dong, Y.; Meng, X.M.; Li, R.; Yang, W.; Hou, F.F.; Lan, H.Y. miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia 2013, 56, 663–674. [Google Scholar] [CrossRef]
- Bhatt, K.; Lanting, L.L.; Jia, Y.; Yadav, S.; Reddy, M.A.; Magilnick, N.; Boldin, M.; Natarajan, R. Anti-Inflammatory Role of MicroRNA-146a in the Pathogenesis of Diabetic Nephropathy. J. Am. Soc. Nephrol. 2016, 27, 2277–2288. [Google Scholar] [CrossRef] [PubMed]
- Faherty, N.; Curran, S.P.; O’Donovan, H.; Martin, F.; Godson, C.; Brazil, D.P.; Crean, J.K. CCN2/CTGF increases expression of miR-302 microRNAs, which target the TGFb type II receptor with implications for nephropathic cell phenotypes. J. Cell Sci. 2012, 125, 5621–5629. [Google Scholar] [CrossRef]
- Pappachan, J.M.; Varughese, G.I.; Sriraman, R.; Arunagirinathan, G. Diabetic cardiomyopathy: Pathophysiology, diagnostic evaluation and management. World J. Diabetes 2013, 4, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.A.; Jin, W.; Villeneuve, L.; Wang, M.; Lanting, L.; Todorov, I.; Kato, M.; Natarajan, R. Pro-inflammatory role of MicroRNA-200 in vascular smooth muscle cells from diabetic mice. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 721–729. [Google Scholar] [CrossRef]
- Kishore, R.; Verma, S.K.; Mackie, A.R.; Vaughan, E.E.; Abramova, T.V.; Aiko, I.; Krishnamurthy, P. Bone marrow progenitor cell therapy-mediated paracrine regulation of cardiac miRNA-155 modulates fibrotic response in diabetic hearts. PLoS ONE 2013, 8, e60161. [Google Scholar] [CrossRef]
- Feng, B.; Chen, S.; George, B.; Feng, Q.; Chakrabarti, S. miR133a regulates cardiomyocyte hypertrophy in diabetes. Diabetes Metab. Res. Rev. 2010, 26, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Abdellatif, M. The Role of MicroRNA-133 in Cardiac Hypertrophy Uncovered. Circ. Res. 2010, 106, 16–18. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Puthanveetil, P.; Feng, B.; Matkovich, S.J.; Dorn, G.W.; Chakrabarti, S. Cardiac miR-133a overexpression prevents early cardiac fibrosis in diabetes. J. Cell Mol. Med. 2014, 18, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Cao, Y.; Chen, S.; Chu, X.; Chu, Y.; Chakrabarti, S. miR-200b mediates endothelial to mesenchymal transition in diabetic cardiomyopathy. Diabetes 2016, 65, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, B.; Lumayag, S.; Cowan, C.; Xu, S. microRNAs in early diabetic retinopathy in streptozotocin-induced diabetic rats. Investig. Ophthalmol. Vis. Sci. 2011, 52, 4402–4409. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.-Z.; Li, E.-H.; Zhang, X.; Xiang, Z.-Y.; Li, G.-C. Effects of miRNA-200b on the development of diabetic retinopathy by targeting VEGFA gene. Biosci. Rep. 2017, 37, BSR20160572. [Google Scholar] [CrossRef]
- Mitra, R.N.; Nichols, C.A.; Guo, J.; Makkia, R.; Cooper, M.J.; Naash, M.I.; Han, Z. Nanoparticle-mediated miR200-b delivery for the treatment of diabetic retinopathy. J. Control. Release 2016, 236, 31–37. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Salta, E.; De Strooper, B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol. 2012, 11, 189–200. [Google Scholar] [CrossRef]
- Wahlestedt, C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat. Rev. Drug Discov. 2013, 12, 433–446. [Google Scholar] [CrossRef]
- Congrains, A.; Kamide, K.; Ohishi, M.; Rakugi, H. ANRIL: Molecular mechanisms and implications in human health. Int. J. Mol. Sci. 2013, 14, 1278–1292. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Dang, V.; Wang, M.; Park, J.T.; Deshpande, S.; Kadam, S.; Mardiros, A.; Zhan, Y.; Oettgen, P.; Putta, S.; et al. TGF-β induces acetylation of chromatin and of Ets-1 to alleviate repression of miR-192 in diabetic nephropathy. Sci. Signal. 2013, 6. [Google Scholar] [CrossRef]
