Human XIST: Origin and Divergence of a cis-Acting Silencing RNA
Abstract
1. Introduction: Sex Chromosomes, the Need for Dosage Compensation and Long Non-Coding RNAs
2. Mammalian X-Chromosome Inactivation
3. XIST Origin: From Ancestral Gene to Mobile Elements
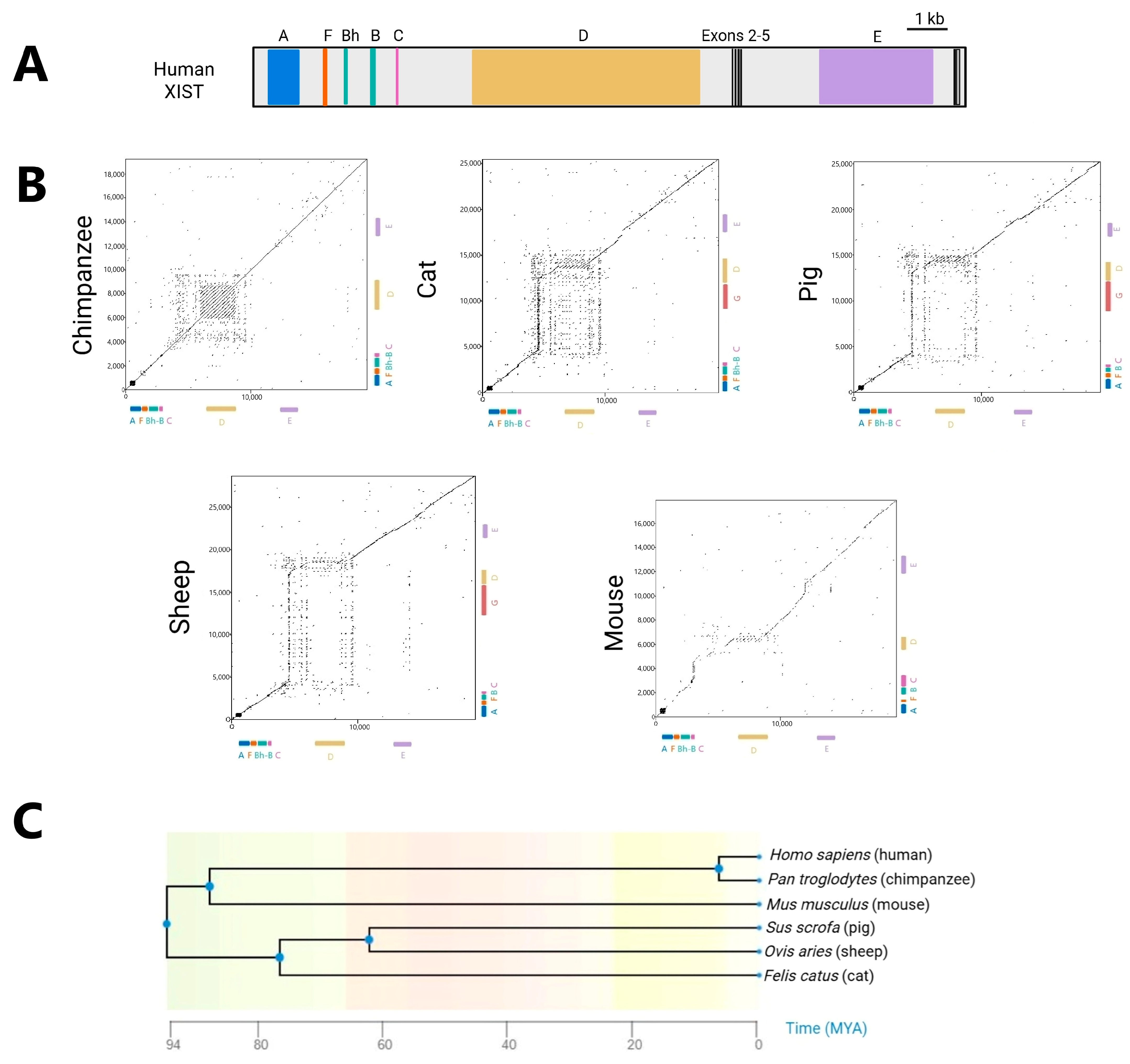
4. XIST Across Eutheria: Conservation and Divergence
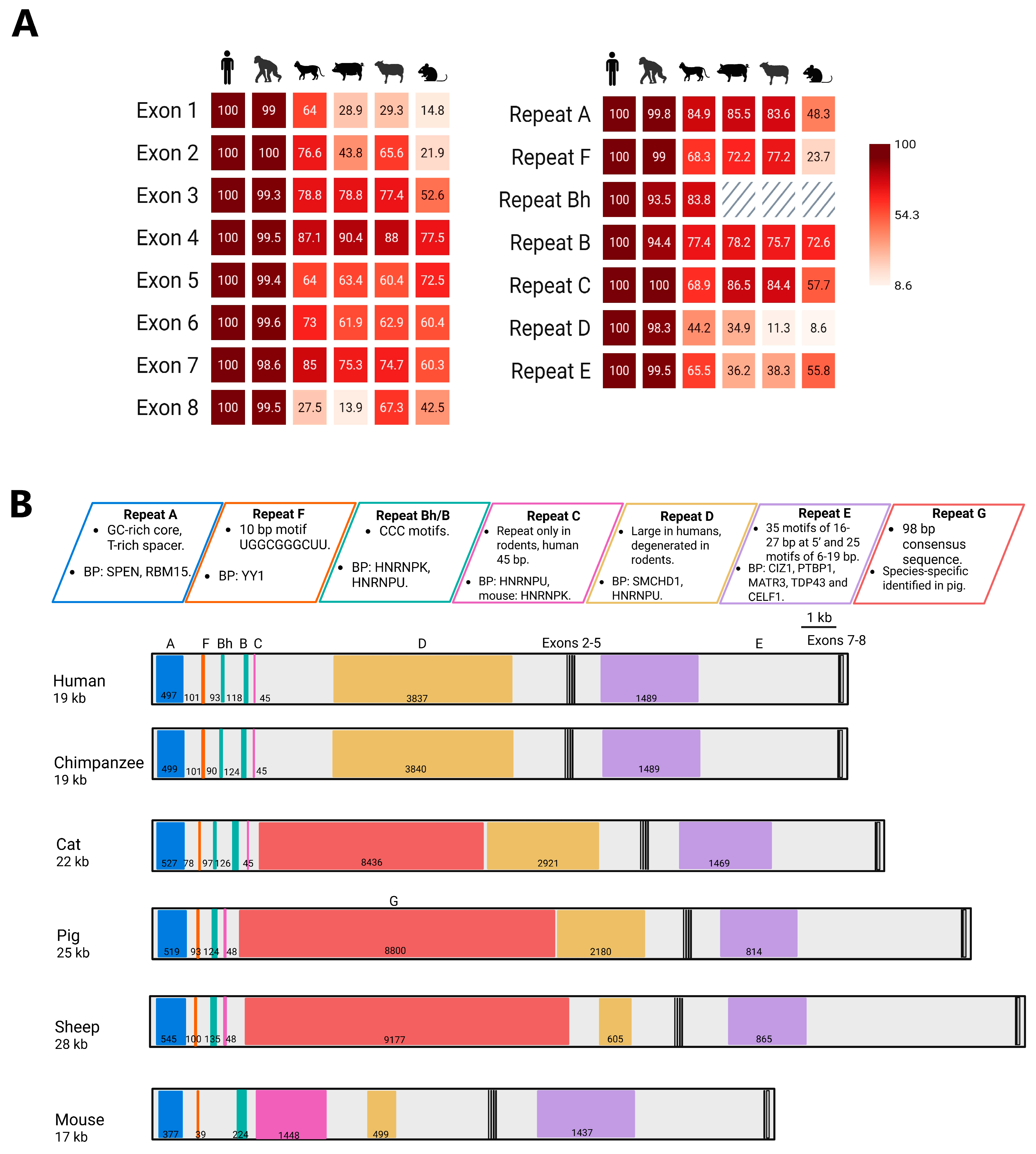
5. Conservation of Exons in XIST Evolution
6. Evolutionary Stability: XIST Repeats Retained Across Species
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ESC | Embryonic stem cells |
| lncRNAs | Long non-coding RNAs |
| MYA | Million years ago |
| XCI | X-chromosome inactivation |
| XIC | X inactivation center |
| Xi | Inactive X |
References
- Zhu, Z.; Younas, L.; Zhou, Q. Evolution and regulation of animal sex chromosomes. Nat. Rev. Genet. 2025, 26, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Kelley, R.L.; Meller, V.H.; Gordadze, P.R.; Roman, G.; Davis, R.L.; Kuroda, M.I. Epigenetic Spreading of the Drosophila Dosage Compensation Complex from roX RNA Genes into Flanking Chromatin. Cell 1999, 98, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Albritton, S.E.; Ercan, S. Caenorhabditis elegans Dosage Compensation: Insights into Condensin-Mediated Gene Regulation. Trends Genet. 2018, 34, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Camilleri-Robles, C.; Amador, R.; Klein, C.C.; Guigó, R.; Corominas, M.; Ruiz-Romero, M. Genomic and functional conservation of lncRNAs: Lessons from flies. Mamm. Genome 2022, 33, 328–342. [Google Scholar] [CrossRef]
- Ohno, S. Sex Chromosomes and Sex-Linked Genes; Springer: Berlin, Heidelberg, 1967. [Google Scholar] [CrossRef]
- Lyon, M.F. Gene action in the X-chromosome of the mouse (Mus. musculus L.). Nature 1961, 190, 373–374. [Google Scholar] [CrossRef]
- Cecalev, D.; Viçoso, B.; Galupa, R. Compensation of gene dosage on the mammalian X. Development 2024, 151, dev202891. [Google Scholar] [CrossRef]
- Russell, L.B. Mammalian X-Chromosome Action: Inactivation Limited in Spread and in Region of Origin. Science 1963, 140, 976–978. [Google Scholar] [CrossRef]
- Brown, C.J.; Ballabio, A.; Rupert, J.L.; Lafreniere, R.G.; Grompe, M.; Tonlorenzi, R.; Willard, H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 1991, 349, 38–44. [Google Scholar] [CrossRef]
- Brown, C.J.; Lafreniere, R.G.; Powers, V.E.; Sebastio, G.; Ballabio, A.; Pettigrew, A.L.; Ledbetter, D.H.; Levy, E.; Craig, I.W.; Willard, H.F. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature 1991, 349, 82–84. [Google Scholar] [CrossRef]
- Borsani, G.; Tonlorenzi, R.; Simmler, M.C.; Dandolo, L.; Arnaud, D.; Capra, V.; Grompe, M.; Pizzuti, A.; Muzny, D.; Lawrence, C.; et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature 1991, 351, 325–329. [Google Scholar] [CrossRef]
