Abstract
As part of their innate immune response against viral infections, mammals activate the expression of type I interferons to prevent viral replication and dissemination. An antiviral RNAi-based response can be also activated in mammals, suggesting that several mechanisms can co-occur in the same cell and that these pathways must interact to enable the best antiviral response. Here, we will review how the classical type I interferon response and the recently described antiviral RNAi pathways interact in mammalian cells. Specifically, we will uncover how the small RNA biogenesis pathway, composed by the nucleases Drosha and Dicer can act as direct antiviral factors, and how the type-I interferon response regulates the function of these. We will also describe how the factors involved in small RNA biogenesis and specific small RNAs impact the activation of the type I interferon response and antiviral activity. With this, we aim to expose the complex and intricate network of interactions between the different antiviral pathways in mammals.
1. Introduction
1.1. The Mammalian Type-I IFN Response
Interferons (IFNs) are the major cytokines expressed during the innate immune response against invading pathogens, such as viruses, bacteria, fungi, and parasites. Besides their role in restricting infections, IFNs also exhibit immunomodulatory functions, and have been implicated in both cancer immunosurveillance and autoimmunity [1,2,3]. IFNs can be produced by virtually all nucleated cells of jawed vertebrates and are classified into three major types: Type I, Type II, and Type III IFNs [4,5]. Type I IFNs are the most diverse family with 8 known subtypes, of which IFN-α and IFN-β can be expressed by nearly every cell type [6,7]. Type II IFN has only one member, IFN-γ, and is mainly expressed by activated natural killer (NK) and T-cells [8,9,10]. Type III IFNs, IFN-λs, have only been recently discovered and are also expressed in multiple cell types [11,12,13,14]. In this review we will focus on the roles of type I IFNs as they are crucial, ubiquitously expressed components of the antiviral response in mammals.
In the context of viral infections, the type I IFN response is activated by sensing the presence of invading viruses. These pathogens pose a particular challenge for detection by the innate immune system due to their small size and constantly evolving surface protein repertoire, therefore, host cells have developed the ability to recognize virus-specific nucleic acid signatures. These sensing mechanisms rely on host proteins termed pattern recognition receptors (PRRs) recognizing specific pathogen features, known as pathogen-associated molecular patterns (PAMPs). During viral infections, PRRs need to appropriately discriminate between non-self, viral-derived nucleic acids and self-derived nucleic acids (Table 1) [15,16]. The binding and recognition of virus-specific nucleic acids by PRRs is necessary for the production and secretion of type I IFNs and pro-inflammatory cytokines. Secreted type I IFN proteins act in an auto- and paracrine fashion by binding to the heterodimeric type I IFN receptors, IFNAR1 and 2, on the surface of the infected and neighboring cells. This initiates the janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathway that activates a second transcriptional response of around 500 IFN-stimulated genes (ISGs), which establishes the antiviral state [17,18]. These processes constitute the first steps of the innate immune response to infections prior to activation of the adaptive immune system. Here we focus on the mechanisms that cells use to detect invasion by viruses.

Table 1.
Pattern recognition receptors (PRRs) implicated in nucleic acid sensing. Different classes of PRRs operate in distinct cellular compartments and recognize ligands that are absent or rare in the host.
1.2. Viral-Derived Nucleic Acids Sensing Mechanisms
1.2.1. TLR-Detection of Viral Nucleic Acids
Toll-like receptors (TLRs) are transmembrane proteins that are located on the cell surface and endosomes. The expression of TLRs is cell specific and each is specialized in the recognition of specific viral-derived PAMPs [19,20]. With the exception of TLR3, which is specialized in dsRNA recognition [21], TLR signaling depends on the adaptor protein myeloid differentiation primary response 88 (MyD88) [20]. MyD88 activates the MAPK pathway, NFκB, and IRF7 transcription factors to induce expression of type I IFNs and pro-inflammatory cytokines [22,23]. TLR3, on the other hand, signals through TRIF to activate the MAPK, NFκB, and IRF3 pathways to induce the expression of similar cytokines [24].
1.2.2. RLR-Detection of Virus-Derived RNA
The presence of cytoplasmic virus-derived RNA is detected by a family of receptors called RIG-I-like receptors (RLRs), which include retinoic-acid-inducible protein 1 (RIG-I), melanoma-differentiation-associated gene 5 (MDA5), laboratory of genetics physiology 2 (LGP2), and signals through the mitochondrial antiviral-signaling protein (MAVS). Activation of RLR-signaling by recognition of virus-derived RNAs also results in type I IFNs and proinflammatory cytokine expression and this, in turn, increases expression of RLRs since these are also ISGs [25,26].
RIG-I and MDA5 are structurally similar, consisting of a central DExD/H RNA helicase domain, two tandem caspase activation and recruitment domains (CARD) at the N-terminal end and a regulatory C-terminal regulatory domain [25]. RIG-I and MDA5 bind a complementary set of viral RNA ligands: MDA5 binds long dsRNA, whereas RIG-I binds 5′tri- and diphosphate short dsRNA [27,28,29]. Binding of ligands occurs at the basic cleft in their C-terminal domains (CTD) which leads to a conformational change to expose the occluded CARD domain [30,31]. This causes RIG-I to form tetramers and short filaments and MDA5 to oligomerize and form long filaments along the length of the dsRNA [32,33]. After filament formation, the CARD domains of both MDA5 and RIG-I interact with the CARD domain of MAVS to promote further signaling [34,35]. MAVS activation causes NF-kB, IRF3, and IRF7 transcription factors to translocate to the nucleus and initiate type I IFNs and pro-inflammatory cytokines expression [36,37] (Figure 1). LGP2, which lacks a CARD domain, seems to regulate both RIG-I and MDA5 activity. LGP2 can enhance the rate of nucleation and consequent filament formation by MDA5 [38,39], whereas it inhibits RIG-I-mediated signaling by competing for the same RNA substrates [40]. Thus, the third RLR can act both as a positive or negative factor for antiviral signaling [41,42,43].
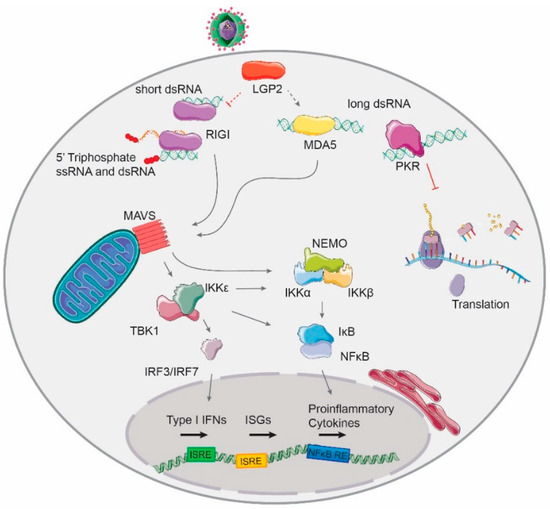
Figure 1.
The typical hallmark of viral replication, dsRNA, is recognized by the RLR family of receptors in the cytoplasm of infected cells. RIG-I recognizes short dsRNA molecules with 5′tri- and diphosphates; MDA5 recognizes long dsRNA molecules. Both MDA5 and RIG-I signal through the mitochondrial-associated factor MAVS to activate the transcription of type I interferons (IFNs) and pro-inflammatory cytokines. The third member of the RLR family, LGP2, modulates the activation of RIG-I and MDA5-mediated signaling pathways. The presence of dsRNA activates the host translational shut-off response through phosphorylation of the translation factor eIF2α by the kinase PKR.
1.2.3. Other Antiviral dsRNA-Activated Pathways
PKR. Viral-derived dsRNA can also be sensed in the cytoplasm by the dsRNA-binding protein kinase PKR [44]. Upon binding to dsRNA, PKR phosphorylates eIF2α at Ser51 causing the sequestration of the guanine nucleotide exchange factor eIF2B which results in cap-dependent translation inhibition [45,46,47]. This process is also known as the host translational shutoff response (Figure 1).
OAS/RNaseL. The presence of cytoplasmic virus-derived dsRNA also activates the OAS/RNase L degradation pathway. The OAS (oligo-adenylate synthase) proteins synthesize 2′,5′-linked adenylates upon binding to dsRNA [48]. These oligomers activate the endoribonuclease RNAse L to cleave ssRNA (single-stranded) in a non-sequence specific manner, preventing viral replication [49,50,51]. RNAse L amplifies IFN signaling further through the production of small RNA cleavage products that can activate RIG-I and MDA5-mediated responses [52].
ADAR. Specific isoforms of the dsRNA binding proteins of the ADAR family have also been implicated in the regulation of the IFN response and are ISGs. ADAR1 and 2 have been shown to prevent activation of the Interferon response by guiding deamination of adenosines to inosines on endogenous dsRNAs. Unlike unmodified dsRNAs, the presence of inosine residues prevents activation of the innate immune response, while still being bound to RLRs. Consequently, the absence of deaminase activity inhibits the cell’s ability to discriminate “self” from “non-self” and results in the undesired activation of a type I IFN response [53,54]. Besides its importance in regulating the IFN response, ADAR proteins have been shown to directly target viral RNA, resulting in pro- and antiviral effects [55].
Additional dsRNA binding factors, such as Drosha and Dicer, classically involved in the biogenesis of small RNAs, have been recently found to provide alternative antiviral activity independent of the IFN response, which will be discussed below.