- Hanson, R.L.; Craig, D.W.; Millis, M.P.; Yeatts, K.A.; Kobes, S.; Pearson, J.V.; Lee, A.M.; Knowler, W.C.; Nelson, R.G.; Wolford, J.K. Identification of PVT1 as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism association study. Diabetes 2007, 56, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Millis, M.P.; Bowen, D.; Kingsley, C.; Watanabe, R.M.; Wolford, J.K. Variants in the plasmacytoma variant translocation gene (PVT1) are associated with end-stage renal disease attributed to type 1 diabetes. Diabetes 2007, 56, 3027–3032. [Google Scholar] [CrossRef]
- Kato, M.; Putta, S.; Wang, M.; Yuan, H.; Lanting, L.; Nair, I.; Gunn, A.; Nakagawa, Y.; Shimano, H.; Todorov, I.; et al. TGF-β activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat. Cell Biol. 2009, 11, 881–889. [Google Scholar] [CrossRef]
- Alvarez, M.L.; DiStefano, J.K. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS ONE 2011, 6. [Google Scholar] [CrossRef]
- Hu, M.; Wang, R.; Li, X.; Fan, M.; Lin, J.; Zhen, J.; Chen, L.; Lv, Z. LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with β-catenin. J. Cell Mol. Med. 2017, 21, 2732–2747. [Google Scholar] [CrossRef]
- Gordon, A.D.; Biswas, S.; Feng, B.; Chakrabarti, S. MALAT1: A regulator of inflammatory cytokines in diabetic complications. Endocrinol. Diabetes Metab. 2018, 1, e00010. [Google Scholar] [CrossRef] [PubMed]
- Puthanveetil, P.; Chen, S.; Feng, B.; Gautam, A.; Chakrabarti, S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J. Cell Mol. Med. 2015, 19, 1418–1425. [Google Scholar] [CrossRef]
- Yi, H.; Peng, R.; Zhang, L.Y.; Sun, Y.; Peng, H.M.; Liu, H.D.; Yu, L.J.; Li, A.L.; Zhang, Y.J.; Jiang, W.H.; et al. LincRNA-Gm4419 knockdown ameliorates NF-κB/NLRP3 inflammasome-mediated inflammation in diabetic nephropathy. Cell Death Dis. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Chen, S.; Xu, J.; Zhu, Q.; Ye, X.; Ding, D.; Yao, W.; Lu, Y. Dysregulation of lncRNAs GM5524 and GM15645 involved in high-glucose-induced podocyte apoptosis and autophagy in diabetic nephropathy. Mol. Med. Rep. 2018, 18, 3657–3664. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, W.; Wang, F.; Guo, C. LncRNA-NR_033515 promotes proliferation, fibrogenesis and epithelial-to-mesenchymal transition by targeting miR-743b-5p in diabetic nephropathy. Biomed. Pharmacother. 2018, 106, 543–552. [Google Scholar] [CrossRef]
- Sun, S.F.; Tang, P.M.; Feng, M.; Xiao, J.; Huang, X.R.; Li, P.; Ma, R.C.; Lan, H.Y. Novel lncRNA Erbb4-IR promotes diabetic kidney injury in db/db mice by targeting miR-29b. Diabetes 2018, 67, 731–744. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.A.; Feng, B.; Chakrabarti, S. ANRIL regulates production of extracellular matrix proteins and vasoactive factors in diabetic complications. Am. J. Physiol. Metab. 2018, 314, E191–E200. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Chen, Z.Y.; Wang, Y.; Liu, Y.; Ma, J.X.; Li, Y.K. Long non coding RNA ASncmtRNA-2 is upregulated in diabetic kidneys and high glucose-treated mesangial cells. Exp. Ther. Med. 2017, 13, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.J.; Ding, M.; Hou, L.J.; Cui, Y.T.; Li, C.J.; Yu, D.M. Long noncoding RNA TUG1 alleviates extracellular matrix accumulation via mediating microRNA-377 targeting of PPARγ in diabetic nephropathy. Biochem. Biophys. Res. Commun. 2017, 484, 598–604. [Google Scholar] [CrossRef] [PubMed]
- Cunard, R.; Sharma, K. The endoplasmic reticulum stress response and diabetic kidney disease. Am. J. Physiol. Physiol. 2011, 300, F1054–F1061. [Google Scholar] [CrossRef]
