Mathematical and Computational Modeling of Poroelastic Cell Scaffolds Used in the Design of an Implantable Bioartificial Pancreas
Abstract
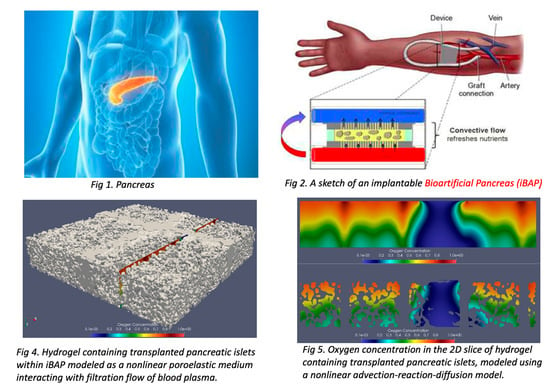
:1. Introduction
- A fluid–structure interaction (FSI) model describing the interaction between the blood plasma modeled by the Navier–Stokes or time-dependent Stokes equations for an incompressible, viscous fluid, and a poroelastic hydrogel containing the cells, modeled by the nonlinear Biot Equations (see Section 2.1). The nonlinearity in the Biot equations comes from the dependence of the hydrogel’s permeability on fluid content/porosity [7];
- Two advection–reaction–diffusion models describing oxygen concentration within the poroelastic hydrogel containing the cells, and oxygen concentration within the gasket containing blood plasma (see Section 2.2). The two models are coupled to the FSI model above through the fluid advection velocity, and through the information about the domain motion. Additionally, the two advection–reaction–diffusion models are coupled among themselves across the interface separating the gasket region from the poroelastic hydrogel scaffold. The coupling conditions describe oxygen transfer from the gasket region to the poroelastic scaffold.
2. The Maco-Scale Mathematical Models
2.1. A Fluid-Structure Interaction Model for Blood Plasma and Poroelastic Scaffold
2.2. Coupled Models for Oxygen Concentration
3. Discretized Problems and Numerical Schemes
3.1. Discretization of the Fluid-Structure Interaction Problem
| Step 1: Given , compute , such that
|
- Polarized identity:
- The discrete trace-inverse inequality:where is a positive constant, uniformly bounded from above with respect to the mesh characteristic size h for a family of shape-regular and quasi-uniform meshes, such as our domains defined in Section 3.1 [37].
3.2. Discretization of the Coupled Advection-Reaction-Diffusion Problem
3.3. Parameter Estimation Using Encoder–Decoder Convolution Neural Networks and Smoothed Particle Hydrodynamics
- Create an ensemble of 100 poroelastic gel matrix geometries with different porosity by using SPH to distribute the solid particles in the hydrogel. The hydrogel is divided into boxes and treated as an image. Every box (cf. pixel) contains the information about the density of the non-moving SPH particles in that box (cf. pixel intensity in terms of image processing).
- Run SPH simulations for each poroelastic matrix geometry to obtain the corresponding filtration flow and pressure, as illustrated in Figure 8a,b.
- Post-processing: At each location in the chamber, compute the local hydraulic permeability tensor using data from step 2 above, see Figure 8c,d, and use it as training data (permeability map) for the Encoder–Decoder CNN.
- Train the Encoder–Decoder CNN with the density data and corresponding permeability map obtained from steps 2 and 3. We use TensorFlow as our platform. The encoder contains several Convolution and Dense layers, and the decoder is just the reflection of those layers in the encoder.
- Feed a new density matrix to the Encoder-decoder CNN and predict the local values of the hydraulic conductivity tensor for a new porous medium chamber.
3.4. Parallel Implementation and Convergence Test
4. Numerical Results
4.1. One Outlet—Current Prototype Design
4.2. Two Outlets versus One Outlet
- The presence of the second outlet improves the flow through the part of the islet chamber closest to that outlet (see Figure 12 right).
- The staggered distribution of islet chamber and membranes underneath the chamber, increases transverse flow through the islet chamber. This is shown by the angled streamlines in Figure 12 right.
4.3. Hydrogel Elasticity
5. Conclusions
- Oxygen concentration and filtration flow through hydrogel scaffolds are significantly affected by the position and number of the ultrafiltrate outlets. The ultrafiltrate outlets should be (equi)distributed to uniformly cover the entire array of cell scaffolds.
- Hydrogel elasticity significantly affects oxygen concentration and filtration flow through scaffolds. Highly elastic scaffolds have a higher capacity for oxygen transfer.
