Catalyzing Transcriptomics Research in Cardiovascular Disease: The CardioRNA COST Action CA17129
Abstract
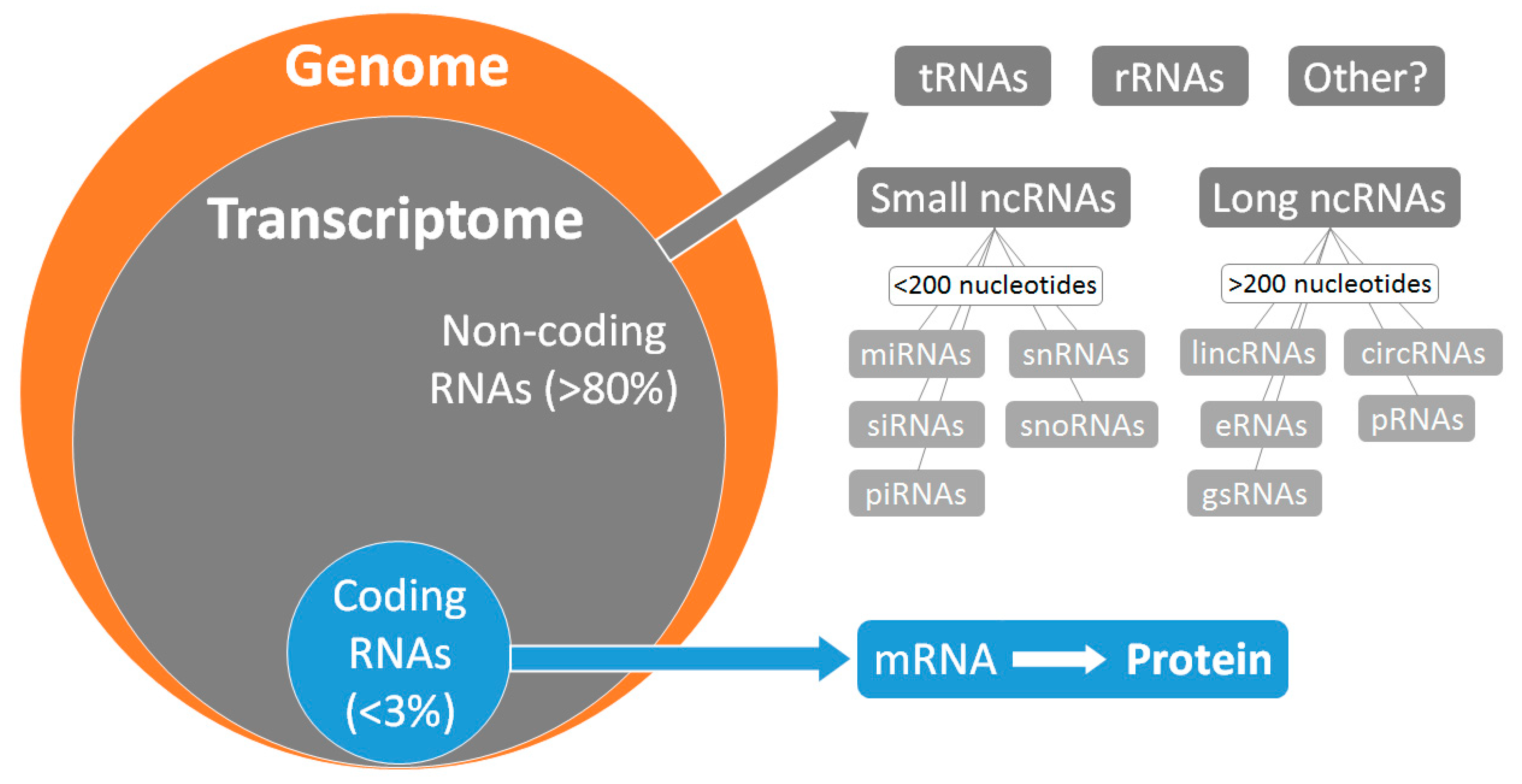
:1. Introduction
2. Relevance and Timeliness
3. Objectives
4. Progress Beyond the State-of-the-Art and Innovation Potential
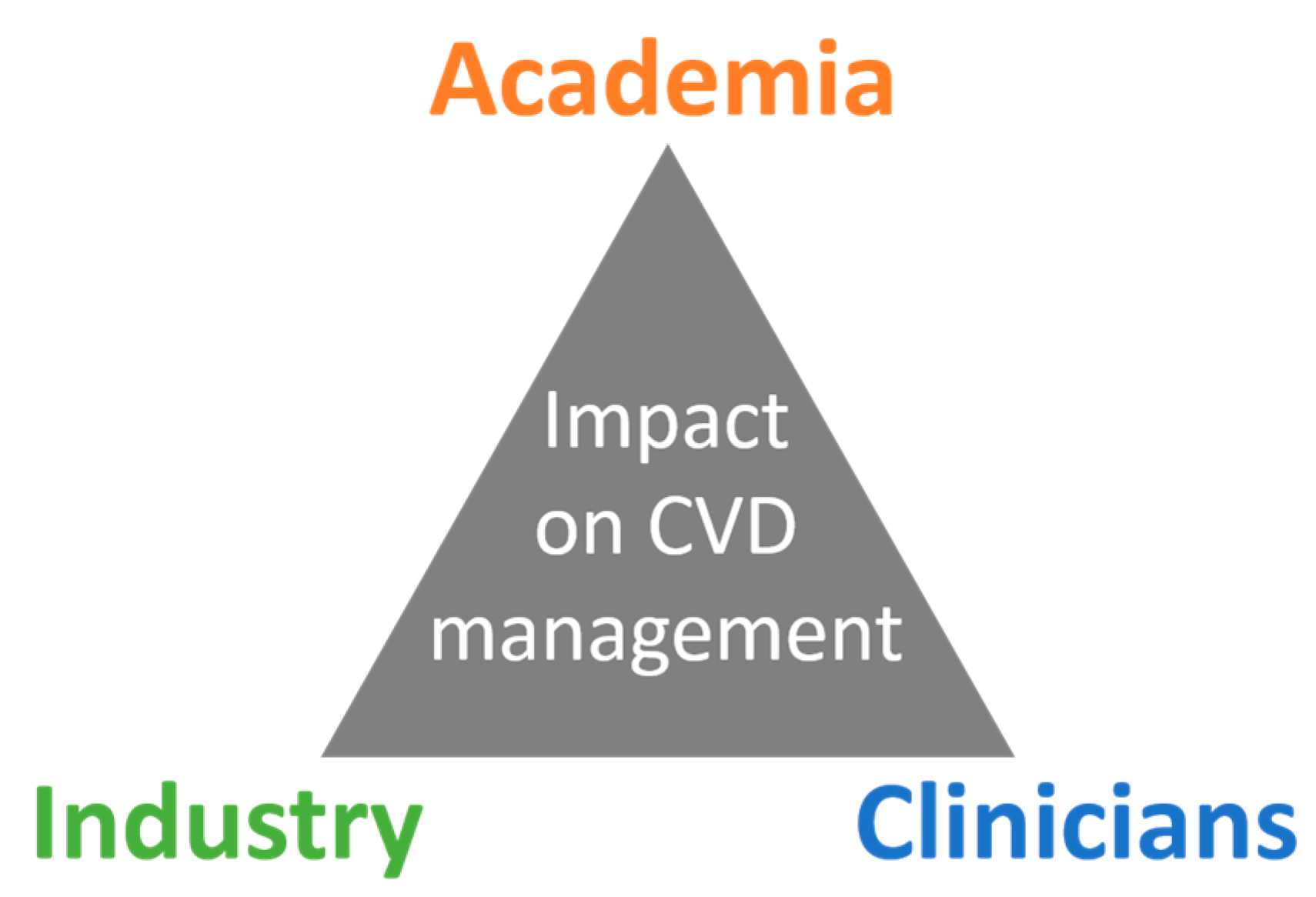
5. Added Value of Networking
6. Expected Impact
- Research collaboration and joint grant applications for future national and European funding;
- Increase the competitiveness of the network participants for European funding and the impact of resulting research;
- Encourage translational research due to the participation of industrial partners in the network;
- Stimulate innovative perspectives to study the transcriptome due to CardioRNA’s multidisciplinary team;
- Provide opportunities and scientific training for ECI and students through workshops, training schools, and STSMs.