- Kato, M.; Wang, M.; Chen, Z.; Bhatt, K.; Oh, H.J.; Lanting, L.; Deshpande, S.; Jia, Y.; Lai, J.Y.; O’Connor, C.L.; et al. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Russo, I.; Frangogiannis, N.G. Diabetes-associated cardiac fibrosis: Cellular effectors, molecular mechanisms and therapeutic opportunities. J. Mol. Cell. Cardiol. 2016, 90, 84–93. [Google Scholar] [CrossRef]
- Qu, X.; Song, X.; Yuan, W.; Shu, Y.; Wang, Y.; Zhao, X.; Gao, M.; Lu, R.; Luo, S.; Zhao, W.; et al. Expression signature of lncRNAs and their potential roles in cardiac fibrosis of post-infarct mice. Biosci. Rep. 2016, 36, e00337. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Pan, Z.; Zhao, X.; Liu, L.; Sun, J.; Su, X.; Xu, C.; Zhou, Y.; Zhao, D.; Xu, B.; et al. LncRNA PFL contributes to cardiac fibrosis by acting as a competing endogenous RNA of let-7d. Theranostics 2018, 8, 1180–1194. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Du, Y.; Shu, Y.; Gao, M.; Sun, F.; Luo, S.; Yang, T.; Zhan, L.; Yuan, Y.; Chu, W.; et al. MIAT is a Pro-fibrotic Long Non-coding RNA Governing Cardiac Fibrosis in Post-infarct Myocardium. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Micheletti, R.; Plaisance, I.; Abraham, B.J.; Sarre, A.; Ting, C.C.; Alexanian, M.; Maric, D.; Maison, D.; Nemir, M.; Young, R.A.; et al. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Piccoli, M.T.; Gupta, S.K.; Viereck, J.; Foinquinos, A.; Samolovac, S.; Kramer, F.L.; Garg, A.; Remke, J.; Zimmer, K.; Batkai, S.; et al. Inhibition of the cardiac fibroblast-enriched lncRNA Meg3 prevents cardiac fibrosis and diastolic dysfunction. Circ. Res. 2017, 121, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Bacci, L.; Barbati, S.A.; Colussi, C.; Aiello, A.; Isidori, A.M.; Grassi, C.; Pontecorvi, A.; Farsetti, A.; Gaetano, C.; Nanni, S. Sildenafil normalizes MALAT1 level in diabetic cardiomyopathy. Endocrine 2018, 62, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gu, H.; Chen, J.; Zhou, X. Involvement of long noncoding RNA MALAT1 in the pathogenesis of diabetic cardiomyopathy. Int. J. Cardiol. 2016, 202, 753–755. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Tao, Z.F.; Li, X.M.; Zhang, H.; Yao, J.; Jiang, Q. Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2014, 55, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Thomas, A.A.; Chen, S.; Aref-Eshghi, E.; Feng, B.; Gonder, J.; Sadikovic, B.; Chakrabarti, S. MALAT1: An Epigenetic Regulator of Inflammation in Diabetic Retinopathy. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Davidovich, C.; Cech, T.R. The recruitment of chromatin modifiers by long noncoding RNAs: Lessons from PRC2. RNA 2015, 21, 2007–2022. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.A.; Biswas, S.; Feng, B.; Chen, S.; Gonder, J.; Chakrabarti, S. lncRNA H19 prevents endothelial—Mesenchymal transition in diabetic retinopathy. Diabetologia 2019. [Google Scholar] [CrossRef]
- Priščáková, P.; Minárik, G.; Repiská, V. Candidate gene studies of diabetic retinopathy in human. Mol. Biol. Rep. 2016, 43, 1327–1345. [Google Scholar] [CrossRef] [PubMed]
- Miao, F.; Chen, Z.; Genuth, S.; Paterson, A.; Zhang, L.; Wu, X.; Li, S.M.; Cleary, P.; Riggs, A.; Harlan, D.M.; et al. Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes 2014, 63, 1748–1762. [Google Scholar] [CrossRef] [PubMed]
- Tewari, S.; Zhong, Q.; Santos, J.M.; Kowluru, R.A. Mitochondria DNA replication and DNA methylation in the metabolic memory associated with continued progression of diabetic retinopathy. Investig. Ophthalmol. Vis. Sci. 2012, 53, 4881–4888. [Google Scholar] [CrossRef] [PubMed]