- Brockdorff, N.; Ashworth, A.; Kay, G.F.; Cooper, P.; Smith, S.; McCabe, V.M.; Norris, D.P.; Penny, G.D.; Patel, D.; Rastan, S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 1991, 351, 329–331. [Google Scholar] [CrossRef]
- Penny, G.D.; Kay, G.F.; Sheardown, S.A.; Rastan, S.; Brockdorff, B.N. Requirement for Xist in X chromosome inactivation. Nature 1996, 379, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Marahrens, Y.; Panning, B.; Dausman, J.; Strauss, W.; Jaenisch, R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes. Dev. 1997, 11, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell Biol. 1990, 10, 28–36. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Brown, C.J.; Hendrich, B.D.; Rupert, J.L. The Human X/ST Gene: Analysis of a 17 kb Inactive X-Specific RNA That Contains Conserved Repeats and Is Highly Localized within the Nucleus. Cell 1992, 71, 527–542. [Google Scholar] [CrossRef] [PubMed]
- Brockdorff, N.; McCabe, M.; Norris, P.; Cooper, J.; Swift, S.; Kay, F. The Product of the Mouse Xist Gene Is a 15 kb Inactive X-Specific Transcript Containing No Conserved ORF and Located in the Nucleus. Cell 1992, 71, 515–5126. [Google Scholar] [CrossRef] [PubMed]
- Clemson, C.M.; McNeil, J.A.; Willard, H.F.; Lawrence, J.B. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 1996, 132, 259–275. [Google Scholar] [CrossRef]
- Lee, J.T. Epigenetic regulation by long noncoding RNAs. Science 2012, 338, 1435–1439. [Google Scholar] [CrossRef] [PubMed]
- Kelsey, A.D.; Yang, C.; Leung, D.; Minks, J.; Dixon-McDougall, T.; Baldry, S.E.L.; Bogutz, A.B.; Lefebvre, L.; Brown, C.J. Impact of flanking chromosomal sequences on localization and silencing by the human non-coding RNA XIST. Genome Biol. 2015, 16, 208. [Google Scholar] [CrossRef]
- Jiang, J.; Jing, Y.; Cost, G.J.; Chiang, J.-C.; Kolpa, H.J.; Cotton, A.M.; Carone, D.M.; Carone, B.R.; Shivak, D.A.; Guschin, D.Y.; et al. Translating dosage compensation to trisomy 21. Nature 2013, 500, 296–300. [Google Scholar] [CrossRef]
- Navarro-Cobos, M.J.; Morales-Guzman, S.I.; Baldry, S.E.L.; Brown, C.J. Derivation of a minimal functional XIST by combining human and mouse interaction domains. Hum. Mol. Genet. 2023, 32, 1289–1300. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Cobos, M.J.; Brown, C.J. Recruitment of chromatin remodelers by XIST B-repeat region is variably dependent on HNRNPK. Hum. Mol. Genet. 2025, 34, 229–238. [Google Scholar] [CrossRef]
- Cortez, D.; Marin, R.; Toledo-Flores, D.; Froidevaux, L.; Liechti, A.; Waters, P.D.; Grützner, F.; Kaessmann, H. Origins and functional evolution of Y chromosomes across mammals. Nature 2014, 508, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A.M.; Koina, E.; Sankovic, N. How the gene content of human sex chromosomes evolved. Curr. Opin. Genet. Dev. 2006, 16, 219–224. [Google Scholar] [CrossRef]