- Expand fundamental knowledge about the role of the transcriptome in CVD to improve its prognostics, diagnostics, and treatment;
- Increase the quality of data generated and reproducibility of studies of the CVD transcriptome, thus raising the quality and impact of publications;
- Establish a leadership of both the network members and the European research axis in the proposed research field;
- Foster personalized medicine and consequently healthcare, through outcomes of public–private collaborations.
7. Potential for Innovation
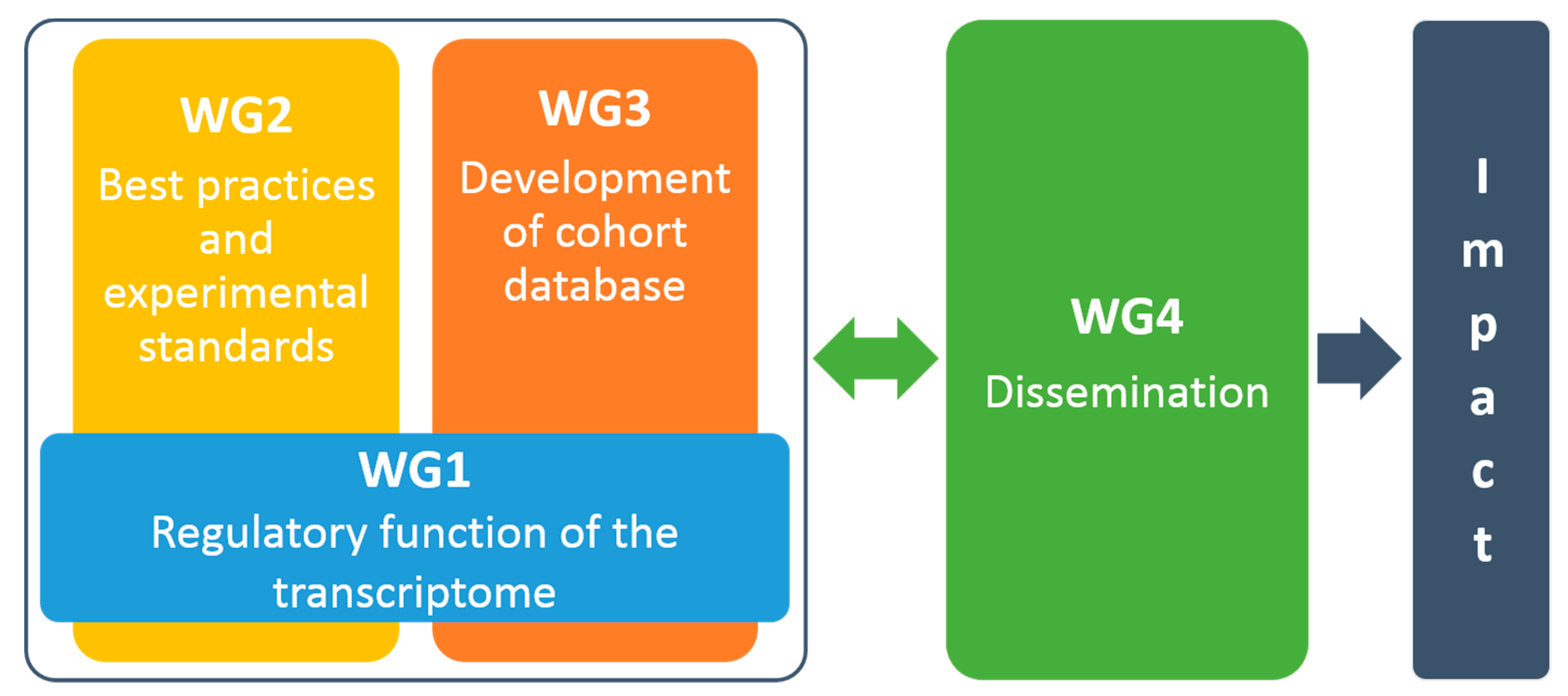
8. Implementation Plan
8.1. WG1: Regulatory Function of the Transcriptome
8.2. WG2: Best Practices and Experimental Standards
8.3. WG3: Development of Cohort Inventory
8.4. WG4: Dissemination
9. Concluding Remarks
Funding
Conflicts of Interest
References
- Mendis, S.; Puska, P.; Norrving, B.; World Health Organization; World Heart Federation; World Stroke Organization. Global Atlas on Cardiovascular Disease Prevention and Control; World Health Organization in collaboration with the World Heart Federation and the World Stroke Organization: Geneva, Switzerland, 2011; p. vi. 155p. [Google Scholar]
- Wilkins, E.; Wilson, L.; Wickramasinghe, K.; Bhatnagar, P.; Leal, J.; Luengo-Fernandez, R.; Burns, R.; Rayner, M.; Townsend, N. European Cardiovascular Disease Statistics 2017; European Heart Network: Brussels, Belgium; European Society of Cardiology: Sophia Antipolis, France, 2017. [Google Scholar]
- Medicine, I.O. Promoting Cardiovascular Health in the Developing World: A Critical Challenge to Achieve Global Health; The National Academies Press: Washington, DC, USA, 2010; p. 482. [Google Scholar] [CrossRef]
- Booth, F.W.; Gordon, S.E.; Carlson, C.J.; Hamilton, M.T. Waging war on modern chronic diseases: Primary prevention through exercise biology. J. Appl. Physiol. 2000, 88, 774–787. [Google Scholar] [CrossRef]
- Kearney, P.M.; Whelton, M.; Reynolds, K.; Muntner, P.; Whelton, P.K.; He, J. Global burden of hypertension: Analysis of worldwide data. Lancet 2005, 365, 217–223. [Google Scholar] [CrossRef]
- Jacquier, A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat. Rev. Genet. 2009, 10, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Papageorgiou, N.; Tslamandris, S.; Giolis, A.; Tousoulis, D. MicroRNAs in Cardiovascular Disease: Perspectives and Reality. Cardiol. Rev. 2016, 24, 110–118. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—The mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef]
- Devaux, Y. Transcriptome of blood cells as a reservoir of cardiovascular biomarkers. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 209–216. [Google Scholar] [CrossRef]
- Goretti, E.; Wagner, D.R.; Devaux, Y. miRNAs as biomarkers of myocardial infarction: A step forward towards personalized medicine? Trends Mol. Med. 2014, 20, 716–725. [Google Scholar] [CrossRef] [PubMed]