- Pirola, L. The DCCT/EDIC study: Epigenetic clues after three decades. Diabetes 2014, 63, 1460–1462. [Google Scholar] [CrossRef]
- Yamamura, S.; Imai-Sumida, M.; Tanaka, Y.; Dahiya, R. Interaction and cross-talk between non-coding RNAs. Cell Mol. Life Sci. 2018, 75, 467–484. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, M.D.; Hatzigeorgiou, A.G. Analyzing MiRNA–LncRNA interactions. Methods Mol. Biol. 2016, 1402, 271–286. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Zhuo, Y.; Shan, B. MicroRNAs, long noncoding RNAs, and their functions in human disease. Methods Mol. Biol. 2017, 1617, 1–25. [Google Scholar] [CrossRef]
- Biswas, S.; Chakrabarti, S. Pathogenetic Mechanisms in Diabetic Retinopathy: From Molecules to Cells to Tissues. Mech. Vasc. Defects Diabetes Mellit. 2017, 209–247. [Google Scholar] [CrossRef]
- Ruiz, M.A.; Feng, B.; Chakrabarti, S. Polycomb repressive complex 2 regulates MiR-200b in retinal endothelial cells: Potential relevance in diabetic retinopathy. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Rice, J.C.; Allis, C.D. Histone methylation versus histone acetylation: New insights into epigenetic regulation. Curr. Opin. Cell Biol. 2001, 13, 263–273. [Google Scholar] [CrossRef]
- Martin, C.; Zhang, Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell. Biol. 2005, 6, 838–849. [Google Scholar] [CrossRef]
- Villeneuve, L.M.; Natarajan, R. The role of epigenetics in the pathology of diabetic complications. AJP Ren. Physiol. 2010, 299, F14–F25. [Google Scholar] [CrossRef] [PubMed]
- Forneris, F.; Binda, C.; Vanoni, M.A.; Battaglioli, E.; Mattevi, A. Human histone demethylase LSD1 reads the histone code. J. Biol. Chem. 2005, 280, 41360–41365. [Google Scholar] [CrossRef] [PubMed]
- O’Meara, M.M.; Simon, J.A. Inner workings and regulatory inputs that control Polycomb repressive complex 2. Chromosoma 2012, 121, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L. LncRNAs: Linking RNA to chromatin. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Zardo, G.; Ciolfi, A.; Vian, L.; Billi, M.; Racanicchi, S.; Grignani, F.; Nervi, C. Transcriptional targeting by microRNA-Polycomb complexes. Cell Cycle 2012, 11, 3543–3549. [Google Scholar] [CrossRef]
- Shang, F.F.; Luo, S.; Liang, X.; Xia, Y. Alterations of circular RNAs in hyperglycemic human endothelial cells. Biochem. Biophys. Res. Commun. 2018, 499, 551–555. [Google Scholar] [CrossRef]
- Henaoui, I.S.; Jacovetti, C.; Mollet, I.G.; Guay, C.; Sobel, J.; Eliasson, L.; Regazzi, R. PIWI-interacting RNAs as novel regulators of pancreatic beta cell function. Diabetologia 2017, 60, 1977–1986. [Google Scholar] [CrossRef]
- Lee, J.; Harris, A.N.; Holley, C.L.; Mahadevan, J.; Pyles, K.D.; Lavagnino, Z.; Scherrer, D.E.; Fujiwara, H.; Sidhu, R.; Zhang, J.; et al. Rpl13a small nucleolar RNAs regulate systemic glucose metabolism. J. Clin. Investig. 2016, 126, 4616–4625. [Google Scholar] [CrossRef]
| miRNA | Cell/Tissue Type(s) | Reported Function(s) | Disease Model(s) |
|---|---|---|---|
| miR-192 * | • Mouse mesangial cells • Human proximal tubule cells and kidneys • Rat tubular epithelial cells • Human podocytes | • Elevated expressions associated with increased Col1a2 expressions; targets SIP1 [108] • Loss of miR-192 expression is associated with increased fibrosis and decreased estimated GFR [111] • Decreased in the diabetic kidney, targets ZEB2, and does not affect extracellular matrix (ECM) protein expressions [112] • Increased in high glucose and diabetic conditions [109,110,142] • Can regulate other transcription factors and miRNAs [109,110] | • DN [108,109,110,111,112,142] |