- Hore, T.A.; Koina, E.; Wakefield, M.J.; Graves, J.A.M. The region homologous to the X-chromosome inactivation centre has been disrupted in marsupial and monotreme mammals. Chromosome Res. 2007, 15, 147–161. [Google Scholar] [CrossRef]
- Grant, J.; Mahadevaiah, S.K.; Khil, P.; Sangrithi, M.N.; Royo, H.; Duckworth, J.; McCarrey, J.R.; VandeBerg, J.L.; Renfree, M.B.; Taylor, W.; et al. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 2012, 487, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Whitworth, D.J.; Pask, A.J. The X factor: X chromosome dosage compensation in the evolutionarily divergent monotremes and marsupials. Semin. Cell Dev. Biol. 2016, 56, 117–121. [Google Scholar] [CrossRef]
- Graves, J.A.M.; Gécz, J. Hameister, Evolution of the human X—A smart and sexy chromosome that controls speciation and development. Cytogenet. Genome Res. 2002, 99, 141–145. [Google Scholar] [CrossRef]
- Balaton, B.P.; Dixon-McDougall, T.; Peeters, S.B.; Brown, C.J. The eXceptional nature of the X chromosome. Hum. Mol. Genet. 2018, 27, R242–R249. [Google Scholar] [CrossRef]
- Lyon, M.F. X-Chromosome inactivation: A repeat hypothesis. Cytogenet. Genome Res. 1998, 80, 133–137. [Google Scholar] [CrossRef]
- Deakin, J.E.; Chaumeil, J.; Hore, T.A.; Marshall Graves, J.A. Unravelling the evolutionary origins of X chromosome inactivation in mammals: Insights from marsupials and monotremes. Chromosome Res. 2009, 17, 671–685. [Google Scholar] [CrossRef] [PubMed]
- Takagi, N.; Sasaki, M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 1975, 256, 640–642. [Google Scholar] [CrossRef] [PubMed]
- Sado, T.; Ferguson-Smith, A.C. Imprinted X inactivation and reprogramming in the preimplantation mouse embryo. Hum. Mol. Genet. 2005, 14, R59–R64. [Google Scholar] [CrossRef]
- Okamoto, I.; Patrat, C.; Thépot, D.; Peynot, N.; Fauque, P.; Daniel, N.; Diabangouaya, P.; Wolf, J.-P.; Renard, J.-P.; Duranthon, V.; et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 2011, 472, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Petropoulos, S.; Edsgärd, D.; Reinius, B.; Deng, Q.; Panula, S.P.; Codeluppi, S.; Plaza, A.; Linnarsson, S.; Sandberg, R.; Lanner, F. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell 2016, 165, 1012–1026. [Google Scholar] [CrossRef]
- Sahakyan, A.; Kim, R.; Chronis, C.; Sabri, S.; Bonora, G.; Theunissen, T.W.; Kuoy, E.; Langerman, J.; Clark, A.T.; Jaenisch, R.; et al. Human Naive Pluripotent Stem Cells Model X Chromosome Dampening and X Inactivation. Cell Stem Cell 2017, 20, 87–101. [Google Scholar] [CrossRef]