1.3. Mammalian Small RNA Biogenesis
Mammalian endogenous small RNAs (20–30 nt long) can be divided into three different categories depending on their biogenesis pathway: micro (mi)RNAs, short-interfering (si)RNAs, and PIWI-interacting (pi)RNAs [56]. mi- and siRNAs associate with Argonaute (Ago) proteins to guide post-transcriptional regulation of target mRNAs expression, whereas piRNAs associate with a subfamily of Ago, PIWI proteins, to target transposable elements in the germline [57].
miRNAs are transcribed in the nucleus by RNA-polymerase II as long precursor molecules (pri-miRNAs) that can be organized in clusters, indicating that a single transcript is sufficient to produce different miRNAs [58,59]. Pri-miRNAs adopt stem-loop structures that are recognized and cleaved by the nuclear microprocessor complex, which consists of the RNAse III endonuclease Drosha associated with two copies of the dsRNA binding protein DiGeorge syndrome chromosomal region 8 (DGCR8) [60,61,62,63,64,65] (Figure 2). DGCR8, interacting with the apical part of the pri-miRNA hairpin, guides Drosha binding and cleavage at the base of the hairpin, releasing a ~60–70 nt hairpin with a 2-nucleotide 3′ overhang [62,65,66,67]. Besides the typical RNA secondary structure of pri-miRNAs, specific sequence motifs determine successful processing. Efficiently processed pri-miRNAs harbor a 5′ UG motif at the basal junction, a UGU motif at the apical loop, a mismatched GHG motif in the 3′ stem region, and a 3′ flanking CNNC motif [65,66,68,69]. Microprocessor processing efficiency is also regulated by additional auxiliary factors that enhance or inhibit its cleavage activity on specific pri-miRNAs [70]. After microprocessor processing, the cleaved hairpin structure (pre-miRNA) is transported to the cytoplasm by Exportin 5 [71,72], where further processing by the RNAse III endonuclease Dicer takes place. Cleavage of pre-miRNAs by Dicer produces the mature miRNA duplex of 20–24 nt length, containing 2-nucleotides 3′ overhangs on both strands [73,74,75,76]. Only one strand of this duplex, the guide strand, is loaded into the RNA-induced silencing complex (RISC) to inhibit expression of the target mRNA [77,78,79] (Figure 2). In mammals, four different Ago proteins are expressed and associate with miRNAs (Ago 1–4) [80,81], of which only Ago2 and Ago3 have retained endonuclease activity [82,83]. The additional co-factors TRBP and PACT in humans, and Loquacious (Loqs) in D. melanogaster, are necessary for successful Dicer-mediated miRNA processing and formation of the RISC complex [84,85,86,87,88,89].
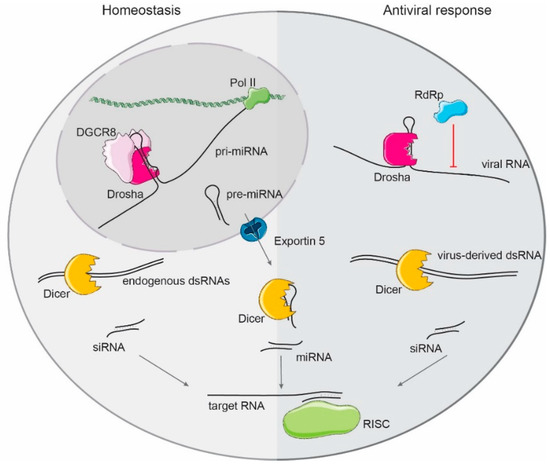
Figure 2.
Primary miRNA (pri-miRNA) precursors are transcribed by RNA-polymerase II, and processed in the nucleus by the microprocessor complex, composed by DGCR8 and Drosha. The released hairpin (pre-miRNA) is exported to the cytoplasm by Exportin-5 to be further processed by Dicer to form mature miRNAs that are loaded into the RISC complex to target complementary mRNAs. Both Drosha and Dicer are also antiviral factors. Drosha binding to RNA secondary structures in the viral genomes blocks the viral RNA-dependent RNA-polymerase and replication of the virus. Dicer can cleave the virus-derived dsRNA intermediates of replication to generate small interfering RNAs (siRNAs) that target and induce the decay of viral RNA molecules.
The binding of the mature miRNA to the target is dependent on partial complementarity to a short (6–7 nt long) sequence in the miRNA, known as the seed sequence, which is enough to mediate translational repression or destabilization of the bound mRNAs [90,91,92,93].
In addition to the canonical miRNA biogenesis pathway, several miRNAs have evolved to use alternative biogenesis routes. Mirtrons are microprocessor-independent miRNAs derived from spliced introns, which are processed by Dicer in the cytoplasm [94,95,96]. Specific miRNAs, such as mir-451, are processed by the microprocessor, followed by an Ago-2 dependent processing step [97,98,99]. Other Ago-associated RNAs, termed Agotrons, bypass both the microprocessor and Dicer processing steps [100,101].
Apart from its role in miRNA biogenesis, Dicer is essential for the production of siRNAs in chordates and non-chordates (Figure 2). Cytoplasmic processing of endogenous dsRNAs derived from sense and antisense transcripts or long stem-loop structures by Dicer generates mature siRNAs that are also loaded into the RISC complex [102,103,104]. Unlike miRNAs, the full complementarity between siRNAs and the target activates Ago2 endonucleolytic activity and degradation of the target RNA [82,83,105]. In mammals, endogenous siRNAs (endo-siRNAs) have been reported in mouse embryonic stem cells and oocytes. In stem cells, endo-siRNAs originate from repetitive elements (SINEs, short interspersed elements) [102], and in oocytes, endo-siRNAs have been shown to control the expression of both mRNAs and retrotransposons [106,107]. In comparison to non-vertebrate organisms such as C. elegans [108,109,110] or D. melanogaster [103,104,111], it is still unclear how widespread the synthesis and function of endo-siRNAs in mammals is. Initially, mammals were not considered to produce siRNAs, since the presence of dsRNAs in the cytoplasm could trigger a type I IFN response. Noticeably, the cellular models where mammalian endo-siRNAs have been reported have an inherently attenuated IFN response [112,113,114,115].
Interestingly, both Drosha and Dicer have been recently shown to have a direct role in controlling viral infections, apart from their classical function in small RNA biogenesis (Figure 2).
3. Type I IFNs Modulate the Activity of The Small RNA Biogenesis Pathway
The relationship between the IFN response and small RNAs is intricate. For instance, the IFN response is highly regulated by miRNAs, but at the same time, the IFN response regulates miRNA expression and RNAi proficiency to ensure the most efficient antiviral state.
During homeostasis, miRNAs regulate a large number of genes involved in the IFN response, which suggests that dysregulation of miRNA expression can lead to incorrect levels of these and other cytokines. Importantly, the unbalanced production of IFNs and pro-inflammatory molecules are at the root of human disorders, including autoimmune disease, inflammation, and cancer [164,165]. To ensure correct regulation of the IFN response, miRNAs target genes involved in different stages of the IFN response pathway, including the PRRs, transduction proteins, and transcription factors [166,167,168]. This homeostatic regulation is considered to act as a general dampening down of the IFN response that needs to be de-repressed following its activation. In agreement, in the absence of miRNAs by depletion of Dicer in microglia, endometrial, and thymic cells, spontaneous expression of IFNs is observed in the absence of infection [169,170,171]. Remarkably, ISGs also seem to be more significantly regulated by miRNAs than housekeeping genes [172]. Besides this global de-repression, there are also a number of miRNAs whose expression is induced by the IFN response. miR-146 and miR-155 are IFN-induced miRNAs that act as negative feedback loop molecules to shut-down the IFN response [173,174]. These and other miRNAs target components of the IFN response, such as TLR-receptors and the signaling molecules TRAF6, IRAK1, and 2 [175,176], but are also found to directly target cytokines such as TNF-α and IFN-β [177,178].
After transcription, the expression of miRNAs can also be post-transcriptionally regulated by the type I IFN response. Stimulation of cells with the viral mimic dsRNA and activation of IFN expression regulates the processing of pri-miRNAs by the microprocessor. This inhibition leads to a transient depletion of specific miRNAs, which is necessary to robustly express IFN- β and initiate the antiviral response [179]. Along these lines, type I IFNs diminish Dicer expression levels [180] and Dicer cleavage activity [181]. However, the exact mechanism by which the type I IFN response impairs both Drosha and Dicer cleavage activities is still unknown. In the case of the microprocessor, the activation of IFN expression reduces the binding affinity of DGCR8 to its substrates [179]. In the case of Dicer, the RLR LGP2 has been found to compete for Dicer binding to its substrates during the antiviral response [182]. Other factors involved in the recognition of dsRNA, such as ADAR, can also modulate the function of Drosha and Dicer proteins. ADAR1 associates to Dicer to enhance its mi-/si-RNA mediated processing [183], whereas ADAR-mediated editing of pri-miRNAs inhibits both Drosha and Dicer cleavage activity [184,185].
Other parts of the small RNA silencing pathway, such as RISC, are also negatively regulated during IFN activation. Activation of the IFN response induces Ago2 poly-ADP-ribosylation, which correlates with diminished miRNA and siRNA activity in cells during infection [172] (Figure 3).
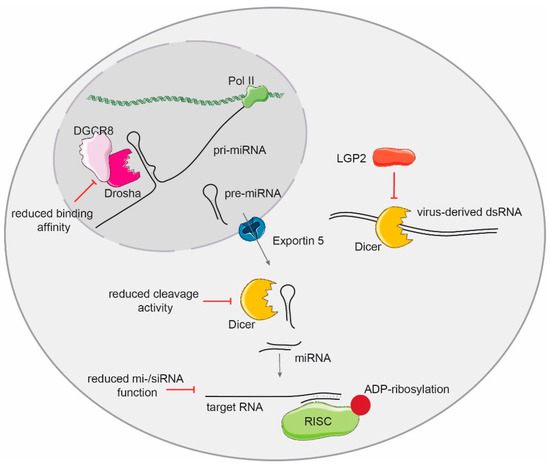
Figure 3.
The activation of the type I IFN response impairs the activity and function of the small RNA biogenesis pathway. The microprocessor complex binding affinity and cleavage activity of pri-miRNA substrates is reduced upon IFN expression activation. IFNs also reduce Dicer protein levels and impair its cleavage activity. The function of the RISC complex is also modulated by the IFN response by poly-ADP-ribosylation of the essential component Ago2. The RLR LGP2 interferes with recognition and cleavage activity of Dicer on dsRNAs.