| miR-215 | • Proximal tubular cells • Rat mesangial cells • Human podocytes | • Decreased in the diabetic kidney • Ectopic expression of miR-215 increases E-cadherin levels by repressing ZEB2 translation | • DN [112] |
| miR-377 | • Human and mouse mesangial cells • Mice kidney tissues | • Up-regulated in hyperglycemic and diabetic conditions • Can indirectly lead to increased fibronectin protein production | • DN [113] |
| miR-21 | • Rat mesangial cells • Mice kidney tissues • Rat tubular epithelial cells | • Expression is increased in DN and can enhance the production of high glucose-induced fibrotic and inflammatory markers | • DN [125] |
| miR-29* | • Human podocytes • Mouse mesangial cells • Mice kidney tissues • Mouse embryonic fibroblasts and tubular epithelial cells • Mice kidney glomeruli, endothelial cells, and podocytes | • Low levels in early DN and fibrosis and can target collagens I and IV [116] • Lost with progressive renal fibrosis, can reduce collagens I and III, and interact with Smad3 [117] • miR-29c is increased in DN, induces cell apoptosis, and increases ECM protein accumulation [118] | • DN [116,117,118] |
| miR-let-7b | • Rat proximal tubular epithelial cells • Mice kidney tissues | • Is reduced in both diabetic and non-diabetic renal fibrosis and can regulate the expression of several ECM genes | • DN [114] |
| miR-93 | • Renal microvascular endothelial cells • Mouse podocytes • Mice kidney tissues | • High glucose and diabetic conditions decrease miR-93 expressions • Can target VEGF and negatively regulate it | • DN [115] |
| miR-200 * | • Human retinal endothelial cells • Mice and rat retinal tissues • Mice kidney tissues • Mouse mesangial cells • Mouse heart endothelial cells, vascular smooth muscle cells, and cardiac tissues | • miR-200b is reduced under hyperglycemic and diabetic conditions [40,179]; can target VEGF [80,179] • Inhibition of miR-200a-3p can provoke renal fibrosis in DN [121] • Increased levels of miR-200b/c detected in diabetic mouse glomeruli; involved in glomerular mesangial hypertrophy [110,120] • miR-200b overexpression shown to prevent diabetes-induced changes in heart structure and function and reduce EndMT markers [134] • miR-200b shown to have a protective role in the diabetic retina [136,137] • Diabetic VSMCs exhibit increased miR-200 levels, which can contribute to inflammation [129] | • DR [40,80,136,137] • DN [110,120,121,179] • DCM [129,134] |
| miR-146a | • Rat and mice retinal tissues • Human umbilical vein endothelial cells • Mice kidney tissues | • miR-146a was shown to be reduced in diabetic tissues; miR-146a mimics can decrease FN expression [55] • miR-146a knockout exacerbates diabetes-induced inflammation and fibrosis in mice kidney tissues [126] • miR-146a mimics can prevent the increased expressions of ECM proteins and inflammatory markers in diabetic tissues [124] | • DR [55] • DCM [55] • DN [124,126] |
| miR-1207-5b | • Human renal proximal tubule epithelial cells, podocytes, and mesangial cells | • Hyperglycemia shown to increase miR-1207-5b levels, which contributes to ECM accumulation in the kidney • Knockdown can decrease levels of TGF-β1, FN1, and PAI-1 | • DN [122] |
| miR-302d | • Mice kidney tissues • Human mesangial cells, proximal tubular epithelial cells, and HEK-293T | • Capable of attenuating TGF-β-induced fibronectin, thrombospondin, vimentin, and N-cadherin expressions • Can regulate TGF-β -induced EMT | • DN [127] |