- Davidson, K.C.; Mason, E.A.; Pera, M.F. The pluripotent state in mouse and human. Development 2015, 142, 3090–3099. [Google Scholar] [CrossRef]
- Vallot, C.; Ouimette, J.-F.; Makhlouf, M.; Féraud, O.; Pontis, J.; Côme, J.; Martinat, C.; Bennaceur-Griscelli, A.; Lalande, M.; Rougeulle, C. Erosion of X Chromosome Inactivation in Human Pluripotent Cells Initiates with XACT Coating and Depends on a Specific Heterochromatin Landscape. Cell Stem Cell 2015, 16, 533–546. [Google Scholar] [CrossRef]
- Patel, S.; Bonora, G.; Sahakyan, A.; Kim, R.; Chronis, C.; Langerman, J.; Fitz-Gibbon, S.; Rubbi, L.; Skelton, R.J.; Ardehali, R.; et al. Human Embryonic Stem Cells Do Not Change Their X Inactivation Status during Differentiation. Cell Reports 2017, 18, 54–67. [Google Scholar] [CrossRef]
- Sarel-Gallily, R.; Benvenisty, N. Large-Scale Analysis of X Inactivation Variations between Primed and Naïve Human Embryonic Stem Cells. Cells 2022, 11, 1729. [Google Scholar] [CrossRef]
- Vallot, C.; Ouimette, J.; Rougeulle, C. Establishment of X chromosome inactivation and epigenomic features of the inactive X depend on cellular contexts. BioEssays 2016, 38, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Loda, A.; Heard, E. Xist RNA in action: Past, present, and future. PLoS Genet. 2019, 15, e1008333. [Google Scholar] [CrossRef] [PubMed]
- Patrat, C.; Ouimette, J.-F.; Rougeulle, C. X chromosome inactivation in human development. Development 2020, 147, dev183095. [Google Scholar] [CrossRef]
- Loda, A.; Collombet, S.; Heard, E. Gene regulation in time and space during X-chromosome inactivation. Nat Rev. Mol Cell Biol. 2022, 23, 231–249. [Google Scholar] [CrossRef]
- Jacobson, E.C.; Pandya-Jones, A.; Plath, K. A lifelong duty: How Xist maintains the inactive X chromosome. Curr. Opin. Genet. Dev. 2022, 75, 101927. [Google Scholar] [CrossRef]
- Lee, J.T. Regulation of X-Chromosome Counting by Tsix and Xite Sequences. Science 2005, 309, 768–771. [Google Scholar] [CrossRef]
- Yin, H.; Wei, C.; Lee, J.T. Revisiting the consequences of deleting the X inactivation center. Proc. Natl. Acad. Sci. USA 2021, 118, e2102683118. [Google Scholar] [CrossRef]
- Lee, J.; Davidow, L.S.; Warshawsky, D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 1999, 21, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Migeon, B.R.; Lee, C.H.; Chowdhury, A.K.; Carpenter, H. Species Differences in TSIX/Tsix Reveal the Roles of These Genes in X-Chromosome Inactivation. Am. J. Hum. Genet. 2002, 71, 286–293. [Google Scholar] [CrossRef]
- Tian, D.; Sun, S.; Lee, J.T. The Long Noncoding RNA, Jpx, Is a Molecular Switch for X Chromosome Inactivation. Cell 2010, 143, 390–403. [Google Scholar] [CrossRef]
- Sun, S.; Del Rosario, B.C.; Szanto, A.; Ogawa, Y.; Jeon, Y.; Lee, J.T. Jpx RNA Activates Xist by Evicting CTCF. Cell 2013, 153, 1537–1551. [Google Scholar] [CrossRef] [PubMed]