Other classically-associated factors of the antiviral response can also modulate Dicer activity. PACT, an activator of PKR function [186], and TRBP, an inhibitor of PKR function [187,188], regulate the endonucleolytic activity and accuracy of Dicer cleavage [58,84,87,189,190,191]. More recently, PACT has also been shown to bind and stimulate the ATPase activity of RIG-I like receptors to initiate the antiviral response [192]. Conversely, overexpression of D. melanogaster Dicer-2 in human cells blocks IFN-β expression and PKR function [153].
A common feature of all the aforementioned factors is their ability to recognize dsRNA [193]. Binding to shared dsRNA molecules may act as a platform for these interactions to occur but also offers an opportunity to compete for binding to the same RNA substrates. Intriguingly, the N-terminal DExD-helicase domain of Dicer shares homology with that of the RIG-I-like family of receptors, highlighting the similarities and evolutionary conservation between the antiviral RNAi and protein-based response in their structural organization [194,195].
Given the growing evidence for a functional incompatibility between these two antiviral systems in mammals, the reason for this becomes more intriguing. Rapid impairment of small RNA biogenesis and RISC function during the activation of the type I IFN response may be necessary for robust ISG expression and establishment of the antiviral state. This is further supported by reports that show mammalian embryonic stem cells and embryonic carcinoma cells have a much better developed RNAi response compared to somatic cells. These cells inherently lack a functional IFN response [196], and are able to process long dsRNA into functional siRNAs [197,198]. Furthermore, RNAi activity in ESCs targeting either endogenous transposable elements [106] or viruses is more pronounced [150]. The shutdown of the IFN response in somatic cells enables the processing of long dsRNA into functional RNAi and inhibits replication of virus with the cognate sequence [199]. However, naturally derived siRNAs can still be detected, to an extent, in IFN-proficient cell lines [158]. All these suggest the presence of a competition between the small RNA biogenesis factors and the type I IFN response when co-occurring in the same cell type.
4. MiRNA-Mediated Regulation of Viruses
Besides regulation of the IFN response, and consequently the antiviral state of cells, miRNAs have been found to directly target viral sequences in human immunodeficiency virus-1 (HIV-1), primate foamy virus-1 (PFV-1), influenza A virus (IAV), hepatitis C virus (HCV), vesicular stomatitis virus (VSV), and the human papilloma virus (HPV) [200,201,202,203,204]. Unlike their endogenous targets, only the minority target the 3′ end of the viral sequences. Although the available experimental data clearly supports a role of miRNAs directly targeting viral genomes or transcripts, its biological significance is still unknown. From an evolutionary point of view, it is difficult to imagine that for very recent infections, such as HIV-1, or low-level infection rates, such as for HCV, there has been enough evolutionary pressure to have developed specific sequences to target these viruses.
Given the importance of miRNAs in the robustness of the IFN response and consequent antiviral state of cells, and the observation of miRNAs directly targeting viral sequences, it is no surprise that there are a number of viruses targeting miRNA regulation. One of the first examples was found in primate foamy virus (PFV-1), which encodes a protein (Tas) that was able to inhibit miRNA function, relieving the suppression of miR-32 [205]. Viral inhibitors of miRNA function are also found in HIV-1 [206,207], Ebola virus [157], Influenza A virus [208], and Vaccinia virus [209], suggesting host miRNAs exert enough pressure to warrant this kind of investment.
The reverse scenario, where viruses seemingly benefit from endogenous miRNAs, is found in the Flaviviridae family, where HCV uses miR-122 to shield its genome from degradation resulting in increased translation [210,211]. This miRNA is highly expressed in the virus’ natural host cells, where it binds the 5′ UTR of the viral genome, resulting in improved viral replication in an AGO2-dependent fashion [212,213]. Recently, miR-21 was identified as a pro-viral factor for the Zika virus, a member of the same Flaviviridae family as HCV, suggesting that utilizing host miRNAs for their own benefit is common in this virus family [214]. Endogenous miRNAs having pro-viral properties have also been reported during HIV-1 and HCMV infections [215,216].
The observation that miRNAs are important for the regulation and robustness of the IFN response, the direct pro- and antiviral effects of miRNAs and the presence of miRNA-regulating activities of viruses makes it extremely difficult to predict the net effects when manipulating the major miRNA biogenesis factors. Despite this inherent complex network of interactions, in the next section we will have a closer look into the results of manipulating the expression of small RNAs in the context of the consequences on the IFN response and viral dissemination.
6. Conclusions
Studies investigating the role of small RNAs and the innate immune response to viral infections have uncovered a complex network of interactions. The overall effect of impairing small RNA biogenesis on the host’s antiviral response is difficult to predict, as small RNAs are responsible for silencing the IFN response, but at the same time are required to establish a robust antiviral state. Another layer of complexity is added by the observation that some miRNAs can directly target viral sequences, whereas others increase viral fitness. Integrating all these processes will be the key in providing a comprehensive understanding of the relationship between small RNAs and the type I IFN response.
Author Contributions
S.F.W., L.I.K., J.W., and S.M. carried out the primary literature research and co-wrote the manuscript. S.F.W. and L.I.K. designed the figures. Figures have been designed using ‘Smart Servier Medical Art’ (https://smart.servier.com) under a CC BY 3.0 (https://creativecommons.org/licenses/by/3.0/).
Funding
This work was supported by the Wellcome Trust (107665/Z/15/Z), L.I.K. is supported by an MRC DTP in Precision Medicine Fellowship.
Conflicts of Interest
The authors declare no conflict of interest.
References
- González-Navajas, J.M.; Lee, J.; David, M.; Raz, E. Immunomodulatory functions of type i interferons. Nat. Rev. Immunol. 2012, 12, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Di Franco, S.; Turdo, A.; Todaro, M.; Stassi, G. Role of type i and ii interferons in colorectal cancer and melanoma. Front. Immunol. 2017, 8, 878. [Google Scholar] [CrossRef]
- Hall, J.C.; Rosen, A. Type i interferons: Crucial participants in disease amplification in autoimmunity. Nat. Rev. Rheumatol. 2010, 6, 40–49. [Google Scholar] [CrossRef]
- Venkatesh, B.; Lee, A.P.; Ravi, V.; Maurya, A.K.; Lian, M.M.; Swann, J.B.; Ohta, Y.; Flajnik, M.F.; Sutoh, Y.; Kasahara, M.; et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature 2014, 505, 174–179. [Google Scholar] [CrossRef]
- Secombes, C.J.; Zou, J. Evolution of interferons and interferon receptors. Front. Immunol. 2017, 8, 209. [Google Scholar] [CrossRef]
- Swiecki, M.; Colonna, M. Type i interferons: Diversity of sources, production pathways and effects on immune responses. Curr. Opin. Virol. 2011, 1, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Pestka, S.; Krause, C.D.; Walter, M.R. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 2004, 202, 8–32. [Google Scholar] [CrossRef]
- Havell, E.A.; Berman, B.; Ogburn, C.A.; Berg, K.; Paucker, K.; Vilcek, J. Two antigenically distinct species of human interferon. Proc. Natl. Acad. Sci. USA 1975, 72, 2185–2187. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.R.; Raulet, D.H.; Pasternack, M.S.; Bevan, M.J. Cytotoxic t lymphocytes produce immune interferon in response to antigen or mitogen. J. Exp. Med. 1982, 155, 1198–1203. [Google Scholar] [CrossRef] [PubMed]
- Scharton, T.M.; Scott, P. Natural killer cells are a source of interferon gamma that drives differentiation of cd4+ t cell subsets and induces early resistance to leishmania major in mice. J. Exp. Med. 1993, 178, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Coccia, E.M.; Severa, M.; Giacomini, E.; Monneron, D.; Remoli, M.E.; Julkunen, I.; Cella, M.; Lande, R.; Uzé, G. Viral infection and toll-like receptor agonists induce a differential expression of type i and λ interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur. J. Immunol. 2004, 34, 796–805. [Google Scholar] [CrossRef]
