Local Tandem Repeat Expansion in Xist RNA as a Model for the Functionalisation of ncRNA
Abstract
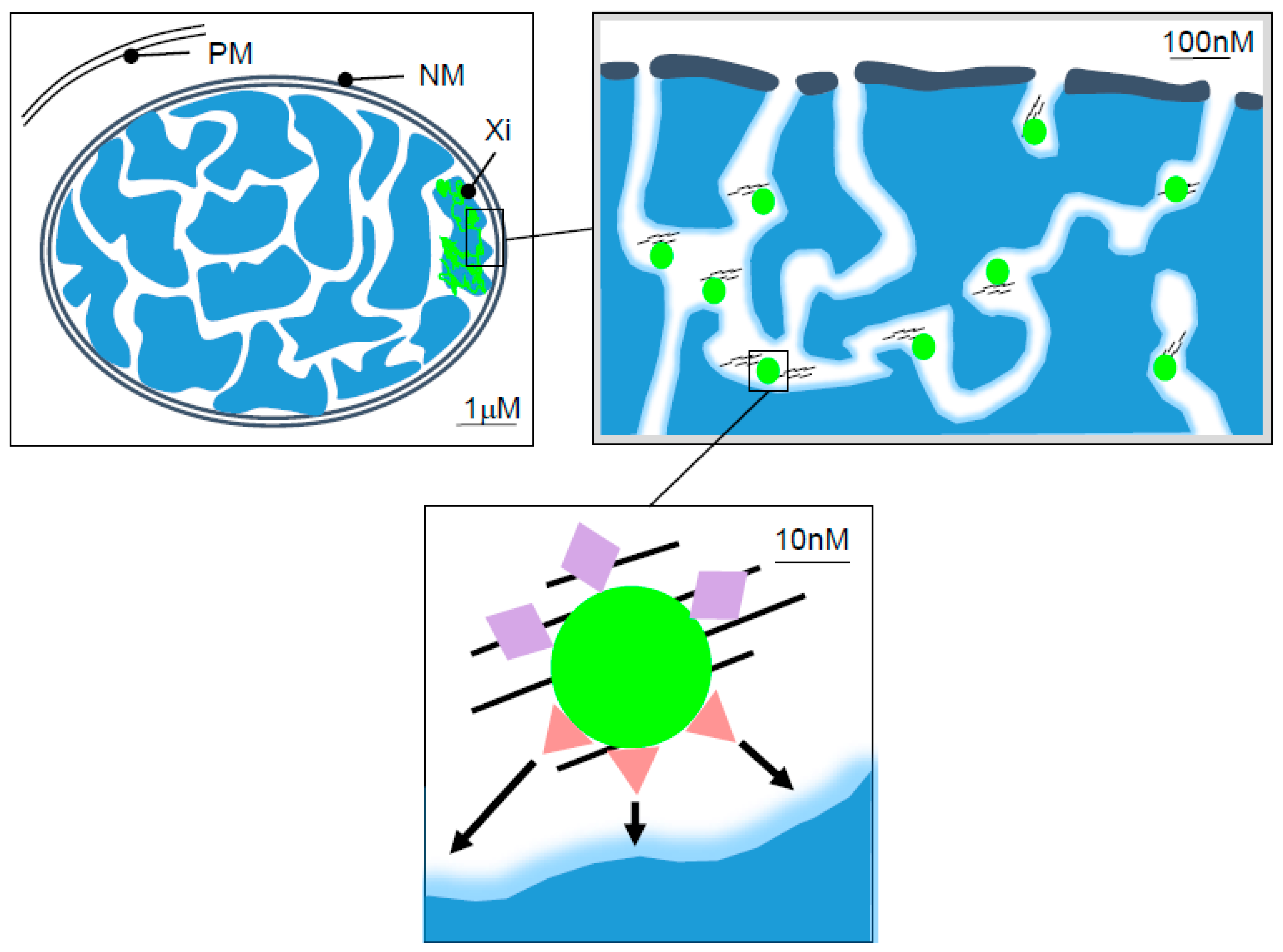
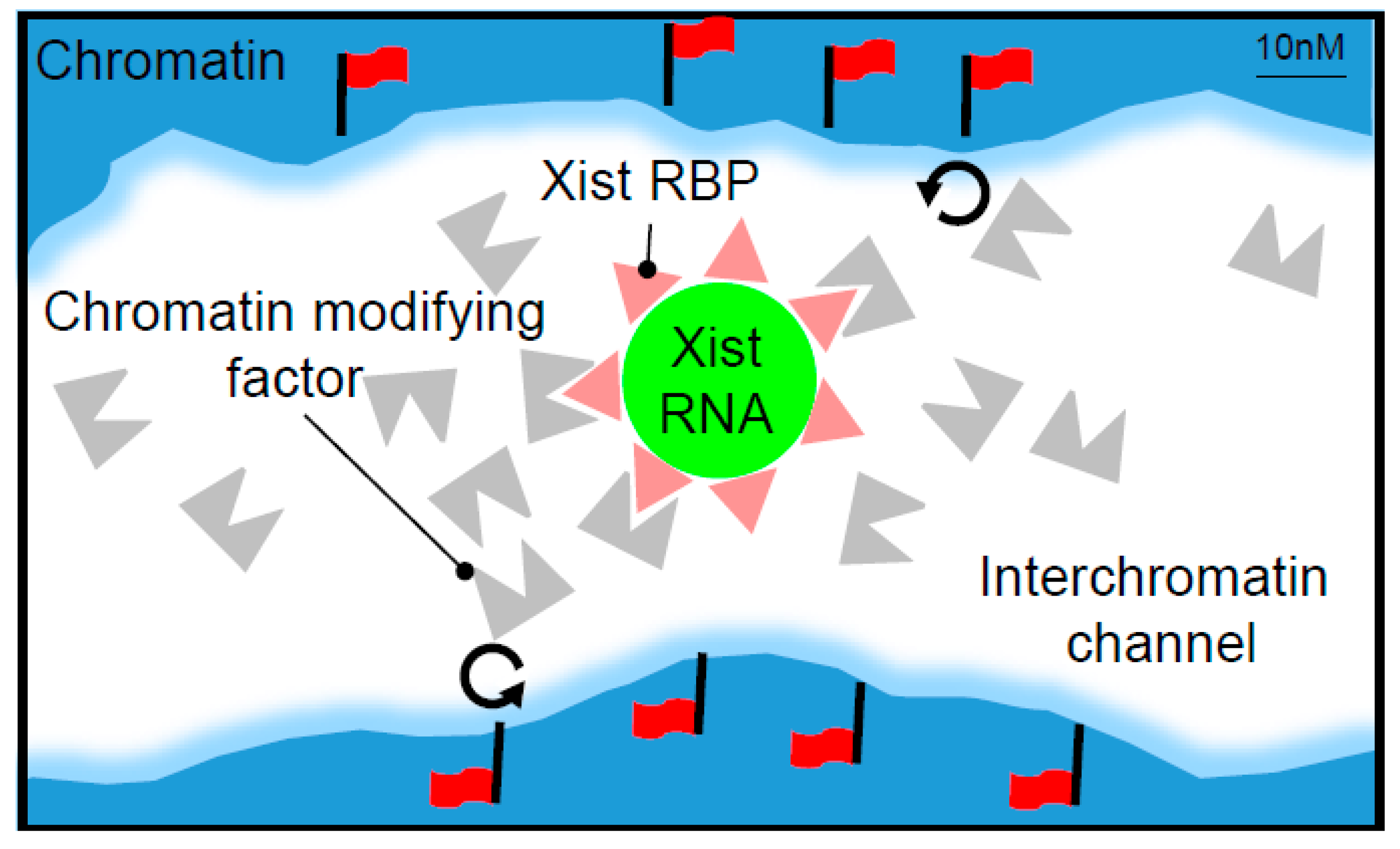
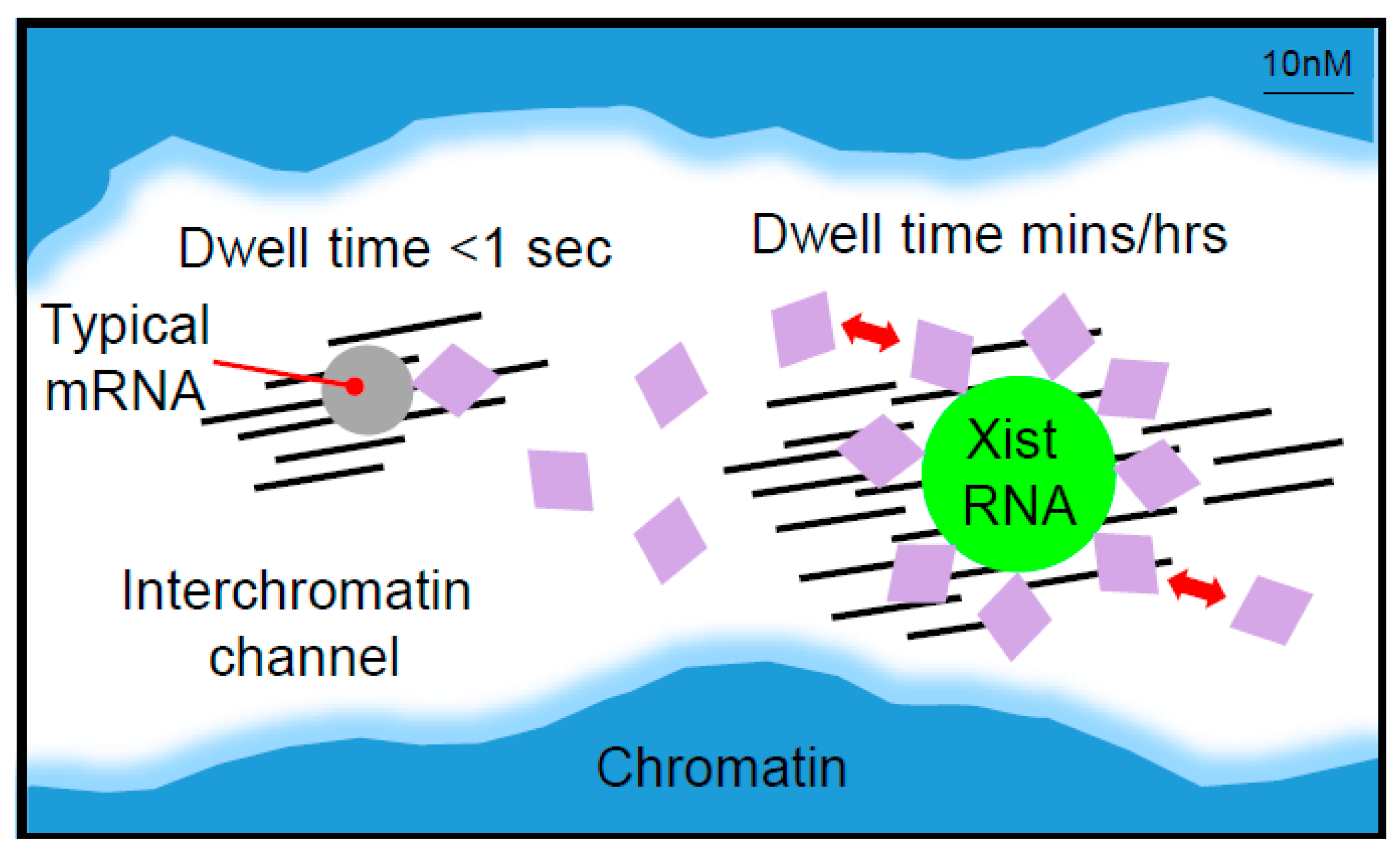
:1. Introduction
2. Interaction of the RBPs Spen and Rbm15 with the Xist A-Repeat Element
3. Recruitment of the Polycomb System by hnRNPK Bound to the Xist B/C-Repeat
4. Ciz1, an Anchoring Factor, Is Recruited by the Xist E-Repeat
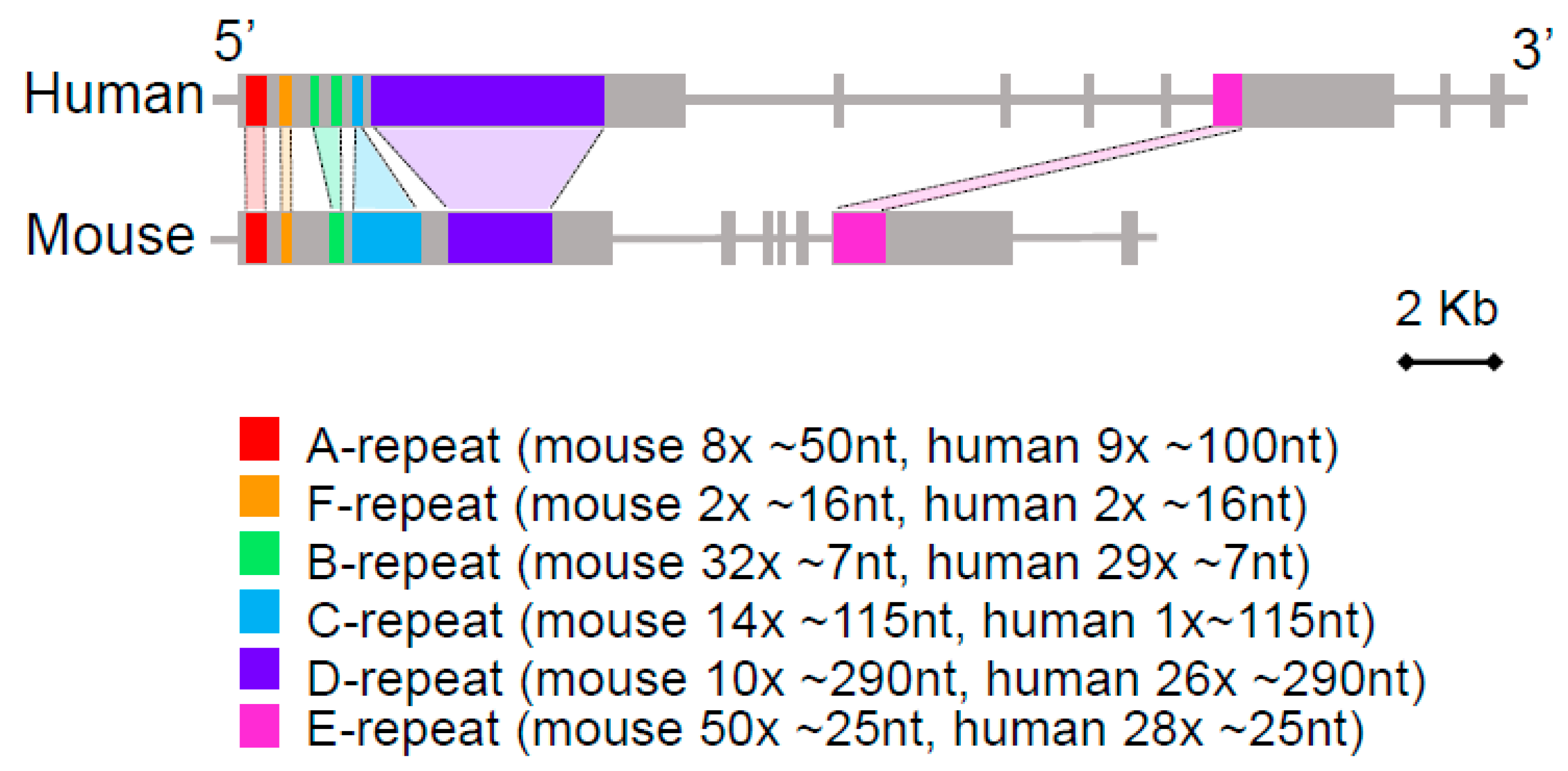
5. Local Tandem Repeat Amplification and the Evolution of Xist RNA Function
6. A Role for Local Tandem Repeat Expansion in Other LncRNAs
7. Summary
Funding
Acknowledgments
Conflicts of Interest
References
- Lyon, M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 1961, 190, 372–373. [Google Scholar] [CrossRef] [PubMed]
- Brockdorff, N.; Ashworth, A.; Kay, G.F.; McCabe, V.M.; Norris, D.P.; Cooper, P.J.; Swift, S.; Rastan, S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 1992, 71, 515–526. [Google Scholar] [CrossRef]
- Penny, G.D.; Kay, G.F.; Sheardown, S.A.; Rastan, S.; Brockdorff, N. Requirement for Xist in X chromosome inactivation. Nature 1996, 379, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.J.; Hendrich, B.D.; Rupert, J.L.; Lafreniere, R.G.; Xing, Y.; Lawrence, J.; Willard, H.F. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 1992, 71, 527–542. [Google Scholar] [CrossRef]
- Lee, J.T.; Jaenisch, R. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature 1997, 386, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Duthie, S.M.; Nesterova, T.B.; Formstone, E.J.; Keohane, A.M.; Turner, B.M.; Zakian, S.M.; Brockdorff, N. Xist RNA exhibits a banded localization on the inactive X chromosome and is excluded from autosomal material in cis. Hum. Mol. Genet. 1999, 8, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Cerase, A.; Pintacuda, G.; Tattermusch, A.; Avner, P. Xist localization and function: New insights from multiple levels. Genome Biol. 2015, 16, 166. [Google Scholar] [CrossRef] [PubMed]