- Chureau, C.; Chantalat, S.; Romito, A.; Galvani, A.; Duret, L.; Avner, P.; Rougeulle, C. Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum. Mol. Genet. 2011, 20, 705–718. [Google Scholar] [CrossRef]
- Rosspopoff, O.; Cazottes, E.; Huret, C.; Loda, A.; Collier, A.J.; Casanova, M.; Rugg-Gunn, P.J.; Heard, E.; Ouimette, J.-F.; Rougeulle, C. Species-specific regulation of XIST by the JPX/FTX orthologs. Nucleic Acids Res. 2023, 51, 2177–2194. [Google Scholar] [CrossRef]
- Flynn, M.; Saha, O.; Young, P. Molecular evolution of the LNX gene family. BMC Evol. Biol. 2011, 11, 235. [Google Scholar] [CrossRef] [PubMed]
- Duret, L.; Chureau, C.; Samain, S.; Weissenbach, J.; Avner, P. The Xist RNA Gene Evolved in Eutherians by Pseudogenization of a Protein-Coding Gene. Science 2006, 312, 1653–1655. [Google Scholar] [CrossRef] [PubMed]
- Elisaphenko, E.A.; Kolesnikov, N.N.; Shevchenko, A.I.; Rogozin, I.B.; Nesterova, T.B.; Brockdorff, N.; Zakian, S.M. Dual Origin of the Xist Gene from a Protein-Coding Gene and a Set of Transposable Elements. PLoS ONE 2008, 3, e2521. [Google Scholar] [CrossRef]
- Shevchenko, A.I.; Zakharova, I.S.; Zakian, S.M. The Evolutionary Pathway of X Chromosome Inactivation in Mammals. Acta Naturae 2013, 5, 40–53. [Google Scholar] [CrossRef]
- Chow, J.C.; Hall, L.L.; Baldry, S.E.L.; Thorogood, N.P.; Lawrence, J.B.; Brown, C.J. Inducible XIST-dependent X-chromosome inactivation in human somatic cells is reversible. Proc. Natl. Acad. Sci. USA 2007, 104, 10104–10109. [Google Scholar] [CrossRef]
- Yue, M.; Ogawa, Y. CRISPR/Cas9-mediated modulation of splicing efficiency reveals short splicing isoform of Xist RNA is sufficient to induce X-chromosome inactivation. Nucleic Acids Res. 2018, 46, e26. [Google Scholar] [CrossRef]
- Lee, H.J.; Gopalappa, R.; Sunwoo, H.; Choi, S.-W.; Ramakrishna, S.; Lee, J.T.; Kim, H.H.; Nam, J.-W. En bloc and segmental deletions of human XIST reveal X chromosome inactivation-involving RNA elements. Nucleic Acids Res. 2019, 47, 3875–3887. [Google Scholar] [CrossRef]
- Caparros, M.-L.; Alexiou, M.; Webster, Z.; Brockdorff, N. Functional analysis of the highly conserved exon IV of Xist RNA. Cytogenet. Genome Res. 2002, 99, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Yen, Z.C.; Meyer, I.M.; Karalic, S.; Brown, C.J. A cross-species comparison of X-chromosome inactivation in Eutheria. Genomics 2007, 90, 453–463. [Google Scholar] [CrossRef]
- Nesterova, T.B.; Slobodyanyuk, S.Y.; Elisaphenko, E.A.; Shevchenko, A.I.; Johnston, C.; Pavlova, M.E.; Rogozin, I.B.; Kolesnikov, N.N.; Brockdorff, N.; Zakian, S.M. Characterization of the Genomic Xist Locus in Rodents Reveals Conservation of Overall Gene Structure and Tandem Repeats but Rapid Evolution of Unique Sequence. Genome Res. 2001, 11, 833–849. [Google Scholar] [CrossRef]