- Karpala, A.J.; Morris, K.R.; Broadway, M.M.; McWaters, P.G.D.; O’Neil, T.E.; Goossens, K.E.; Lowenthal, J.W.; Bean, A.G.D. Molecular cloning, expression, and characterization of chicken ifn -λ. J. Interferon Cytokine Res. 2008, 28, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Kotenko, S.V.; Gallagher, G.; Baurin, V.V.; Lewis-Antes, A.; Shen, M.; Shah, N.K.; Langer, J.A.; Sheikh, F.; Dickensheets, H.; Donnelly, R.P. Ifn-λs mediate antiviral protection through a distinct class ii cytokine receptor complex. Nat. Immunol. 2003, 4, 69–77. [Google Scholar] [CrossRef]
- Spann, K.M.; Tran, K.-C.; Chi, B.; Rabin, R.L.; Collins, P.L. Suppression of the induction of alpha, beta, and lambda interferons by the ns1 and ns2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J. Virol. 2004, 78, 4363–4369. [Google Scholar] [CrossRef]
- Gürtler, C.; Bowie, A.G. Innate immune detection of microbial nucleic acids. Trends Microbiol. 2013, 21, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Roers, A.; Hiller, B.; Hornung, V. Recognition of endogenous nucleic acids by the innate immune system. Immunity 2016, 44, 739–754. [Google Scholar] [CrossRef] [PubMed]
- Novick, D.; Cohen, B.; Rubinstein, M. The human interferon alpha/beta receptor: Characterization and molecular cloning. Cell 1994, 77, 391–400. [Google Scholar] [CrossRef]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Lester, S.N.; Li, K. Toll-like receptors in antiviral innate immunity. J. Mol. Biol. 2014, 426, 1246–1264. [Google Scholar] [CrossRef]
- Takeda, K.; Akira, S. Toll-like receptors in innate immunity. Int. Immunol. 2004, 17, 1–14. [Google Scholar] [CrossRef]
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R.A. Recognition of double-stranded RNA and activation of nf-κb by toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar] [CrossRef]
- Deguine, J.; Barton, G.M. Myd88: A central player in innate immune signaling. F1000Prime Rep. 2014, 6, 97. [Google Scholar] [CrossRef]
- Honda, K.; Yanai, H.; Mizutani, T.; Negishi, H.; Shimada, N.; Suzuki, N.; Ohba, Y.; Takaoka, A.; Yeh, W.-C.; Taniguchi, T. Role of a transductional-transcriptional processor complex involving myd88 and irf-7 in toll-like receptor signaling. Proc. Natl. Acad. Sci. USA 2004, 101, 15416–15421. [Google Scholar] [CrossRef]
- Vercammen, E.; Staal, J.; Beyaert, R. Sensing of viral infection and activation of innate immunity by toll-like receptor 3. Clin. Microbiol. Rev. 2008, 21, 13–25. [Google Scholar] [CrossRef]
- Yoneyama, M.; Kikuchi, M.; Matsumoto, K.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Foy, E.; Loo, Y.M.; Gale, M.; Akira, S.; et al. Shared and unique functions of the dexd/h-box helicases rig-i, mda5, and lgp2 in antiviral innate immunity. J. Immunol. 2005, 175, 2851–2858. [Google Scholar] [CrossRef]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagishi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA helicase rig-i has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004, 5, 730–737. [Google Scholar] [CrossRef]
- Hornung, V.; Ellegast, J.; Kim, S.; Brzózka, K.; Jung, A.; Kato, H.; Poeck, H.; Akira, S.; Conzelmann, K.-K.; Schlee, M.; et al. 5’-triphosphate RNA is the ligand for rig-i. Science 2006, 314, 994–997. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Mikamo-Satoh, E.; Hirai, R.; Kawai, T.; Matsushita, K.; Hiiragi, A.; Dermody, T.S.; Fujita, T.; Akira, S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-i and melanoma differentiation-associated gene 5. J. Exp. Med. 2008, 205, 1601–1610. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of mda5 and rig-i helicases in the recognition of Rna viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef]
- Jiang, F.; Ramanathan, A.; Miller, M.T.; Tang, G.-Q.Q.; Gale, M.; Patel, S.S.; Marcotrigiano, J. Structural basis of RNA recognition and activation by innate immune receptor rig-i. Nature 2011, 479, 423–427. [Google Scholar] [CrossRef]
- Takahasi, K.; Yoneyama, M.; Nishihori, T.; Hirai, R.; Kumeta, H.; Narita, R.; Gale, M.; Inagaki, F.; Fujita, T. Nonself RNA-sensing mechanism of rig-i helicase and activation of antiviral immune responses. Mol. Cell 2008, 29, 428–440. [Google Scholar] [CrossRef]
- Peisley, A.; Lin, C.; Wu, B.; Orme-Johnson, M.; Liu, M.; Walz, T.; Hur, S. Cooperative assembly and dynamic disassembly of mda5 filaments for viral dsRNA recognition. Proc. Natl. Acad. Sci. USA 2011, 108, 21010–21015. [Google Scholar] [CrossRef]
- Peisley, A.; Wu, B.; Xu, H.; Chen, Z.J.; Hur, S. Structural basis for ubiquitin-mediated antiviral signal activation by rig-i. Nature 2014, 509, 110–114. [Google Scholar] [CrossRef]
- Hou, F.; Sun, L.; Zheng, H.; Skaug, B.; Jiang, Q.-X.; Chen, Z.J. Mavs forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 2011, 146, 448–461. [Google Scholar] [CrossRef]
- Kawai, T.; Takahashi, K.; Sato, S.; Coban, C.; Kumar, H.; Kato, H.; Ishii, K.J.; Takeuchi, O.; Akira, S. Ips-1, an adaptor triggering rig-i- and mda5-mediated type i interferon induction. Nat. Immunol. 2005, 6, 981–988. [Google Scholar] [CrossRef]
- Hiscott, J. Triggering the innate antiviral response through irf-3 activation. J. Biol. Chem. 2007, 282, 15325–15329. [Google Scholar] [CrossRef]
- Reikine, S.; Nguyen, J.B.; Modis, Y. Pattern recognition and signaling mechanisms of rig-i and mda5. Front. Immunol. 2014, 5, 342. [Google Scholar] [CrossRef]
- Bruns, A.M.; Leser, G.P.; Lamb, R.A.; Horvath, C.M. The innate immune sensor lgp2 activates antiviral signaling by regulating mda5-RNA interaction and filament assembly. Mol. Cell 2014, 55, 771–781. [Google Scholar] [CrossRef]
- Satoh, T.; Kato, H.; Kumagai, Y.; Yoneyama, M.; Sato, S.; Matsushita, K.; Tsujimura, T.; Fujita, T.; Akira, S.; Takeuchi, O. Lgp2 is a positive regulator of rig-i- and mda5-mediated antiviral responses. Proc. Natl. Acad. Sci. USA 2010, 107, 1512–1517. [Google Scholar] [CrossRef]
- Rothenfusser, S.; Goutagny, N.; DiPerna, G.; Gong, M.; Monks, B.G.; Schoenemeyer, A.; Yamamoto, M.; Akira, S.; Fitzgerald, K.A. The RNA helicase lgp2 inhibits tlr-independent sensing of viral replication by retinoic acid-inducible gene-i. J. Immunol. 2005, 175, 5260–5268. [Google Scholar] [CrossRef]
- Bruns, A.M.; Horvath, C.M. Lgp2 synergy with mda5 in rlr-mediated RNA recognition and antiviral signaling. Cytokine 2015, 74, 198–206. [Google Scholar] [CrossRef]
- Komuro, A.; Horvath, C.M. Rna- and virus-independent inhibition of antiviral signaling by RNA helicase lgp2. J. Virol. 2006, 80, 12332–12342. [Google Scholar] [CrossRef]
- Vitour, D.; Meurs, E.F. Regulation of interferon production by rig-i and lgp2: A lesson in self-control. Sci. Signal. 2007, 2007, pe20. [Google Scholar] [CrossRef]
- Meurs, E.; Chong, K.; Galabru, J.; Thomas, N.S.B.; Kerr, I.M.; Williams, B.R.G.; Hovanessian, A.G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 1990, 62, 379–390. [Google Scholar] [CrossRef]
- Gross, M.; Wing, M.; Rundquist, C.; Rubino, S. Evidence that phosphorylation of eif-2 (4 prevents the eif-2b-mediated dissociation of eif-z * gdp from the 60 s subunit of complete initiation complexes. J. Biol. Chem. 1987, 262, 6899–6907. [Google Scholar]
- Meurs, E.F.; Watanabe, Y.; Kadereit, S.; Barber, G.N.; Katze, M.G.; Chong, K.; Williams, B.R.; Hovanessian, A.G. Constitutive expression of human double-stranded RNA-activated p68 kinase in murine cells mediates phosphorylation of eukaryotic initiation factor 2 and partial resistance to encephalomyocarditis virus growth. J. Virol. 1992, 66, 5805–5814. [Google Scholar]
- Sudhakar, A.; Ramachandran, A.; Ghosh, S.; Hasnain, S.E.; Kaufman, R.J.; Ramaiah, K.V.A. Phosphorylation of serine 51 in initiation factor 2α (eif2α) promotes complex formation between eif2α(p) and eif2b and causes inhibition in the guanine nucleotide exchange activity of eif2b. Biochemistry 2000, 39, 12929–12938. [Google Scholar] [CrossRef]
- Hovanessian, A.G.; Justesen, J. The human 2′-5′oligoadenylate synthetase family: Unique interferon-inducible enzymes catalyzing 2′-5′ instead of 3′-5′ phosphodiester bond formation. Biochimie 2007, 89, 779–788. [Google Scholar] [CrossRef]
- Floyd-Smith, G.; Slattery, E.; Lengyel, P. Interferon action: Rna cleavage pattern of a (2’-5’) oligoadenylate--dependent endonuclease. Science 1981, 212, 1030–1032. [Google Scholar] [CrossRef]
- Nakanishi, M.; Goto, Y.; Kitade, Y. 2-5a induces a conformational change in the ankyrin-repeat domain of RNAse L. Proteins: Struct. Funct. Bioinform. 2005, 60, 131–138. [Google Scholar] [CrossRef]