- Oxygen concentration is largest near the flow inlet into the scaffold, and near the drilled ultrafiltrate channels.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shaheen, R.; Gurlin, R.E.; Gologorsky, R.; Blaha, C.; Munnangi, P.; Santandreu, A.; Nair, G.; Roy, S. Superporous agarose scaffolds for encapsulation of adult human islets and human stem-cell-derived β cells for intravascular bioartificial pancreas applications. J. Biomed. Mater. Res. 2021, 109, 2438–2448. [Google Scholar] [CrossRef] [PubMed]
- Fissell, W.H.; Dubnisheva, A.; Eldridge, A.N.; Fleischman, A.J.; Zydney, A.L.; Roy, S. High-performance silicon nanopore hemofiltration membranes. J. Membr. Sci. 2009, 32, 58–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanani, D.M.; Fissell, W.H.; Roy, S.; Dubnisheva, A.; Fleischman, A.; Zydney, A.L. Permeability-selectivity analysis for ultrafiltration: Effect of pore geometry. J. Memb. Sci. 2010, 349, 405–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Feinberg, B.; Kant, R.; Chui, B.; Goldman, K.; Park, J.; Moses, W.; Blaha, C.; Iqbal, Z.; Chow, C. Diffusive silicon nanopore membranes for hemodialysis applications. PLoS ONE 2016, 11, e0159526. [Google Scholar] [CrossRef] [Green Version]
- Song, S.; Faleo, G.; Yeung, R.; Kant, R.; Posselt, A.M.; Desai, T.A.; Tang, Q.; Roy, S. Silicon nanopore membrane (SNM) for islet encapsulation and immunoisolation under convective transport. Nat. Sci. Rep. 2016, 6, 1–9. [Google Scholar]
- Desai, T.; Shea, L. Advances in islet encapsulation technologies. Nat. Rev. Drug Discov. 2017, 16, 338–351. [Google Scholar] [CrossRef]
- Etzold, M.A.; Linden, P.F.; Worster, M.G. Transpiration through hydrogels. J. Fluid Mech. 2021, 925, A8-1–A8-31. [Google Scholar] [CrossRef]
- Buchwald, P. Fem-based oxygen consumption and cell viability models for avascular pancreatic islets. Theor. Biol. Med. Model. 2009, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Han, E.X.; Wang, J.; Kural, M.; Jiang, B.; Leiby, K.L.; Chowdhury, N.; Niklason, L.E. Development of a bioartificial vascular pancreas. J. Tissue Eng. 2021, 12, 20417314211027714. [Google Scholar] [CrossRef]
- Fernandez, S.A.; Champion, K.S.; Danielczak, L.; Gasparrini, M.; Paraskevas, S.; Leask, R.L.; Hoesli, C.A. Engineering vascularized islet macroencapsulation devices: An in vitro platform to study oxygen transport in perfused immobilized pancreatic beta cell cultures. Front. Bioeng. Biotechnol. 2022, 10. [Google Scholar] [CrossRef]
- Buchwald, P.; Cechin, S.R. Glucose-stimulated insulin secretion in isolated pancreatic islets: Multiphysics fem model calculations compared to results of perifusion experiments with human islets. J. Biomed. Sci. Eng. 2013, 6, 26–35. [Google Scholar] [CrossRef] [Green Version]
- Buchwald, P.; Cechin, S.R.; Weaver, J.D.; Stabler, C.L. Experimental evaluation and computational modeling of the effects of encapsulation on the time-profile of glucose-stimulated insulin release of pancreatic islets. Biomed. Eng. 2015, 14, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchwald, P.; Tamayo-Garcia, A.; Manzoli, V.; Tomei, A.A.; Stabler, C.L. Glucose-stimulated insulin release: Parallel perifusion studies of free and hydrogel encapsulated human pancreatic islets. Biotechnol. Bioeng. 2018, 115, 232–245. [Google Scholar] [CrossRef] [PubMed]
- Ambartsumyan, I.; Ervin, V.J.; Nguyen, T.; Yotov, I. A nonlinear stokes–biot model for the interaction of a non-newtonian fluid with poroelastic media. ESAIM Math. Model. Numer. Anal. 2019, 53, 1915–1955. [Google Scholar] [CrossRef]
- Ambartsumyan, I.; Khattatov, E.; Yotov, I.; Zunino, P. A Lagrange multiplier method for a Stokes–Biot fluid-poroelastic structure interaction model. Numer. Math. 2018, 140, 513–553. [Google Scholar] [CrossRef] [Green Version]
- Bergkamp, E.; Verhoosel, C.; Remmers, J.; Smeulders, D. A staggered finite element procedure for the coupled stokes-biot system with fluid entry resistance. Comput. Geosci. 2020, 24, 1497–1522. [Google Scholar] [CrossRef]