- Hwang, J.Y.; Kim, E.B.; Ka, H.; Lee, C.-K. Identification of the Porcine XIST Gene and Its Differential CpG Methylation Status in Male and Female Pig Cells. PLoS ONE 2013, 8, e73677. [Google Scholar] [CrossRef] [PubMed]
- Chureau, C.; Prissette, M.; Bourdet, A.; Barbe, V.; Cattolico, L.; Jones, L.; Eggen, A.; Avner, P.; Duret, L. Comparative Sequence Analysis of the X-Inactivation Center Region in Mouse, Human, and Bovine. Genome Res. 2002, 12, 894–908. [Google Scholar] [CrossRef]
- Liang, M.; Zhang, L.; Lai, L.; Li, Z. Unraveling the role of Xist in X chromosome inactivation: Insights from rabbit model and deletion analysis of exons and repeat A. Cell Mol. Life Sci. 2024, 81, 156. [Google Scholar] [CrossRef]
- Yamada, N.; Hasegawa, Y.; Yue, M.; Hamada, T.; Nakagawa, S.; Ogawa, Y. Xist Exon 7 Contributes to the Stable Localization of Xist RNA on the Inactive X-Chromosome. PLoS Genet. 2015, 11, e1005430. [Google Scholar] [CrossRef]
- Czermiński, J.T.; Lawrence, J.B. Silencing Trisomy 21 with XIST in Neural Stem Cells Promotes Neuronal Differentiation. Dev. Cell 2020, 52, 294–308.e3. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Moss, W.N.; Rutenberg-Schoenberg, M.; Simon, M.D. Probing Xist RNA Structure in Cells Using Targeted Structure-Seq. PLoS Genet. 2015, 11, e1005668. [Google Scholar] [CrossRef]
- Lu, Z.; Zhang, Q.C.; Lee, B.; Flynn, R.A.; Smith, M.A.; Robinson, J.T.; Davidovich, C.; Gooding, A.R.; Goodrich, K.J.; Mattick, J.S.; et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 2016, 165, 1267–1279. [Google Scholar] [CrossRef]
- Lu, Z.; Guo, J.K.; Wei, Y.; Dou, D.R.; Zarnegar, B.; Ma, Q.; Li, R.; Zhao, Y.; Liu, F.; Choudhry, H.; et al. Structural modularity of the XIST ribonucleoprotein complex. Nat. Commun. 2020, 11, 6163. [Google Scholar] [CrossRef] [PubMed]
- Raposo, A.C.; Casanova, M.; Gendrel, A.-V.; Da Rocha, S.T. The tandem repeat modules of Xist lncRNA: A swiss army knife for the control of X-chromosome inactivation. Biochem. Soc. Trans. 2021, 49, 2549–2560. [Google Scholar] [CrossRef]
- Minks, J.; Baldry, S.E.; Yang, C.; Cotton, A.M.; Brown, C.J. XIST-induced silencing of flanking genes is achieved by additive action of repeat a monomers in human somatic cells. Epigenetics Chromatin 2013, 6, 23. [Google Scholar] [CrossRef]
- Hoki, Y.; Kimura, N.; Kanbayashi, M.; Amakawa, Y.; Ohhata, T.; Sasaki, H.; Sado, T. A proximal conserved repeat in the Xist gene is essential as a genomic element for X-inactivation in mouse. Development 2009, 136, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhong, Y.; Zhou, Y.; Tanaseichuk, O.; Li, Z.; Zhao, J.C. Identification of a Xist silencing domain by Tiling CRISPR. Sci. Rep. 2019, 9, 2408. [Google Scholar] [CrossRef]