- Wreschner, D.H.; McCauley, J.W.; Skehel, J.J.; Kerr, I.M. Interferon action—sequence specificity of the ppp(a2′p)na-dependent ribonuclease. Nature 1981, 289, 414–417. [Google Scholar] [CrossRef] [PubMed]
- Malathi, K.; Dong, B.; Gale, M.; Silverman, R.H. Small self-RNA generated by RNAse l amplifies antiviral innate immunity. Nature 2007, 448, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Vitali, P.; Scadden, A.D. Double-stranded RNAs containing multiple iu pairs are sufficient to suppress interferon induction and apoptosis. Nat. Struct Mol. Biol. 2010, 17, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Mannion, N.M.; Greenwood, S.M.; Young, R.; Cox, S.; Brindle, J.; Read, D.; Nellaker, C.; Vesely, C.; Ponting, C.P.; McLaughlin, P.J. , et al. The RNA-editing enzyme adar1 controls innate immune responses to RNA. Cell Rep. 2014, 9, 1482–1494. [Google Scholar] [CrossRef] [PubMed]
- Samuel, C.E. Adenosine deaminases acting on RNA (adars) are both antiviral and proviral. Virology 2011, 411, 180–193. [Google Scholar] [CrossRef]
- Farazi, T.A.; Juranek, S.A.; Tuschl, T. Development. Development 2008, 132, 4645–4652. [Google Scholar]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. Piwi-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef]
- Lee, Y.; Jeon, K.; Lee, J.-T.; Kim, S.; Kim, V.N.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase ii. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.B.J.; Plasterk, R.H.A.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Gregory, R.I.; Yan, K.-P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The drosha-dgcr8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Landthaler, M.; Yalcin, A.; Tuschl, T. The human digeorge syndrome critical region gene 8 and its d. Melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004, 14, 2162–2167. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNAse iii drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.A.; Jo, M.H.; Choi, Y.-G.; Park, J.; Kwon, S.C.; Hohng, S.; Kim, V.N.; Woo, J.-S. Functional anatomy of the human microprocessor. Cell 2015, 161, 1374–1387. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.C.; Baek, S.C.; Choi, Y.-G.; Yang, J.; Lee, Y.-S.; Woo, J.-S.; Kim, V.N. Molecular basis for the single-nucleotide precision of primary microRNA processing. Mol. Cell 2018, 73, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Yi, R.; Cullen, B.R. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme drosha. EMBO J. 2005, 24, 138–148. [Google Scholar] [CrossRef]
- Auyeung, V.C.; Ulitsky, I.; McGeary, S.E.; Bartel, D.P. Beyond secondary structure: Primary-sequence determinants license pri-miRNA hairpins for processing. Cell 2013, 152, 844–858. [Google Scholar] [CrossRef]
- Fang, W.; Bartel, D.P. The menu of features that define primary microRNAs and enable de novo design of microRNA genes. Mol. Cell 2015, 60, 131–145. [Google Scholar] [CrossRef]
- Michlewski, G.; Cáceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2019, 25, 1–16. [Google Scholar] [CrossRef]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a rangtp-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef]
- Ketting, R.F.; Fischer, S.E.; Bernstein, E.; Sijen, T.; Hannon, G.J.; Plasterk, R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in c. Elegans. Genes Dev. 2001, 15, 2654–2659. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human dicer and bacterial RNAse iii. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef]
- Iwasaki, S.; Kobayashi, M.; Yoda, M.; Sakaguchi, Y.; Katsuma, S.; Suzuki, T.; Tomari, Y. Hsc70/hsp90 chaperone machinery mediates atp-dependent risc loading of small RNA duplexes. Mol. Cell 2010, 39, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Hutvágner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Dueck, A.; Ziegler, C.; Eichner, A.; Berezikov, E.; Meister, G. MicroRNAs associated with the different human argonaute proteins. Nucleic Acids Res. 2012, 40, 9850–9862. [Google Scholar] [CrossRef]
- Su, H.; Trombly, M.I.; Chen, J.; Wang, X. Essential and overlapping functions for mammalian argonautes in microRNA silencing. Genes Dev. 2009, 23, 304–317. [Google Scholar] [CrossRef]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.-J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar] [CrossRef]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. Trbp recruits the dicer complex to ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef]
- Fukunaga, R.; Han, B.W.; Hung, J.H.; Xu, J.; Weng, Z.; Zamore, P.D. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell 2012, 151, 533–546. [Google Scholar] [CrossRef]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human risc couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef]
- Haase, A.D.; Jaskiewicz, L.; Zhang, H.; Lainé, S.; Sack, R.; Gatignol, A.; Filipowicz, W. Trbp, a regulator of cellular pkr and hiv-1 virus expression, interacts with dicer and functions in RNA silencing. EMBO Rep. 2005, 6, 961–967. [Google Scholar] [CrossRef]
- MacRae, I.J.; Ma, E.; Zhou, M.; Robinson, C.V.; Doudna, J.A. In vitro reconstitution of the human risc-loading complex. Proc. Natl. Acad. Sci. USA 2008, 105, 512–517. [Google Scholar] [CrossRef]
- Maniataki, E.; Mourelatos, Z. A human, atp-independent, risc assembly machine fueled by pre-miRNA. Genes Dev. 2005, 19, 2979–2990. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Lewis, B.P.; Shih, I.-h.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The mirtron pathway generates microRNA-class regulatory RNAs in drosophila. Cell 2007, 130, 89–100. [Google Scholar] [CrossRef]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef]
- Westholm, J.O.; Lai, E.C. Mirtrons: MicroRNA biogenesis via splicing. Biochimie 2011, 93, 1897–1904. [Google Scholar] [CrossRef]
- Cheloufi, S.; Dos Santos, C.O.; Chong, M.M.W.; Hannon, G.J. A dicer-independent miRNA biogenesis pathway that requires ago catalysis. Nature 2010, 465, 584–589. [Google Scholar] [CrossRef]
- Cifuentes, D.; Xue, H.; Taylor, D.W.; Patnode, H.; Mishima, Y.; Cheloufi, S.; Ma, E.; Mane, S.; Hannon, G.J.; Lawson, N.D.; et al. A novel miRNA processing pathway independent of dicer requires argonaute2 catalytic activity. Science 2010, 328, 1694–1698. [Google Scholar] [CrossRef]
- Yang, J.-S.; Maurin, T.; Robine, N.; Rasmussen, K.D.; Jeffrey, K.L.; Chandwani, R.; Papapetrou, E.P.; Sadelain, M.; O’Carroll, D.; Lai, E.C. Conserved vertebrate mir-451 provides a platform for dicer-independent, ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 15163–15168. [Google Scholar] [CrossRef]
- Daugaard, I.; Hansen, T.B. Biogenesis and function of ago-associated RNAs. Trends Genet. 2017, 33. [Google Scholar] [CrossRef]
- Hansen, T.B.; Venø, M.T.; Jensen, T.I.; Schaefer, A.; Damgaard, C.K.; Kjems, J. Argonaute-associated short introns are a novel class of gene regulators. Nat. Commun. 2016, 7, 11538. [Google Scholar] [CrossRef]
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse es cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, dicer-dependent small RNAs. Genes Dev. 2008, 22, 2773–2785. [Google Scholar] [CrossRef]
- Chung, W.-J.; Okamura, K.; Martin, R.; Lai, E.C. Endogenous RNA interference provides a somatic defense against drosophila transposons. Curr. Biol. 2008, 18, 795–802. [Google Scholar] [CrossRef]
- Czech, B.; Malone, C.D.; Zhou, R.; Stark, A.; Schlingeheyde, C.; Dus, M.; Perrimon, N.; Kellis, M.; Wohlschlegel, J.A.; Sachidanandam, R.; et al. An endogenous small interfering RNA pathway in drosophila. Nature 2008, 453, 798–802. [Google Scholar] [CrossRef]
- Song, J.-J.; Smith, S.K.; Hannon, G.J.; Joshua-Tor, L. Crystal structure of argonaute and its implications for risc slicer activity. Science 2004, 305, 1434–1437. [Google Scholar] [CrossRef]
- Tam, O.H.; Aravin, A.A.; Stein, P.; Girard, A.; Murchison, E.P.; Cheloufi, S.; Hodges, E.; Anger, M.; Sachidanandam, R.; Schultz, R.M.; et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 2008, 453, 534–538. [Google Scholar] [CrossRef]
- Watanabe, T.; Totoki, Y.; Toyoda, A.; Kaneda, M.; Kuramochi-Miyagawa, S.; Obata, Y.; Chiba, H.; Kohara, Y.; Kono, T.; Nakano, T.; et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 2008, 453, 539–543. [Google Scholar] [CrossRef]
- Gu, W.; Shirayama, M.; Conte, D.; Vasale, J.; Batista, P.J.; Claycomb, J.M.; Moresco, J.J.; Youngman, E.M.; Keys, J.; Stoltz, M.J.; et al. Distinct argonaute-mediated 22g-RNA pathways direct genome surveillance in the c. Elegans germline. Mol. Cell 2009, 36, 231–244. [Google Scholar] [CrossRef]
- Pak, J.; Fire, A. Distinct populations of primary and secondary effectors during RNAi in c. Elegans. Science 2007, 315, 241–244. [Google Scholar] [CrossRef]