- Cesmelioglu, A.; Chidyagwai, P. Numerical analysis of the coupling of free fluid with a poroelastic material. Numer. Methods Partial. Differ. Eq. 2020, 36, 463–494. [Google Scholar] [CrossRef]
- Rauch, A.D.; Vuong, A.T.; Yoshihara, L.; Wall, W.A. A coupled approach for fluid saturated poroelastic media and immersed solids for modeling cell-tissue interactions. Int. J. Numer. Methods Biomed. Eng. 2018, 34, e3139. [Google Scholar]
- Ruiz-Baier, R.; Taffetani, M.; Westermeyer, H.D.; Yotov, I. The biot—Stokes coupling using total pressure: Formulation, analysis and application to interfacial flow in the eye. Comput. Methods Appl. Mech. Eng. 2022, 389, 114384. [Google Scholar] [CrossRef]
- Taffetani, M.; Ruiz-Baier, R.; Waters, S. Coupling stokes flow with inhomogeneous poroelasticity. Q. J. Mech. Appl. Math. 2021, 74, 411–439. [Google Scholar] [CrossRef]
- Wen, J.; He, Y. A strongly conservative finite element method for the coupled stokes–biot model. Comput. Math. Appl. 2020, 80, 1421–1442. [Google Scholar] [CrossRef]
- Wen, J.; Su, J.; He, Y.; Chen, H. Discontinuous galerkin method for the coupled stokes-biot model. Numer. Methods Partial. Differ. Eq. 2021, 37, 383–405. [Google Scholar] [CrossRef]
- Song, S.; Blaha, C.; Moses, W.; Park, J.; Wright, N.; Groszek, J.W.; Fissell, S.; Vartanian, A.; Posselt, M.; Roy, S. An intravascular bioartificial pancreas device (iBAP) with silicon nanopore membranes (SNM) for islet encapsulation under convective mass transport. Lab Chip 2018, 17, 1778–1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Huang, R.; Ravi-Chandar, K. Linear and nonlinear poroelastic analysis of swelling and drying behavior of gelatin-based hydrogels. Int. J. Solids Struct. 2020, 195, 43–56. [Google Scholar] [CrossRef]
- Yoon, J.; Cai, S.; Suo, Z.; Hayward, R.C. Poroelastic swelling kinetics of thin hydrogel layers: Comparison of theory and experiment. Soft Matter 2010, 6, 6004–6012. [Google Scholar] [CrossRef]
- Iritani, E.; Katagiri, N.; Yamaguchi, K.; Cho, J.H. Compression-permeability properties of compressed bed of superabsorbent hydrogel particles. Dry. Technol. 2006, 24, 1243–1249. [Google Scholar] [CrossRef]
- Bukač, M.; Yotov, I.; Zakerzadeh, R.; Zunino, P. Partitioning strategies for the interaction of a fluid with a poroelastic material based on a Nitsche’s coupling approach. Comput. Methods Appl. Mech. Eng. 2015, 292, 138–170. [Google Scholar] [CrossRef] [Green Version]
- Bukac, M.; Yotov, I.; Zunino, P. An operator splitting approach for the interaction between a fluid and a multilayered poroelastic structure. Numer. Methods Partial. Differ. Eq. 2015, 31, 1054–1100. [Google Scholar] [CrossRef]
- Jager, W.; Mikelic, A. On the boundary conditions at the contact interface between a porous medium and a free fluid. Ann. Scuola Norm. Sup. Pisa Cl. Sci. 1996, 23, 403–465. [Google Scholar]
- Jager, W.; Mikelic, A. On the interface boundary condition of Beavers, Joseph, and Saffman. SIAM J. Appl. Math. 2000, 60, 1111–1127. [Google Scholar]
- Čanić, S.; Wang, Y.; Bukač, M. A next-generation mathematical model for drug-eluting stents. SIAM J. Appl. Math. 2021, 81, 1503–1529. [Google Scholar] [CrossRef]
- Beard, D.; Bassingthwaighte, J. Modeling advection and diffusion of oxygen in complex vascular networks. Annas Biomed. Eng. 2001, 29, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Buchwald, P. A local glucose-and oxygen concentration-based insulin secretion model for pancreatic islets. Theor. Biol. Med. Model. 2011, 8, 1–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, J.A.; Rudenski, A.; Gibson, J.; Howard, L.; O’Driscoll, R. Relating Oxygen Partial Pressure, Saturation and Content: The Haemoglobin-Oxygen Dissociation Curve; Breathe: Sheffield, UK, 2015; pp. 194–201. [Google Scholar]
- Burkardt, J.; Trenchea, C. Refactorization of the midpoint rule. Appl. Math. Lett. 2020, 107, 106438. [Google Scholar] [CrossRef]
- Masud, A.; Hughes, T. A stabilized mixed finite element method for Darcy flow. Comput. Methods Appl. Mech. Eng. 2002, 191, 4341–4370. [Google Scholar] [CrossRef]