- Wutz, A.; Rasmussen, T.P.; Jaenisch, R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002, 30, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Brockdorff, N.; Kawano, S.; Tsutui, K.; Tsutui, K.; Nakagawa, S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell 2010, 19, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Ridings-Figueroa, R.; Stewart, E.R.; Nesterova, T.B.; Coker, H.; Pintacuda, G.; Godwin, J.; Wilson, R.; Haslam, A.; Lilley, F.; Ruigrok, R.; et al. The nuclear matrix protein CIZ1 facilitates localization of Xist RNA to the inactive X-chromosome territory. Genes Dev. 2017, 31, 876–888. [Google Scholar] [CrossRef] [PubMed]
- Sunwoo, H.; Colognori, D.; Froberg, J.E.; Jeon, Y.; Lee, J.T. Repeat E anchors Xist RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1). Proc. Natl. Acad. Sci. USA 2017, 114, 10654–10659. [Google Scholar] [CrossRef] [PubMed]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Zhang, Q.C.; da Rocha, S.T.; Flynn, R.A.; Bharadwaj, M.; Calabrese, J.M.; Magnuson, T.; Heard, E.; Chang, H.Y. Systematic discovery of Xist RNA binding proteins. Cell 2015, 161, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Moindrot, B.; Cerase, A.; Coker, H.; Masui, O.; Grijzenhout, A.; Pintacuda, G.; Schermelleh, L.; Nesterova, T.B.; Brockdorff, N. A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep. 2015, 12, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Monfort, A.; Di Minin, G.; Postlmayr, A.; Freimann, R.; Arieti, F.; Thore, S.; Wutz, A. Identification of Spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 2015, 12, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Minajigi, A.; Froberg, J.E.; Wei, C.; Sunwoo, H.; Kesner, B.; Colognori, D.; Lessing, D.; Payer, B.; Boukhali, M.; Haas, W.; et al. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 2015, 349. [Google Scholar] [CrossRef] [PubMed]
- Pintacuda, G.; Wei, G.; Roustan, C.; Kirmizitas, B.A.; Solcan, N.; Cerase, A.; Castello, A.; Mohammed, S.; Moindrot, B.; Nesterova, T.B.; et al. hnRNPK recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol. Cell 2017, 68, 955–969. [Google Scholar] [CrossRef] [PubMed]
- Clemson, C.M.; McNeil, J.A.; Willard, H.F.; Lawrence, J.B. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 1996, 132, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Cerase, A.; Smeets, D.; Tang, Y.A.; Gdula, M.; Kraus, F.; Spivakov, M.; Moindrot, B.; Leleu, M.; Tattermusch, A.; Demmerle, J.; et al. Spatial separation of Xist RNA and polycomb proteins revealed by superresolution microscopy. Proc. Natl. Acad. Sci. USA 2014, 111, 2235–2240. [Google Scholar] [CrossRef] [PubMed]
- Smeets, D.; Markaki, Y.; Schmid, V.J.; Kraus, F.; Tattermusch, A.; Cerase, A.; Sterr, M.; Fiedler, S.; Demmerle, J.; Popken, J.; et al. Three-dimensional super-resolution microscopy of the inactive X chromosome territory reveals a collapse of its active nuclear compartment harboring distinct Xist RNA foci. Epigenetics Chromatin 2014, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Sunwoo, H.; Wu, J.Y.; Lee, J.T. The Xist RNA-PRC2 complex at 20-nm resolution reveals a low Xist stoichiometry and suggests a hit-and-run mechanism in mouse cells. Proc. Natl. Acad. Sci. USA 2015, 112, E4216–E4225. [Google Scholar] [CrossRef] [PubMed]