- Sjien, T.; Steiner, F.A.; Thijssen, K.L.; Plasterk, R.H.A.; Sijen, T.; Steiner, F.A.; Thijssen, K.L.; Plasterk, R.H.A. Secondary siRNAs result from form a distinct class. Science 2007, 315, 2005–2008. [Google Scholar]
- Ghildiyal, M.; Seitz, H.; Horwich, M.D.; Li, C.; Du, T.; Lee, S.; Xu, J.; Kittler, E.L.W.; Zapp, M.L.; Weng, Z.; et al. Endogenous siRNAs derived from transposons and mRNAs in drosophila somatic cells. Science 2008, 320, 1077–1081. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L.; Yang, L.; Carmichael, G.G. Molecular basis for an attenuated cytoplasmic dsRNA response in human embryonic stem cells. Cell Cycle 2010, 9, 3552–3564. [Google Scholar] [CrossRef]
- Nejepinska, J.; Malik, R.; Filkowski, J.; Flemr, M.; Filipowicz, W.; Svoboda, P. DsRNA expression in the mouse elicits RNAi in oocytes and low adenosine deamination in somatic cells. Nucleic Acids Res. 2012, 40, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Stein, P.; Zeng, F.; Pan, H.; Schultz, R.M. Absence of non-specific effects of RNA interference triggered by long double-stranded RNA in mouse oocytes. Dev. Biol. 2005, 286, 464–471. [Google Scholar] [CrossRef]
- Wang, R.; Wang, J.; Paul, A.M.; Acharya, D.; Bai, F.; Huang, F.; Guo, Y.-L. Mouse embryonic stem cells are deficient in type i interferon expression in response to viral infections and double-stranded RNA. J. Biol. Chem. 2013, 288, 15926–15936. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Shin, C. Emerging roles of drosha beyond primary microRNA processing. RNA Biol. 2018, 15, 186–193. [Google Scholar] [CrossRef]
- Macias, S.; Cordiner, R.A.; Cáceres, J.F. Cellular functions of the microprocessor. Biochem. Soc. Trans. 2013, 41, 838–843. [Google Scholar] [CrossRef]
- Macias, S.; Plass, M.; Stajuda, A.; Michlewski, G.; Eyras, E.; Cáceres, J.F. Dgcr8 hits-clip reveals novel functions for the microprocessor. Nat. Struct. Mol. Biol. 2012, 19, 760–766. [Google Scholar] [CrossRef]
- Han, J.; Pedersen, J.S.; Kwon, S.C.; Belair, C.D.; Kim, Y.-K.; Yeom, K.-H.; Yang, W.-Y.; Haussler, D.; Blelloch, R.; Kim, V.N. Posttranscriptional crossregulation between drosha and dgcr8. Cell 2009, 136, 75–84. [Google Scholar] [CrossRef]
- Kadener, S.; Rodriguez, J.; Abruzzi, K.C.; Khodor, Y.L.; Sugino, K.; Marr, M.T.; Nelson, S.; Rosbash, M. Genome-wide identification of targets of the drosha-pasha/dgcr8 complex. RNA 2009, 15, 537–545. [Google Scholar] [CrossRef]
- Triboulet, R.; Chang, H.-M.; LaPierre, R.J.; Gregory, R.I. Post-transcriptional control of dgcr8 expression by the microprocessor. RNA 2009, 15, 1005–1011. [Google Scholar] [CrossRef]
- Heras, S.R.; Macias, S.; Plass, M.; Fernandez, N.; Cano, D.; Eyras, E.; Garcia-Perez, J.L.; Cáceres, J.F. The microprocessor controls the activity of mammalian retrotransposons. Nat. Struct. Mol. Biol. 2013, 20, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Rouha, H.; Thurner, C.; Mandl, C.W. Functional microRNA generated from a cytoplasmic RNA virus. Nucleic Acids Res. 2010, 38, 8328–8337. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.S.; Langlois, R.A.; Pham, A.M.; Tenoever, B.R. Evidence for a cytoplasmic microprocessor of pri-miRNAs. RNA 2012, 18, 1338–1346. [Google Scholar] [CrossRef]
- Shapiro, J.S.; Schmid, S.; Aguado, L.C.; Sabin, L.R.; Yasunaga, A.; Shim, J.V.; Sachs, D.; Cherry, S.; TenOever, B.R. Drosha as an interferon-independent antiviral factor. Proc. Natl. Acad. Sci. USA 2014, 111, 7108–7113. [Google Scholar] [CrossRef]
- Shapiro, J.S.; Varble, A.; Pham, A.M.; Tenoever, B.R. Noncanonical cytoplasmic processing of viral microRNAs. RNA 2010, 16, 2068–2074. [Google Scholar] [CrossRef] [PubMed]
- Varble, A.; Chua, M.A.; Perez, J.T.; Manicassamy, B.; Garcia-Sastre, A.; tenOever, B.R. Engineered RNA viral synthesis of microRNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 11519–11524. [Google Scholar] [CrossRef] [PubMed]
- Park, H.H.; Triboulet, R.; Bentler, M.; Guda, S.; Du, P.; Xu, H.; Gregory, R.I.; Brendel, C.; Williams, D.A. Drosha knockout leads to enhancement of viral titers for vectors encoding miRNA-adapted shRNAs. Mol. Ther. Nucleic Acids 2018, 12, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Aguado, L.C.; Schmid, S.; May, J.; Sabin, L.R.; Panis, M.; Blanco-Melo, D.; Shim, J.V.; Sachs, D.; Cherry, S.; Simon, A.E.; et al. Rnase iii nucleases from diverse kingdoms serve as antiviral effectors. Nature 2017, 547, 114–117. [Google Scholar] [CrossRef]
- Feldman, E.R.; Kara, M.; Coleman, C.B.; Grau, K.R.; Oko, L.M.; Krueger, B.J.; Renne, R.; van Dyk, L.F.; Tibbetts, S.A. Virus-encoded microRNAs facilitate gammaherpesvirus latency and pathogenesis in vivo. MBio 2014, 5, e00981-14. [Google Scholar] [CrossRef]
- Guo, Y.E.; Oei, T.; Steitz, J.A. Herpesvirus saimiri microRNAs preferentially target host cell cycle regulators. J. Virol. 2015, 89, 10901–10911. [Google Scholar] [CrossRef] [PubMed]
- Lo, A.K.; To, K.F.; Lo, K.W.; Lung, R.W.; Hui, J.W.; Liao, G.; Hayward, S.D. Modulation of lmp1 protein expression by ebv-encoded microRNAs. Proc. Natl. Acad. Sci. USA 2007, 104, 16164–16169. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Zavolan, M.; Grasser, F.A.; Chien, M.; Russo, J.J.; Ju, J.; John, B.; Enright, A.J.; Marks, D.; Sander, C.; et al. Identification of virus-encoded microRNAs. Science 2004, 304, 734–736. [Google Scholar] [CrossRef] [PubMed]
- Riley, K.J.; Rabinowitz, G.S.; Steitz, J.A. Comprehensive analysis of rhesus lymphocryptovirus microRNA expression. J. Virol. 2010, 84, 5148–5157. [Google Scholar] [CrossRef] [PubMed]
- Umbach, J.L.; Kramer, M.F.; Jurak, I.; Karnowski, H.W.; Coen, D.M.; Cullen, B.R. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 2008, 454, 780–783. [Google Scholar] [CrossRef] [PubMed]
- Bogerd, H.P.; Karnowski, H.W.; Cai, X.; Shin, J.; Pohlers, M.; Cullen, B.R. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral microRNAs. Mol. Cell 2010, 37, 135–142. [Google Scholar] [CrossRef]
- Cazalla, D.; Xie, M.; Steitz, J.A. A primate herpesvirus uses the integrator complex to generate viral microRNAs. Mol. Cell 2011, 43, 982–992. [Google Scholar] [CrossRef]
- Lin, Y.T.; Sullivan, C.S. Expanding the role of drosha to the regulation of viral gene expression. Proc. Natl. Acad. Sci. USA 2011, 108, 11229–11234. [Google Scholar] [CrossRef]
- Xing, L.; Kieff, E. Epstein-barr virus bhrf1 micro- and stable RNAs during latency iii and after induction of replication. J. Virol. 2007, 81, 9967–9975. [Google Scholar] [CrossRef]
- Dang, Y.; Yang, Q.; Xue, Z.; Liu, Y. Rna interference in fungi: Pathways, functions, and applications. Eukaryot. Cell 2011, 10, 1148–1155. [Google Scholar] [CrossRef]
- Lee, Y.S.; Nakahara, K.; Pham, J.W.; Kim, K.; He, Z.; Sontheimer, E.J.; Carthew, R.W. Distinct roles for drosophila dicer-1 and dicer-2 in the siRNA/miRNA silencing pathways. Cell 2004, 117, 69–81. [Google Scholar] [CrossRef]
- Margis, R.; Fusaro, A.F.; Smith, N.A.; Curtin, S.J.; Watson, J.M.; Finnegan, E.J.; Waterhouse, P.M. The evolution and diversification of dicers in plants. FEBS Lett. 2006, 580, 2442–2450. [Google Scholar] [CrossRef]
- Backes, S.; Langlois, R.A.; Schmid, S.; Varble, A.; Shim, J.V.; Sachs, D.; tenOever, B.R. The mammalian response to virus infection is independent of small RNA silencing. Cell Rep. 2014, 8, 114–125. [Google Scholar] [CrossRef]
- Bogerd, H.P.; Skalsky, R.L.; Kennedy, E.M.; Furuse, Y.; Whisnant, A.W.; Flores, O.; Schultz, K.L.; Putnam, N.; Barrows, N.J.; Sherry, B.; et al. Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. J. Virol. 2014, 88, 8065–8076. [Google Scholar] [CrossRef]
- Girardi, E.; Chane-Woon-Ming, B.; Messmer, M.; Kaukinen, P.; Pfeffer, S. Identification of RNAse l-dependent, 3’-end-modified, viral small RNAs in sindbis virus-infected mammalian cells. MBio 2013, 4, e00698-13. [Google Scholar] [CrossRef]
- Kennedy, E.M.; Whisnant, A.W.; Kornepati, A.V.; Marshall, J.B.; Bogerd, H.P.; Cullen, B.R. Production of functional small interfering RNAs by an amino-terminal deletion mutant of human dicer. Proc. Natl. Acad. Sci. USA 2015, 112, E6945–E6954. [Google Scholar] [CrossRef]
- Parameswaran, P.; Sklan, E.; Wilkins, C.; Burgon, T.; Samuel, M.A.; Lu, R.; Ansel, K.M.; Heissmeyer, V.; Einav, S.; Jackson, W.; et al. Six RNA viruses and forty-one hosts: Viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog 2010, 6, e1000764. [Google Scholar] [CrossRef]
- Qiu, Y.; Xu, Y.; Zhang, Y.; Zhou, H.; Deng, Y.Q.; Li, X.F.; Miao, M.; Zhang, Q.; Zhong, B.; Hu, Y.; et al. Human virus-derived small RNAs can confer antiviral immunity in mammals. Immunity 2017, 46, 992–1004. [Google Scholar] [CrossRef]