| miR-216a | • Primary mouse mesangial cells (MMCs) | • Upregulated by TGF-β in MMCs • Also increased in isolated renal glomeruli from type 1 and type 2 diabetic mice • Inhibiting miR-216a in MMCs reverses the effects of TGF-β on Pten and P-Akt levels | • DN [145] |
| miR-217 | • Primary mouse mesangial cells | • Upregulated by TGF-β in MMCs • Also increased in diabetic mice kidneys • Along with miR-216a, miR-217 mimics can induce hypertrophy in MMCs | • DN [145] |
| miR-133a | • Mice cardiac tissues • Neonatal rat myocytes | • Downregulated in diabetic cardiomyopathy [131,132,133] • Mediates glucose-induced cardiomyocytes hypertrophy [131,132,133] • Cardiac-specific overexpression of miR-133a can significantly decrease cardiac fibrosis [133] | • DCM [131,132,133] |
| miR-155 | • Mouse cardiac fibroblasts • Mouse bone marrow progenitor cells (BMPCs) | • Increased expression of miR-155 in MI mice • Transplantation of BMPCs in MI mice can decrease miR-155 expressions and in association, show decreased cardiac fibrosis expressions | • DCM [130] |
| lncRNA | Cell/Tissue Type(s) | Reported Function(s) | Disease Model |
|---|---|---|---|
| PVT1 | • Humanmesangial cells | • Upregulated by glucose treatment in mesangial cells • PVT1 knockdown can significantly reduce the levels of major ECM proteins (FN and COL4A1) | • DN [146] |
| MALAT1 | • Mice kidney tissues • Mouse podocytes • Mice and rat cardiac tissues • HRECs • Mice retinal tissues • RF/6A cells • Aqueous and vitreous humors | • MALAT1 levels are increased in the kidney cortices of STZ-induced diabetic mice [147] • MALAT1 regulates diabetes-induced inflammatory gene expressions in the heart and kidneys [148,149] • MALAT1 is upregulated in HG-treated RF/6A cells, aqueous humor samples and in fibrovascular membranes of diabetic patients [167] • MALAT1 has a pathogenetic role in the heart [165,166] • MALAT1 can regulate inflammation through its association with other epigenetic mechanisms in DR [168] | • DN [147,148,149] • DR [167,168] • DCM [148,165,166] |
| GM4419 | • Mouse mesangial cells (MMCs) | • Upregulated in MMCs following high glucose culture • Knockdown of GM4419 inhibits the glucose-induced expressions of pro-inflammatory cytokines and renal fibrosis markers • GM4419 can regulate NF-κB signalling | • DN [150] |
| GM5524 | • Mice kidney tissues • Mouse podocytes | • Expressions are significantly upregulated in DN • May regulate podocytes apoptosis and autophagy during DN | • DN [151] |
| GM15645 | • Mice kidney tissues • Mouse podocytes | • Downregulated in DN • Similar to GM5524, GM15645 may also regulate podocytes apoptosis and autophagy during DN | • DN [151] |
| ANRIL | • Human retinal endothelial cells (HRECs) • Mice retinal tissues • Mice kidney and cardiac tissues | • High glucose and diabetic conditions upregulate ANRIL expressions [81,154] • ANRIL can regulate VEGF and ECM expressions through several epigenetic mechanisms (i.e., p300 and PRC2) [81,154] | • DN [154] • DR [81] • DCM [154] |
| NR_033515 | • Human blood • HEK293-T • MMCs | • Significantly increased in the serum of DN patients • Overexpression of NR_033515 can accelerate TGF-β1-induced EMT • Promotes cell proliferation and fibrogenesis in high glucose conditions | • DN [152] |
| Erbb4-IR | • Mice kidney tissues • Mouse embryonic fibroblasts • MMCs • Mouse tubular epithelial cells | • Significantly upregulated in the kidneys of diabetic mice • A Smad3-dependent lncRNA that promotes renal fibrosis in type 2 DN • Can negatively regulate miR-29b • Kidney-specific silencing of Erbb4-IR shown to prevent renal injury in diabetic mice | • DN [153] |