- Nesterova, T.B.; Slobodyanyuk, S.Y.; Elisaphenko, E.A.; Shevchenko, A.I.; Johnston, C.; Pavlova, M.E.; Rogozin, I.B.; Kolesnikov, N.N.; Brockdorff, N.; Zakian, S.M. Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res. 2001, 11, 833–849. [Google Scholar] [CrossRef] [PubMed]
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Oswald, F.; Kostezka, U.; Astrahantseff, K.; Bourteele, S.; Dillinger, K.; Zechner, U.; Ludwig, L.; Wilda, M.; Hameister, H.; Knochel, W.; et al. SHARP is a novel component of the Notch/RBP-Jκ signalling pathway. EMBO J. 2002, 21, 5417–5426. [Google Scholar] [CrossRef] [PubMed]
- Kuang, B.; Wu, S.C.; Shin, Y.; Luo, L.; Kolodziej, P. Split ends encodes large nuclear proteins that regulate neuronal cell fate and axon extension in the Drosophila embryo. Development 2000, 127, 1517–1529. [Google Scholar] [PubMed]
- Ariyoshi, M.; Schwabe, J.W. A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes Dev. 2003, 17, 1909–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Downes, M.; Xie, W.; Kao, H.Y.; Ordentlich, P.; Tsai, C.C.; Hon, M.; Evans, R.M. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001, 15, 1140–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Z.; Zhang, Q.C.; Lee, B.; Flynn, R.A.; Smith, M.A.; Robinson, J.T.; Davidovich, C.; Gooding, A.R.; Goodrich, K.J.; Mattick, J.S.; et al. RNA duplex map in living cells reveals higher order transcriptome structure. Cell 2016, 165, 1267–1279. [Google Scholar] [CrossRef] [PubMed]
- Mercher, T.; Coniat, M.B.; Monni, R.; Mauchauffe, M.; Nguyen Khac, F.; Gressin, L.; Mugneret, F.; Leblanc, T.; Dastugue, N.; Berger, R.; et al. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc. Natl. Acad. Sci. USA 2001, 98, 5776–5779. [Google Scholar] [CrossRef] [PubMed]