- Antman, E.M.; Loscalzo, J. Precision medicine in cardiology. Nat. Rev. Cardiol. 2016, 13, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Duffy, D.J. Problems, challenges and promises: Perspectives on precision medicine. Brief. Bioinform. 2016, 17, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.A.; Moore, J.H.; Ritchie, M.D. Embracing Complex Associations in Common Traits: Critical Considerations for Precision Medicine. Trends Genet. 2016, 32, 470–484. [Google Scholar] [CrossRef] [PubMed]
- Byron, S.A.; Van Keuren-Jensen, K.R.; Engelthaler, D.M.; Carpten, J.D.; Craig, D.W. Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nat. Rev. Genet. 2016, 17, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Devaux, Y.; Zangrando, J.; Schroen, B.; Creemers, E.E.; Pedrazzini, T.; Chang, C.P.; Dorn, G.W., 2nd; Thum, T.; Heymans, S.; Cardiolinc, N. Long noncoding RNAs in cardiac development and ageing. Nat. Rev. Cardiol. 2015, 12, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Sullenger, B.A.; Nair, S. From the RNA world to the clinic. Science 2016, 352, 1417–1420. [Google Scholar] [CrossRef]
- Pedrotty, D.M.; Morley, M.P.; Cappola, T.P. Transcriptomic biomarkers of cardiovascular disease. Prog. Cardiovasc. Dis. 2012, 55, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Boon, R.A.; Jae, N.; Holdt, L.; Dimmeler, S. Long Noncoding RNAs: From Clinical Genetics to Therapeutic Targets? J. Am. Coll. Cardiol. 2016, 67, 1214–1226. [Google Scholar] [CrossRef] [PubMed]
- Schnabel, R.B.; Baccarelli, A.; Lin, H.; Ellinor, P.T.; Benjamin, E.J. Next steps in cardiovascular disease genomic research--sequencing, epigenetics, and transcriptomics. Clin. Chem. 2012, 58, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Gurha, P.; Marian, A.J. Noncoding RNAs in cardiovascular biology and disease. Circ. Res. 2013, 113, e115–e120. [Google Scholar] [CrossRef] [PubMed]
- Philippen, L.E.; Dirkx, E.; da Costa-Martins, P.A.; De Windt, L.J. Non-coding RNA in control of gene regulatory programs in cardiac development and disease. J. Mol. Cell Cardiol. 2015, 89, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.B.; Katwadi, K.; Kwek, X.Y.; Ismail, N.I.; Chinda, K.; Ong, S.G.; Hausenloy, D.J. Non-coding RNAs as therapeutic targets for preventing myocardial ischemia-reperfusion injury. Expert Opin. Therap. Targets 2018, 22, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Ozsolak, F.; Milos, P.M. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Wang, K.; Yuan, Y.; Cho, J.H.; McClarty, S.; Baxter, D.; Galas, D.J. Comparing the MicroRNA spectrum between serum and plasma. PLoS ONE 2012, 7, e41561. [Google Scholar] [CrossRef]
| Specific | Measurable | Achievable | Relevant | Timely |
|---|---|---|---|---|
| Further the understanding of the transcriptome’s role in CVD by establishing a network | Number of institutions and investigators joining the network | Active and increasing collaborative research activity in the field as seen in the biomedical literature | Increased knowledge on the subject has noteworthy implications for the healthcare of patients with CVD | Expansion of the network will be continuous (month 1–48) |