- Colognori, D.; Sunwoo, H.; Wang, D.; Wang, C.-Y.; Lee, J.T. Xist Repeat A contributes to early recruitment of Polycomb complexes during X-chromosome inactivation. Dev. Cell 2021, 56, 1236–1237. [Google Scholar] [CrossRef] [PubMed]
- Wutz, A.; Rasmussen, T.P.; Jaenisch, R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002, 30, 167–174. [Google Scholar] [CrossRef]
- Maenner, S.; Blaud, M.; Fouillen, L.; Savoye, A.; Marchand, V.; Dubois, A.; Sanglier-Cianférani, S.; Van Dorsselaer, A.; Clerc, P.; Avner, P.; et al. 2-D Structure of the A Region of Xist RNA and Its Implication for PRC2 Association. PLoS Biol. 2010, 8, e1000276. [Google Scholar] [CrossRef]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef]
- Patil, D.P.; Chen, C.-K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef]
- Carter, A.C.; Xu, J.; Nakamoto, M.Y.; Wei, Y.; Zarnegar, B.J.; Shi, Q.; Broughton, J.P.; Ransom, R.C.; Salhotra, A.; Nagaraja, S.D.; et al. Spen links RNA-mediated endogenous retrovirus silencing and X chromosome inactivation. eLife 2020, 9, e54508. [Google Scholar] [CrossRef]
- Makhlouf, M.; Ouimette, J.-F.; Oldfield, A.; Navarro, P.; Neuillet, D.; Rougeulle, C. A prominent and conserved role for YY1 in Xist transcriptional activation. Nat. Commun. 2014, 5, 5–4878. [Google Scholar] [CrossRef]
- Chapman, A.G.; Cotton, A.M.; Kelsey, A.D.; Brown, C.J. Differentially methylated CpG island within human XIST mediates alternative P2 transcription and YY1 binding. BMC Genet. 2014, 15, 89. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-K.; Blanco, M.; Jackson, C.; Aznauryan, E.; Ollikainen, N.; Surka, C.; Chow, A.; Cerase, A.; McDonel, P.; Guttman, M. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 2016, 354, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Bousard, A.; Raposo, A.C.; Żylicz, J.J.; Picard, C.; Pires, V.B.; Qi, Y.; Gil, C.; Syx, L.; Chang, H.Y.; Heard, E.; et al. The role of Xist -mediated Polycomb recruitment in the initiation of X-chromosome inactivation. EMBO Rep. 2019, 20, e48019. [Google Scholar] [CrossRef] [PubMed]
- Nesterova, T.B.; Wei, G.; Coker, H.; Pintacuda, G.; Bowness, J.S.; Zhang, T.; Almeida, M.; Bloechl, B.; Moindrot, B.; Carter, E.J.; et al. Systematic allelic analysis defines the interplay of key pathways in X chromosome inactivation. Nat. Commun. 2019, 10, 3129. [Google Scholar] [CrossRef]
- Dixon-McDougall, T.; Brown, C.J. Independent domains for recruitment of PRC1 and PRC2 by human XIST. PLoS Genet. 2021, 17, e1009123. [Google Scholar] [CrossRef]