- Bomsztyk, K.; Denisenko, O.; Ostrowski, J. hnRNP K: One protein multiple processes. Bioessays 2004, 26, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.; Pintacuda, G.; Masui, O.; Koseki, Y.; Gdula, M.; Cerase, A.; Brown, D.; Mould, A.; Innocent, C.; Nakayama, M.; et al. PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science 2017, 356, 1081–1084. [Google Scholar] [CrossRef] [PubMed]
- Cirillo, D.; Blanco, M.; Armaos, A.; Buness, A.; Avner, P.; Guttman, M.; Cerase, A.; Tartaglia, G.G. Quantitative predictions of protein interactions with long noncoding RNAs. Nat. Methods 2016, 14, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Paziewska, A.; Wyrwicz, L.S.; Bujnicki, J.M.; Bomsztyk, K.; Ostrowski, J. Cooperative binding of the hnRNP K three KH domains to mRNA targets. FEBS Lett. 2004, 577, 134–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moritz, B.; Lilie, H.; Naarmann-de Vries, I.S.; Urlaub, H.; Wahle, E.; Ostareck-Lederer, A.; Ostareck, D.H. Biophysical and biochemical analysis of hnRNP K: Arginine methylation, reversible aggregation and combinatorial binding to nucleic acids. Biol. Chem. 2014, 395, 837–853. [Google Scholar] [CrossRef] [PubMed]
- Marks, H.; Chow, J.C.; Denissov, S.; Francoijs, K.J.; Brockdorff, N.; Heard, E.; Stunnenberg, H.G. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res. 2009, 19, 1361–1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calabrese, J.M.; Sun, W.; Song, L.; Mugford, J.W.; Williams, L.; Yee, D.; Starmer, J.; Mieczkowski, P.; Crawford, G.E.; Magnuson, T. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell 2012, 151, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Pinter, S.F.; Sadreyev, R.I.; Yildirim, E.; Jeon, Y.; Ohsumi, T.K.; Borowsky, M.; Lee, J.T. Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res. 2012, 22, 1864–1876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendenhall, E.M.; Koche, R.P.; Truong, T.; Zhou, V.W.; Issac, B.; Chi, A.S.; Ku, M.; Bernstein, B.E. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010, 6, e1001244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakaguchi, T.; Hasegawa, Y.; Brockdorff, N.; Tsutsui, K.; Tsutsui, K.M.; Sado, T.; Nakagawa, S. Control of chromosomal localization of Xist by hnRNP U family molecules. Dev. Cell 2016, 39, 11–12. [Google Scholar] [CrossRef] [PubMed]
- Kolpa, H.J.; Fackelmayer, F.O.; Lawrence, J.B. SAF-A Requirement in anchoring XIST RNA to chromatin varies in transformed and primary cells. Dev. Cell 2016, 39, 9–10. [Google Scholar] [CrossRef] [PubMed]
- Pauzaite, T.; Thacker, U.; Tollitt, J.; Copeland, N.A. Emerging roles for Ciz1 in cell cycle regulation and as a driver of tumorigenesis. Biomolecules 2016, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Ainscough, J.F.; Rahman, F.A.; Sercombe, H.; Sedo, A.; Gerlach, B.; Coverley, D. C-terminal domains deliver the DNA replication factor Ciz1 to the nuclear matrix. J. Cell Sci. 2007, 120, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, R.S.; Boteva, L.; Soares, D.C.; Naughton, C.; Dun, A.R.; Buckle, A.; Ramsahoye, B.; Bruton, P.C.; Saleeb, R.S.; Arnedo, M.; et al. SAF-A regulates interphase chromosome structure through oligomerization with chromatin-associated RNAs. Cell 2017, 169, 1214–1227.e18. [Google Scholar] [CrossRef] [PubMed]
- Beletskii, A.; Hong, Y.K.; Pehrson, J.; Egholm, M.; Strauss, W.M. PNA interference mapping demonstrates functional domains in the noncoding RNA Xist. Proc. Natl. Acad. Sci. USA 2001, 98, 9215–9220. [Google Scholar] [CrossRef] [PubMed]