- Li, Y.; Lu, J.; Han, Y.; Fan, X.; Ding, S.-W.W. Rna interference functions as an antiviral immunity mechanism in mammals. Science 2013, 342, 231–234. [Google Scholar] [CrossRef]
- Maillard, P.V.; Ciaudo, C.; Marchais, A.; Li, Y.; Jay, F.; Ding, S.W.; Voinnet, O. Antiviral RNA interference in mammalian cells. Science 2013, 342, 235–238. [Google Scholar] [CrossRef]
- Rossi, J.J. Mammalian dicer finds a partner. EMBO Rep. 2005, 6, 927–929. [Google Scholar] [CrossRef] [PubMed]
- Provost, P.; Dishart, D.; Doucet, J.; Frendewey, D.; Samuelsson, B.; Rådmark, O. Ribonuclease activity and RNA binding of recombinant human dicer. EMBO J. 2002, 21, 5864–5874. [Google Scholar] [CrossRef] [PubMed]
- Girardi, E.; Lefèvre, M.; Chane-Woon-Ming, B.; Paro, S.; Claydon, B.; Imler, J.-L.; Meignin, C.; Pfeffer, S. Cross-species comparative analysis of dicer proteins during sindbis virus infection. Sci. Rep. 2015, 5, 10693. [Google Scholar] [CrossRef] [PubMed]
- Ma, E.; MacRae, I.J.; Kirsch, J.F.; Doudna, J.A. Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol 2008, 380, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Flemr, M.; Malik, R.; Franke, V.; Nejepinska, J.; Sedlacek, R.; Vlahovicek, K.; Svoboda, P. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell 2013, 155, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Wang, H.; Ji, Y.; Yang, J.; Xu, S.; Huang, X.; Wang, Z.; Qin, L.; Tien, P.; Zhou, X.; et al. The nucleocapsid protein of coronaviruses acts as a viral suppressor of RNA silencing in mammalian cells. J. Virol 2015, 89, 9029–9043. [Google Scholar] [CrossRef] [PubMed]
- Haasnoot, J.; de Vries, W.; Geutjes, E.J.; Prins, M.; de Haan, P.; Berkhout, B. The ebola virus vp35 protein is a suppressor of RNA silencing. PLoS Pathog 2007, 3, e86. [Google Scholar] [CrossRef]
- Li, Y.; Basavappa, M.; Lu, J.; Dong, S.; Cronkite, D.A.; Prior, J.T.; Reinecker, H.C.; Hertzog, P.; Han, Y.; Li, W.X.; et al. Induction and suppression of antiviral RNA interference by influenza a virus in mammalian cells. Nat. Microbiol 2016, 2, 16250. [Google Scholar] [CrossRef]
- Andersson, M.G.; Haasnoot, P.C.J.; Xu, N.; Berenjian, S.; Berkhout, B.; Akusjärvi, G. Suppression of RNA interference by adenovirus virus-associated RNA. J. Virol. 2005, 79, 9556–9565. [Google Scholar] [CrossRef]
- Lu, S.; Cullen, B.R. Adenovirus va1 noncoding RNA can inhibit small interfering RNA and microRNA biogenesis. J. Virol. 2004, 78, 12868–12876. [Google Scholar] [CrossRef]
- Kitajewski, J.; Schneider, R.J.; Safer, B.; Munemitsu, S.M.; Samuel, C.E.; Thimmappaya, B.; Shenk, T. Adenovirus vai RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eif-2 alpha kinase. Cell 1986, 45, 195–200. [Google Scholar] [CrossRef]
- O’Malley, R.P.; Mariano, T.M.; Siekierka, J.; Mathews, M.B. A mechanism for the control of protein synthesis by adenovirus va RNAi. Cell 1986, 44, 391–400. [Google Scholar] [CrossRef]
- Machitani, M.; Sakurai, F.; Wakabayashi, K.; Tomita, K.; Tachibana, M.; Mizuguchi, H. Dicer functions as an antiviral system against human adenoviruses via cleavage of adenovirus-encoded noncoding RNA. Sci Rep. 2016, 6, 27598. [Google Scholar] [CrossRef]
- Ablasser, A.; Gulen, M.F. The role of cgas in innate immunity and beyond. J. Mol. Med. 2016, 94, 1085–1093. [Google Scholar] [CrossRef]
- Lee-Kirsch, M.A. The type i interferonopathies. Annu Rev. Med. 2017, 68, 297–315. [Google Scholar] [CrossRef]
- Forster, S.C.; Tate, M.D.; Hertzog, P.J. MicroRNA as type i interferon-regulated transcripts and modulators of the innate immune response. Front. Immunol 2015, 6, 334. [Google Scholar] [CrossRef]
- Li, Y.; Shi, X. MicroRNAs in the regulation of tlr and rig-i pathways. Cell Mol. Immunol 2013, 10, 65–71. [Google Scholar] [CrossRef]
- Momen-Heravi, F.; Bala, S. MiRNA regulation of innate immunity. J. Leukoc Biol 2018. [Google Scholar] [CrossRef]
- Chiappinelli, K.B.; Haynes, B.C.; Brent, M.R.; Goodfellow, P.J. Reduced dicer1 elicits an interferon response in endometrial cancer cells. Mol. Cancer Res. 2012, 10, 316–325. [Google Scholar] [CrossRef]
- Papadopoulou, A.S.; Dooley, J.; Linterman, M.A.; Pierson, W.; Ucar, O.; Kyewski, B.; Zuklys, S.; Hollander, G.A.; Matthys, P.; Gray, D.H.D.; et al. The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via mir-29a mediated suppression of the ifn-α receptor. Nat. Immunol. 2012, 13, 181–187. [Google Scholar] [CrossRef]
- Varol, D.; Mildner, A.; Blank, T.; Maggio, N.; Prinz, M.; Jung, S. Dicer deficiency differentially impacts microglia of the developing and adult brain. Immunity 2017, 46, 1030–1044. [Google Scholar] [CrossRef]
- Seo, G.J.; Kincaid, R.P.; Phanaksri, T.; Burke, J.M.; Pare, J.M.; Cox, J.E.; Hsiang, T.Y.; Krug, R.M.; Sullivan, C.S. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe 2013, 14, 435–445. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.; Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. Nf-kappab-dependent induction of microRNA mir-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, J.; Wang, H.; Shi, J.; Wu, K.; Liu, S.; Liu, Y.; Wu, J. Hcv-induced mir-21 contributes to evasion of host immune system by targeting myd88 and irak1. PLoS Pathog 2013, 9, e1003248. [Google Scholar] [CrossRef]
- Hou, J.; Wang, P.; Lin, L.; Liu, X.; Ma, F.; An, H.; Wang, Z.; Cao, X. MicroRNA-146a feedback inhibits rig-i-dependent type i ifn production in macrophages by targeting traf6, irak1, and irak2. J. Immunol 2009, 183, 2150–2158. [Google Scholar] [CrossRef]
- Tili, E.; Michaille, J.J.; Cimino, A.; Costinean, S.; Dumitru, C.D.; Adair, B.; Fabbri, M.; Alder, H.; Liu, C.G.; Calin, G.A.; et al. Modulation of mir-155 and mir-125b levels following lipopolysaccharide/tnf-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol 2007, 179, 5082–5089. [Google Scholar] [CrossRef]
- Witwer, K.W.; Sisk, J.M.; Gama, L.; Clements, J.E. MicroRNA regulation of ifn-beta protein expression: Rapid and sensitive modulation of the innate immune response. J. Immunol. 2010, 184, 2369–2376. [Google Scholar] [CrossRef]
- Witteveldt, J.; Ivens, A.; Macias, S. Inhibition of microprocessor function during the activation of the type i interferon response. Cell Rep. 2018, 23, 3275–3285. [Google Scholar] [CrossRef]
- Wiesen, J.L.; Tomasi, T.B. Dicer is regulated by cellular stresses and interferons. Mol. Immunol. 2009, 46, 1222–1228. [Google Scholar] [CrossRef]
- Machitani, M.; Sakurai, F.; Wakabayashi, K.; Takayama, K.; Tachibana, M.; Mizuguchi, H. Type i interferons impede short hairpin RNA-mediated RNAi via inhibition of dicer-mediated processing to small interfering RNA. Mol. Ther. Nucleic Acids 2017, 6, 173–182. [Google Scholar] [CrossRef]
- van der Veen, A.G.; Maillard, P.V.; Schmidt, J.M.; Lee, S.A.; Deddouche-Grass, S.; Borg, A.; Kjaer, S.; Snijders, A.P.; Reis e Sousa, C. The rig-i-like receptor lgp2 inhibits dicer-dependent processing of long double-stranded RNA and blocks RNA interference in mammalian cells. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Ota, H.; Sakurai, M.; Gupta, R.; Valente, L.; Wulff, B.E.; Ariyoshi, K.; Iizasa, H.; Davuluri, R.V.; Nishikura, K. Adar1 forms a complex with dicer to promote microRNA processing and RNA-induced gene silencing. Cell 2013, 153, 575–589. [Google Scholar] [CrossRef]
- Kawahara, Y.; Zinshteyn, B.; Chendrimada, T.P.; Shiekhattar, R.; Nishikura, K. Rna editing of the microRNA-151 precursor blocks cleavage by the dicer-trbp complex. EMBO Rep. 2007, 8, 763–769. [Google Scholar] [CrossRef]
- Yang, W.; Chendrimada, T.P.; Wang, Q.; Higuchi, M.; Seeburg, P.H.; Shiekhattar, R.; Nishikura, K. Modulation of microRNA processing and expression through RNA editing by adar deaminases. Nat. Struct Mol. Biol 2006, 13, 13–21. [Google Scholar] [CrossRef]
- Patel, R.C.; Sen, G.C. Pact, a protein activator of the interferon-induced protein kinase, pkr. EMBO J. 1998, 17, 4379–4390. [Google Scholar] [CrossRef]
- Park, H.; Davies, M.V.; Langland, J.O.; Chang, H.W.; Nam, Y.S.; Tartaglia, J.; Paoletti, E.; Jacobs, B.L.; Kaufman, R.J.; Venkatesan, S. Tar RNA-binding protein is an inhibitor of the interferon-induced protein kinase pkr. Proc. Natl. Acad. Sci. USA 1994, 91, 4713–4717. [Google Scholar] [CrossRef]
- Sanghvi, V.R.; Steel, L.F. The cellular tar RNA binding protein, trbp, promotes hiv-1 replication primarily by inhibiting the activation of double-stranded RNA-dependent kinase pkr. J. Virol. 2011, 85, 12614–12621. [Google Scholar] [CrossRef]
- Chakravarthy, S.; Sternberg, S.H.; Kellenberger, C.A.; Doudna, J.A. Substrate-specific kinetics of dicer-catalyzed RNA processing. J. Mol. Biol. 2010, 404, 392–402. [Google Scholar] [CrossRef]
- Kok, K.H.; Ng, M.-H.J.; Ching, Y.-P.; Jin, D.-Y. Human trbp and pact directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J. Biol. Chem. 2007, 282, 17649–17657. [Google Scholar] [CrossRef]