- Thomee, V. Galerkin Finite Element Methods for Parabolic Problems; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Monaghan, J.J. Smoothed particle hydrodynamics. Annu. Rev. Astron. Astrophys. 1992, 30, 543–574. [Google Scholar] [CrossRef]
- Monaghan, J.J. Simulating free surface flows with SPH. J. Comput. Phys. 1994, 110, 399–406. [Google Scholar] [CrossRef]
- Morris, J.P.; Fox, P.J.; Zhu, Y. Modeling low Reynolds number incompressible flows using SPH. J. Comput. Phys. 1997, 136, 214–226. [Google Scholar] [CrossRef]
- Wang, Y.; Canic, S.; Kokot, G.; Snezhko, A.; Aranson, I.S. Quantifying the role of hydrodynamic interactions on the onset of collective states in ensembles of magnetic colloidal spinners and rollers. Phys. Rev. Fluids 2019, 4, 013701. [Google Scholar] [CrossRef]
- Baymani, M. Artificial neural network method for solving the Navier-Stokes equations. Neural Comput. Appl. 2015, 26, 765–773. [Google Scholar] [CrossRef]
- Lähivaara, T.; Kärkkäinen, L.; Huttunen, J.M.; Hesthaven, J.S. Deep convolutional neural networks for estimating porous material parameters with ultrasound tomography. J. Acoust. Soc. Am. 2018, 143, 1148–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raissi, M.; Yazdani, A.; Karniadakis, G.E. Hidden fluid mechanics: A Navier-Stokes informed deep learning framework for assimilating flow visualization data. arXiv 2018, arXiv:abs/1808.04327. [Google Scholar]
- FEniCS. Open Source Software Developed by a Global Community of Scientists and Software Developers. Available online: https://fenicsproject.org/ (accessed on 23 May 2022).
- Alonzo, M.; Kumar, S.A.; Allen, S.; Delgado, M.; Alvarez-Primo, F.; Suggs, L.; Joddar, B. Hydrogel scaffolds with elasticity-mimicking embryonic substrates promote cardiac cellular network formation. Prog. Biomater. 2020, 9, 125–137. [Google Scholar] [CrossRef] [PubMed]




















| Parameter | Value |
|---|---|
| Blood inlet pressure (Average) (mmHg) | 46 |
| Blood outlet pressure (Average) (mmHg) | 20 |
| Channel height (cm) | 0.3 |
| Channel length (cm) | 6.5 |
| Channel width (cm) | 0.7 |
| Fluid density (g/cm) | 1 |
| Fluid viscosity (cm/s) | 0.04 |
| Poroelastic structure density (g/cm) | 1.2 |
| Pressure storage coefficient | 1 |
| Permeability | |
| Young’s modulus E (d y n e s/cm) | |
| Poisson’s ratio | 0.49 |
| Biot-Willis parameter |
| Parameters | Value (Units) |
|---|---|
| Concentration of oxygen at fluid inlet | (mol · cm) [34] |
| Diffusion coefficient in fluid channel | |
| Diffusion coefficient in hydrogel | |
| Maximum oxygen consumption rate | (mol · cm) |
| Critical oxygen concentration | (mol · cm) |
| The Michaelis–Menten constant | (mol · cm) |
| Rate | Rate | Rate | ||||
|---|---|---|---|---|---|---|
| - | - | 0.3127 | - | |||
| 2.78 | 2.11 | 0.0707295 | 2.14 | |||
| 1.99 | 0.96 | 0.0252249 | 1.49 |
| Rate | Rate | Rate | ||||
|---|---|---|---|---|---|---|
| - | - | - | ||||
| 2.16 | 2.16 | 2.05 | ||||
| 1.92 | 1.92 | 2.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Čanić, S.; Bukač, M.; Blaha, C.; Roy, S. Mathematical and Computational Modeling of Poroelastic Cell Scaffolds Used in the Design of an Implantable Bioartificial Pancreas. Fluids 2022, 7, 222. https://doi.org/10.3390/fluids7070222
Wang Y, Čanić S, Bukač M, Blaha C, Roy S. Mathematical and Computational Modeling of Poroelastic Cell Scaffolds Used in the Design of an Implantable Bioartificial Pancreas. Fluids. 2022; 7(7):222. https://doi.org/10.3390/fluids7070222
Chicago/Turabian StyleWang, Yifan, Sunčica Čanić, Martina Bukač, Charles Blaha, and Shuvo Roy. 2022. "Mathematical and Computational Modeling of Poroelastic Cell Scaffolds Used in the Design of an Implantable Bioartificial Pancreas" Fluids 7, no. 7: 222. https://doi.org/10.3390/fluids7070222
APA StyleWang, Y., Čanić, S., Bukač, M., Blaha, C., & Roy, S. (2022). Mathematical and Computational Modeling of Poroelastic Cell Scaffolds Used in the Design of an Implantable Bioartificial Pancreas. Fluids, 7(7), 222. https://doi.org/10.3390/fluids7070222