| Foster collaborative initiatives in CVD transcriptomics | Number of Action outputs such as projects submitted for funding and scientific publications | Collaborations will allow optimization and standardization of protocols, thus helping to deal with the complexity of studies | Join expertise to aid European research groups improve and consolidate their research capacity and leadership in this field | Project application to different funding bodies per year (month 12–48) |
| Develop improved guidelines for best practices and experimental standards that offer the greatest potential for cardiovascular transcriptomics studies | Production of specific documents (peer-reviewed publications and others) | Different guidelines and techniques are successfully used in this field of research | Increasing standardization will facilitate comparison and reproducibility of results, allowing faster interpretation of results and development of new tools to the clinic | Publish documents on (1) best practices on collection and processing of biological material (month 24); (2) experimental standards for RNA analysis in CVD (month 40) |
| Stimulate development and optimization of RNA-based products for prognostic, diagnostic and therapy for CVD management | Number of projects that target translational research and personalized medicine | Partnerships from the network will facilitate study designs on the translation of research knowledge into medicinal products | Regards healthcare improvement of patients with CVD | Projects on clinical products development (month 48) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomes, C.P.d.C.; Ágg, B.; Andova, A.; Arslan, S.; Baker, A.; Barteková, M.; Beis, D.; Betsou, F.; Bezzina Wettinger, S.; Bugarski, B.; et al. Catalyzing Transcriptomics Research in Cardiovascular Disease: The CardioRNA COST Action CA17129. Non-Coding RNA 2019, 5, 31. https://doi.org/10.3390/ncrna5020031
Gomes CPdC, Ágg B, Andova A, Arslan S, Baker A, Barteková M, Beis D, Betsou F, Bezzina Wettinger S, Bugarski B, et al. Catalyzing Transcriptomics Research in Cardiovascular Disease: The CardioRNA COST Action CA17129. Non-Coding RNA. 2019; 5(2):31. https://doi.org/10.3390/ncrna5020031
Chicago/Turabian StyleGomes, Clarissa Pedrosa da Costa, Bence Ágg, Andrejaana Andova, Serdal Arslan, Andrew Baker, Monika Barteková, Dimitris Beis, Fay Betsou, Stephanie Bezzina Wettinger, Branko Bugarski, and et al. 2019. "Catalyzing Transcriptomics Research in Cardiovascular Disease: The CardioRNA COST Action CA17129" Non-Coding RNA 5, no. 2: 31. https://doi.org/10.3390/ncrna5020031
APA StyleGomes, C. P. d. C., Ágg, B., Andova, A., Arslan, S., Baker, A., Barteková, M., Beis, D., Betsou, F., Bezzina Wettinger, S., Bugarski, B., Condorelli, G., da Silva, G. J. J., Danilin, S., de Gonzalo-Calvo, D., Buil, A., Carmo-Fonseca, M., Enguita, F. J., Felekkis, K., Ferdinandy, P., ... Devaux, Y. (2019). Catalyzing Transcriptomics Research in Cardiovascular Disease: The CardioRNA COST Action CA17129. Non-Coding RNA, 5(2), 31. https://doi.org/10.3390/ncrna5020031






