- Sarma, K.; Levasseur, P.; Aristarkhov, A.; Lee, J.T. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc. Natl. Acad. Sci. USA 2010, 107, 22196–22201. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A. Evolution of vertebrate sex chromosomes and dosage compensation. Nat. Rev. Genet. 2016, 17, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Lubelsky, Y.; Ulitsky, I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature 2018, 555, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.; Mahadevaiah, S.K.; Khil, P.; Sangrithi, M.N.; Royo, H.; Duckworth, J.; McCarrey, J.R.; VandeBerg, J.L.; Renfree, M.B.; Taylor, W.; et al. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 2012, 487, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Hacisuleyman, E.; Shukla, C.J.; Weiner, C.L.; Rinn, J.L. Function and evolution of local repeats in the Firre locus. Nat. Commun. 2016, 7, 11021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hacisuleyman, E.; Goff, L.A.; Trapnell, C.; Williams, A.; Henao-Mejia, J.; Sun, L.; McClanahan, P.; Hendrickson, D.G.; Sauvageau, M.; Kelley, D.R.; et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014, 21, 198–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Kopp, F.; Chang, T.C.; Sataluri, A.; Chen, B.; Sivakumar, S.; Yu, H.; Xie, Y.; Mendell, J.T. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 2016, 164, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Tichon, A.; Gil, N.; Lubelsky, Y.; Havkin Solomon, T.; Lemze, D.; Itzkovitz, S.; Stern-Ginossar, N.; Ulitsky, I. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat. Commun. 2016, 7, 12209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Souquere, S.; Chujo, T.; Kobelke, S.; Chong, Y.S.; Fox, A.H.; Bond, C.S.; Nakagawa, S.; Pierron, G.; Hirose, T. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell 2018, 70, 1038–1053. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Sleutels, F.; Zwart, R.; Barlow, D.P. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 2002, 415, 810–813. [Google Scholar] [CrossRef] [PubMed]