- Lee, H.Y.; Zhou, K.; Smith, A.M.; Noland, C.L.; Doudna, J.A. Differential roles of human dicer-binding proteins trbp and pact in small RNA processing. Nucleic Acids Res. 2013, 41, 6568–6576. [Google Scholar] [CrossRef]
- Kok, K.-H.; Lui, P.-Y.; Ng, M.-H.J.; Siu, K.-L.; Au, S.W.N.; Jin, D.-Y. The double-stranded RNA-binding protein pact functions as a cellular activator of rig-i to facilitate innate antiviral response. Cell Host Microbe 2011, 9, 299–309. [Google Scholar] [CrossRef]
- Saunders, L.R.; Barber, G.N. The dsRNA binding protein family: Critical roles, diverse cellular functions. FASEB J. 2003, 17, 961–983. [Google Scholar] [CrossRef]
- Kolakofsky, D.; Kowalinski, E.; Cusack, S. A structure-based model of rig-i activation. RNA 2012, 18, 2118–2127. [Google Scholar] [CrossRef]
- Rawling, D.C.; Pyle, A.M. Parts, assembly and operation of the rig-i family of motors. Curr. Opin. Struct. Biol. 2014, 25, 25–33. [Google Scholar] [CrossRef]
- Burke, D.C.; Graham, C.F.; Lehman, J.M. Appearance of interferon inducibility and sensitivity during differentiation of murine teratocarcinoma cells in vitro. Cell 1978, 13, 243–248. [Google Scholar] [CrossRef]
- Billy, E.; Brondani, V.; Zhang, H.; Muller, U.; Filipowicz, W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl. Acad. Sci. USA 2001, 98, 14428–14433. [Google Scholar] [CrossRef]
- Paddison, P.J.; Caudy, A.A.; Hannon, G.J. Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl. Acad. Sci. USA 2002, 99, 1443–1448. [Google Scholar] [CrossRef]
- Maillard, P.V.; Van der Veen, A.G.; Deddouche-Grass, S.; Rogers, N.C.; Merits, A.; e Sousa, C.R. Inactivation of the type i interferon pathway reveals long double-stranded RNA-mediated RNA interference in mammalian cells. EMBO J. 2016, 35, 2505–2518. [Google Scholar] [CrossRef]
- Hou, W.; Tian, Q.; Zheng, J.; Bonkovsky, H.L. MicroRNA-196 represses bach1 protein and hepatitis c virus gene expression in human hepatoma cells expressing hepatitis c viral proteins. Hepatology 2010, 51, 1494–1504. [Google Scholar] [CrossRef]
- Murakami, Y.; Aly, H.H.; Tajima, A.; Inoue, I.; Shimotohno, K. Regulation of the hepatitis c virus genome replication by mir-199a. J. Hepatol 2009, 50, 453–460. [Google Scholar] [CrossRef]
- Pedersen, I.M.; Cheng, G.; Wieland, S.; Volinia, S.; Croce, C.M.; Chisari, F.V.; David, M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 2007, 449, 919–922. [Google Scholar] [CrossRef]
- Russo, A.; Potenza, N. Antiviral effects of human microRNAs and conservation of their target sites. FEBS Lett 2011, 585, 2551–2555. [Google Scholar] [CrossRef]
- Swaminathan, G.; Martin-Garcia, J.; Navas-Martin, S. Rna viruses and microRNAs: Challenging discoveries for the 21st century. Physiol Genom. 2013, 45, 1035–1048. [Google Scholar] [CrossRef]
- Lecellier, C.H.; Dunoyer, P.; Arar, K.; Lehmann-Che, J.; Eyquem, S.; Himber, C.; Saib, A.; Voinnet, O. A cellular microRNA mediates antiviral defense in human cells. Science 2005, 308, 557–560. [Google Scholar] [CrossRef]
- Bennasser, Y.; Le, S.-Y.; Benkirane, M.; Jeang, K.-T. Evidence that hiv-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 2005, 22, 607–619. [Google Scholar] [CrossRef]
- Bennasser, Y.; Yeung, M.L.; Jeang, K.T. Hiv-1 tar RNA subverts RNA interference in transfected cells through sequestration of tar RNA-binding protein, trbp. J. Biol Chem 2006, 281, 27674–27678. [Google Scholar] [CrossRef]
- de Vries, W.; Haasnoot, J.; Fouchier, R.; de Haan, P.; Berkhout, B. Differential RNA silencing suppression activity of ns1 proteins from different influenza a virus strains. J. Gen. Virol 2009, 90, 1916–1922. [Google Scholar] [CrossRef]
- Backes, S.; Shapiro, J.S.; Sabin, L.R.; Pham, A.M.; Reyes, I.; Moss, B.; Cherry, S.; tenOever, B.R. Degradation of host microRNAs by poxvirus poly(a) polymerase reveals terminal RNA methylation as a protective antiviral mechanism. Cell Host Microbe 2012, 12, 200–210. [Google Scholar] [CrossRef]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis c virus RNA abundance by a liver-specific microRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef]
- Sedano, C.D.; Sarnow, P. Hepatitis c virus subverts liver-specific mir-122 to protect the viral genome from exoribonuclease xrn2. Cell Host Microbe 2014, 16, 257–264. [Google Scholar] [CrossRef]
- Jopling, C.L. Regulation of hepatitis c virus by microRNA-122. Biochem Soc. Trans. 2008, 36, 1220–1223. [Google Scholar] [CrossRef]
- Wilson, J.A.; Zhang, C.; Huys, A.; Richardson, C.D. Human ago2 is required for efficient microRNA 122 regulation of hepatitis c virus RNA accumulation and translation. J. Virol 2011, 85, 2342–2350. [Google Scholar] [CrossRef]
- Ziv, O.; Gabryelska, M.M.; Lun, A.T.L.; Gebert, L.F.R.; Sheu-Gruttadauria, J.; Meredith, L.W.; Liu, Z.Y.; Kwok, C.K.; Qin, C.F.; MacRae, I.J.; et al. Comrades determines in vivo RNA structures and interactions. Nat. Methods 2018, 15, 785–788. [Google Scholar] [CrossRef]
- Chiang, K.; Liu, H.; Rice, A.P. Mir-132 enhances hiv-1 replication. Virology 2013, 438, 1–4. [Google Scholar] [CrossRef]
- O’Connor, C.M.; Vanicek, J.; Murphy, E.A. Host microRNA regulation of human cytomegalovirus immediate early protein translation promotes viral latency. J. Virol 2014, 88, 5524–5532. [Google Scholar] [CrossRef]
- White, E.; Schlackow, M.; Kamieniarz-Gdula, K.; Proudfoot, N.J.; Gullerova, M. Human nuclear dicer restricts the deleterious accumulation of endogenous double-stranded RNA. Nat. Struct Mol. Biol 2014, 21, 552–559. [Google Scholar] [CrossRef]
- Matskevich, A.A.; Moelling, K. Dicer is involved in protection against influenza a virus infection. J. Gen. Virol. 2007, 88, 2627–2635. [Google Scholar] [CrossRef]
- Bucher, E.; Hemmes, H.; de Haan, P.; Goldbach, R.; Prins, M. The influenza a virus ns1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J. Gen. Virol 2004, 85, 983–991. [Google Scholar] [CrossRef]
- Ma, Y.; Ouyang, J.; Wei, J.; Maarouf, M.; Chen, J.L. Involvement of host non-coding RNAs in the pathogenesis of the influenza virus. Int. J. Mol. Sci 2016, 18, 39. [Google Scholar] [CrossRef]
- Chen, J.S.; Li, H.C.; Lin, S.I.; Yang, C.H.; Chien, W.Y.; Syu, C.L.; Lo, S.Y. Cleavage of dicer protein by i7 protease during vaccinia virus infection. PLoS ONE 2015, 10, e0120390. [Google Scholar] [CrossRef]
- Bernstein, E.; Kim, S.Y.; Carmell, M.A.; Murchison, E.P.; Alcorn, H.; Li, M.Z.; Mills, A.A.; Elledge, S.J.; Anderson, K.V.; Hannon, G.J. Dicer is essential for mouse development. Nat. Genet. 2003, 35, 215–217. [Google Scholar] [CrossRef]
- Otsuka, M.; Jing, Q.; Georgel, P.; New, L.; Chen, J.; Mols, J.; Kang, Y.J.; Jiang, Z.; Du, X.; Cook, R.; et al. Hypersusceptibility to vesicular stomatitis virus infection in dicer1-deficient mice is due to impaired mir24 and mir93 expression. Immunity 2007, 27, 123–134. [Google Scholar] [CrossRef]
- Ostermann, E.; Tuddenham, L.; Macquin, C.; Alsaleh, G.; Schreiber-Becker, J.; Tanguy, M.; Bahram, S.; Pfeffer, S.; Georgel, P. Deregulation of type i ifn-dependent genes correlates with increased susceptibility to cytomegalovirus acute infection of dicer mutant mice. PLoS ONE 2012, 7, e43744. [Google Scholar] [CrossRef]
- Aguado, L.C.; Schmid, S.; Sachs, D.; Shim, J.V.; Lim, J.K.; tenOever, B.R. MicroRNA function is limited to cytokine control in the acute response to virus infection. Cell Host Microbe 2015, 18, 714–722. [Google Scholar] [CrossRef]
- Bogerd, H.P.; Whisnant, A.W.; Kennedy, E.M.; Flores, O.; Cullen, B.R. Derivation and characterization of dicer- and microRNA-deficient human cells. RNA 2014, 20, 923–937. [Google Scholar] [CrossRef]
- Barlow, D.P.; Randle, B.J.; Burke, D.C. Interferon synthesis in the early post-implantation mouse embryo. Differentiation 1984, 27, 229–235. [Google Scholar] [CrossRef]
- Murchison, E.P.; Partridge, J.F.; Tam, O.H.; Cheloufi, S.; Hannon, G.J. Characterization of dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA 2005, 102, 12135–12140. [Google Scholar] [CrossRef]
- Bodak, M.; Cirera-Salinas, D.; Yu, J.; Ngondo, R.P.; Ciaudo, C. Dicer, a new regulator of pluripotency exit and line-1 elements in mouse embryonic stem cells. FEBS Open Bio 2017, 7, 204–220. [Google Scholar] [CrossRef]
- Calabrese, J.M.; Seila, A.C.; Yeo, G.W.; Sharp, P.A. Rna sequence analysis defines dicer’s role in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 18097–18102. [Google Scholar] [CrossRef]
- Witteveldt, J.; Knol, L.I.; Macias, S. MicroRNA-deficient embryonic stem cells acquire a functional interferon response. bioRxiv 2018, 501254. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).