- Pintacuda, G.; Wei, G.; Roustan, C.; Kirmizitas, B.A.; Solcan, N.; Cerase, A.; Castello, A.; Mohammed, S.; Moindrot, B.; Nesterova, T.B.; et al. hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-Repeat to Establish Polycomb-Mediated Chromosomal Silencing. Mol. Cell 2017, 68, 955–969.e10. [Google Scholar] [CrossRef]
- Sarma, K.; Levasseur, P.; Aristarkhov, A.; Lee, J.T. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc. Natl. Acad. Sci. USA 2010, 107, 22196–22201. [Google Scholar] [CrossRef]
- Hasegawa, Y.; Brockdorff, N.; Kawano, S.; Tsutui, K.; Tsutui, K.; Nakagawa, S. The Matrix Protein hnRNP U Is Required for Chromosomal Localization of Xist RNA. Dev. Cell 2010, 19, 469–476. [Google Scholar] [CrossRef]
- Colognori, D.; Sunwoo, H.; Kriz, A.J.; Wang, C.-Y.; Lee, J.T. Xist Deletional Analysis Reveals an Interdependency between Xist RNA and Polycomb Complexes for Spreading along the Inactive X. Mol. Cell 2019, 74, 101–117.e10. [Google Scholar] [CrossRef] [PubMed]
- Smola, M.J.; Christy, T.W.; Inoue, K.; Nicholson, C.O.; Friedersdorf, M.; Keene, J.D.; Lee, D.M.; Calabrese, J.M.; Weeks, K.M. Weeks, SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl. Acad. Sci. USA 2016, 113, 10322–10327. [Google Scholar] [CrossRef] [PubMed]
- Sunwoo, H.; Colognori, D.; Froberg, J.E.; Jeon, Y.; Lee, J.T. Repeat E anchors Xist RNA to the inactive X chromosomal compartment through CDKN1A-interacting protein (CIZ1). Proc. Natl. Acad. Sci. USA 2017, 114, 10654–10659. [Google Scholar] [CrossRef] [PubMed]
- Pandya-Jones, A.; Markaki, Y.; Serizay, J.; Chitiashvili, T.; Mancia Leon, W.R.; Damianov, A.; Chronis, C.; Papp, B.; Chen, C.K.; McKee, R.; et al. A protein assembly mediates Xist localization and gene silencing. Nature 2020, 587, 145–151. [Google Scholar] [CrossRef]
- Ding, M.; Wang, D.; Chen, H.; Kesner, B.; Grimm, N.-B.; Weissbein, U.; Lappala, A.; Jiang, J.; Rivera, C.; Lou, J.; et al. Biophysical basis for the spreading behavior and limited diffusion of Xist. Cell 2025, 188, 978–997.e25. [Google Scholar] [CrossRef]
- Michaels, T.C.T.; Wutz, A. Phase separation paints Xi with Xist. Cell Res. 2025, 14, 1–2. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navarro-Cobos, M.J.; Brown, C.J. Human XIST: Origin and Divergence of a cis-Acting Silencing RNA. Non-Coding RNA 2025, 11, 35. https://doi.org/10.3390/ncrna11030035
Navarro-Cobos MJ, Brown CJ. Human XIST: Origin and Divergence of a cis-Acting Silencing RNA. Non-Coding RNA. 2025; 11(3):35. https://doi.org/10.3390/ncrna11030035
Chicago/Turabian StyleNavarro-Cobos, Maria Jose, and Carolyn J. Brown. 2025. "Human XIST: Origin and Divergence of a cis-Acting Silencing RNA" Non-Coding RNA 11, no. 3: 35. https://doi.org/10.3390/ncrna11030035
APA StyleNavarro-Cobos, M. J., & Brown, C. J. (2025). Human XIST: Origin and Divergence of a cis-Acting Silencing RNA. Non-Coding RNA, 11(3), 35. https://doi.org/10.3390/ncrna11030035