- Redrup, L.; Branco, M.R.; Perdeaux, E.R.; Krueger, C.; Lewis, A.; Santos, F.; Nagano, T.; Cobb, B.S.; Fraser, P.; Reik, W. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development 2009, 136, 525–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Martens, J.H.; O’Sullivan, R.J.; Braunschweig, U.; Opravil, S.; Radolf, M.; Steinlein, P.; Jenuwein, T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005, 24, 800–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delihas, N. Formation of a family of long intergenic noncoding RNA genes with an embedded translocation breakpoint motif in human chromosomal low copy repeats of 22q11.2-Some surprises and questions. Non-Coding RNA 2018, 4, 16. [Google Scholar] [CrossRef] [PubMed]
- Kirk, J.M.; Kim, S.O.; Inoue, K.; Smola, M.J.; Lee, D.M.; Schertzer, M.D.; Wooten, J.S.; Baker, A.R.; Sprague, D.; Collins, D.W.; et al. Functional classification of long non-coding RNAs by k-mer content. Nat. Genet. 2018, 50, 1474–1482. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brockdorff, N. Local Tandem Repeat Expansion in Xist RNA as a Model for the Functionalisation of ncRNA. Non-Coding RNA 2018, 4, 28. https://doi.org/10.3390/ncrna4040028
Brockdorff N. Local Tandem Repeat Expansion in Xist RNA as a Model for the Functionalisation of ncRNA. Non-Coding RNA. 2018; 4(4):28. https://doi.org/10.3390/ncrna4040028
Chicago/Turabian StyleBrockdorff, Neil. 2018. "Local Tandem Repeat Expansion in Xist RNA as a Model for the Functionalisation of ncRNA" Non-Coding RNA 4, no. 4: 28. https://doi.org/10.3390/ncrna4040028
APA StyleBrockdorff, N. (2018). Local Tandem Repeat Expansion in Xist RNA as a Model for the Functionalisation of ncRNA. Non-Coding RNA, 4(4), 28. https://doi.org/10.3390/ncrna4040028