| ASncmtRNA-2 | • Mice kidney tissues • Human mesangial cells | • Expressions are significantly heightened in diabetic mice kidneys and mesangial cells treated with high glucose • May promote glomerular fibrosis in DN | • DN [155] |
| Tug1 | • Mice kidney tissues • MMCs | • Suppresses the proliferation of mesangial cells and decreases the expression of ECM-associated proteins in DN • Functions as an endogenous sponge of miR-377, which directly targets PPARγ | • DN [156] |
| NONMMUT022554 | • Mice cardiac tissues | • Upregulated in cardiac fibrosis and positively correlated with 6 upregulated genes involved in ECM–receptor interactions and the PI3K–Akt signalling pathway | • Cardiac fibrosis/MI [160] |
| PFL (NONMMUT022555) | • Mice cardiac tissues • Mice cardiac fibroblasts and cardiomyocytes | • Upregulated in the hearts of MI mice • Knockdown of PFL can attenuate cardiac interstitial fibrosis and improve cardiac function • Overexpression of PFL promotes proliferation, fibroblast-myofibroblast transition, and mice cardiac fibroblasts • Acts as a competitive endogenous RNA of let-7d | • Cardiac fibrosis/MI [161] |
| MIAT | • Mice cardiac tissues • Mouse cardiac fibroblasts | • Significantly upregulated in the infarcted myocardium of mice • Knockdown of MIAT reduces cardiac fibrosis and improves cardiac function • Functions as a sponge of miR-24 in cardiac fibroblasts | • Cardiac fibrosis/MI [162] |
| Wisper | • Mice and human cardiac tissues • Mouse cardiac fibroblasts and cardiomyocytes • Human fibroblasts | • Expression of Wisper is strongly correlated with cardiac fibrosis in both animal and human heart tissues • Wisper knockdown affects cardiac fibroblast survival, migration, and proliferation • In vivo depletion of Wisper inhibits cardiac fibrosis and improves function | • Cardiac fibrosis/MI [163] |
| Meg3 | • Mice cardiac tissues • Mouse cardiac fibroblasts | • Strongly expressed in adult cardiac fibroblasts • Regulates the production of MMP-2 in vitro • In vivo inhibition of Meg3 after transverse aortic constriction decreases cardiac fibrosis and improves diastolic function | • Cardiac fibrosis [164] |
| H19 | • HRECs • Mice retinal tissues • Human vitreous humors | • Downregulated in HG-treated endothelial cells and in the vitreous humors of diabetic patients • Capable of regulating EndMT in vitro and in vivo | • DR [170] |
| Lnc-MGC | • Mice kidney tissues • Mouse and human mesangial cells • Human renal biopsies | • Elevated levels of lnc-MGC present in the kidneys during diabetes • Host to a megacluster of miRNAs • CHOP (an ER-stress transcription factor) regulates lnc-MGC expressions • Inhibition of lnc-MGC results in reduced cluster miRNAs, ECM accumulation, and glomerular hypertrophy | • DN [158] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biswas, S.; Chakrabarti, S. Increased Extracellular Matrix Protein Production in Chronic Diabetic Complications: Implications of Non-Coding RNAs. Non-Coding RNA 2019, 5, 30. https://doi.org/10.3390/ncrna5010030
Biswas S, Chakrabarti S. Increased Extracellular Matrix Protein Production in Chronic Diabetic Complications: Implications of Non-Coding RNAs. Non-Coding RNA. 2019; 5(1):30. https://doi.org/10.3390/ncrna5010030
Chicago/Turabian StyleBiswas, Saumik, and Subrata Chakrabarti. 2019. "Increased Extracellular Matrix Protein Production in Chronic Diabetic Complications: Implications of Non-Coding RNAs" Non-Coding RNA 5, no. 1: 30. https://doi.org/10.3390/ncrna5010030
APA StyleBiswas, S., & Chakrabarti, S. (2019). Increased Extracellular Matrix Protein Production in Chronic Diabetic Complications: Implications of Non-Coding RNAs. Non-Coding RNA, 5(1), 30. https://doi.org/10.3390/ncrna5010030





