Reconstruction of Gut Bacteria in Spodoptera frugiperda Infected by Beauveria bassiana Affects the Survival of Host Pest
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Insects
2.2. Molecular Identification of the Strains
2.3. The Virulence of Fungi against S. frugiperda
2.4. Analysis and Identification of the Intestinal Bacterial Diversity in the Insects
2.5. Determination of the Influence of the Gut Micro-Organisms of S. frugiperda against B. bassiana
2.6. Data Analysis
3. Results
3.1. Isolation and Identification of B. bassiana
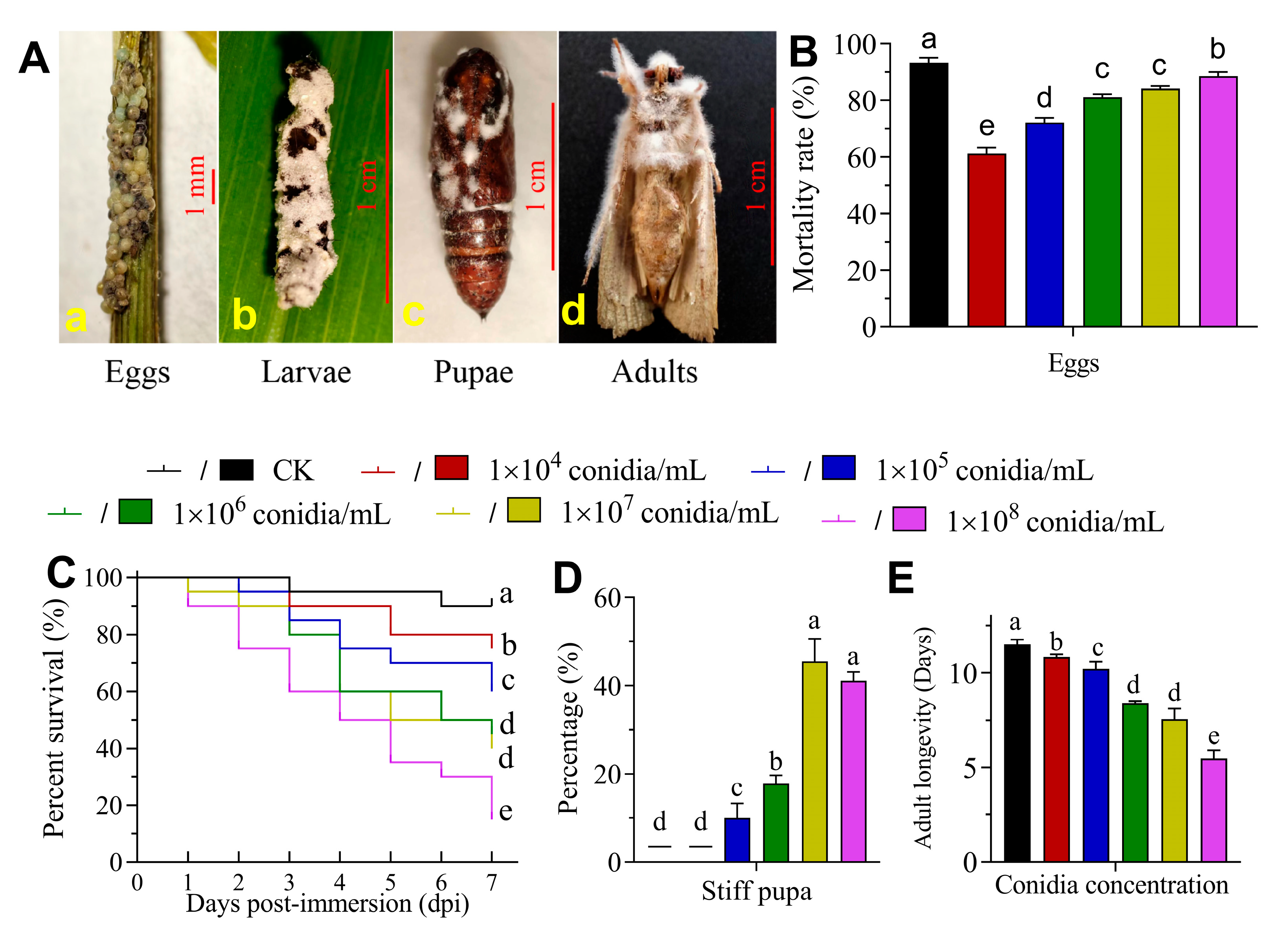
3.2. The Virulence of B. bassiana to S. frugiperda
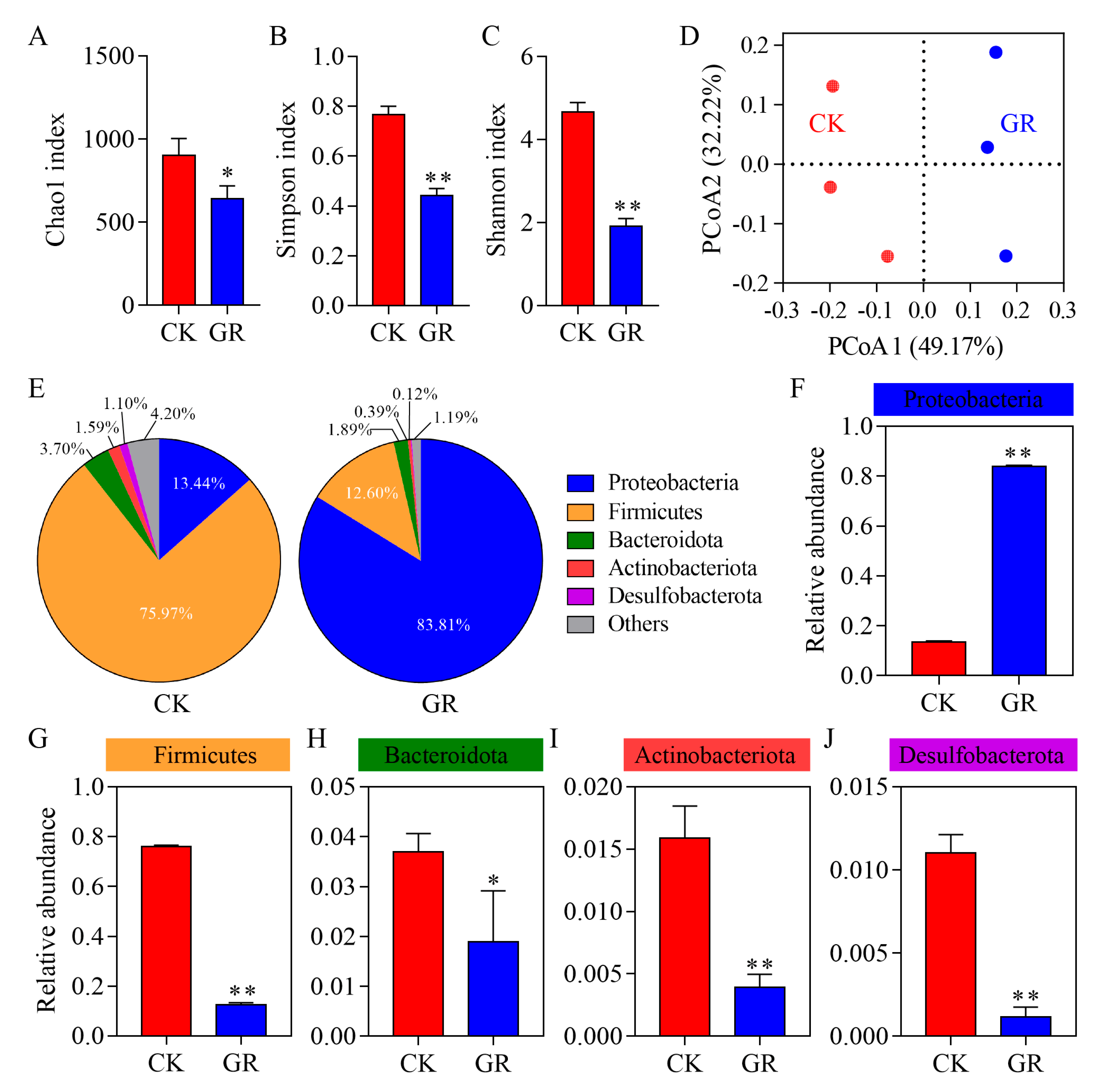
3.3. Analysis of the Abundance and Diversity of the Gut Bacteria in S. frugiperda
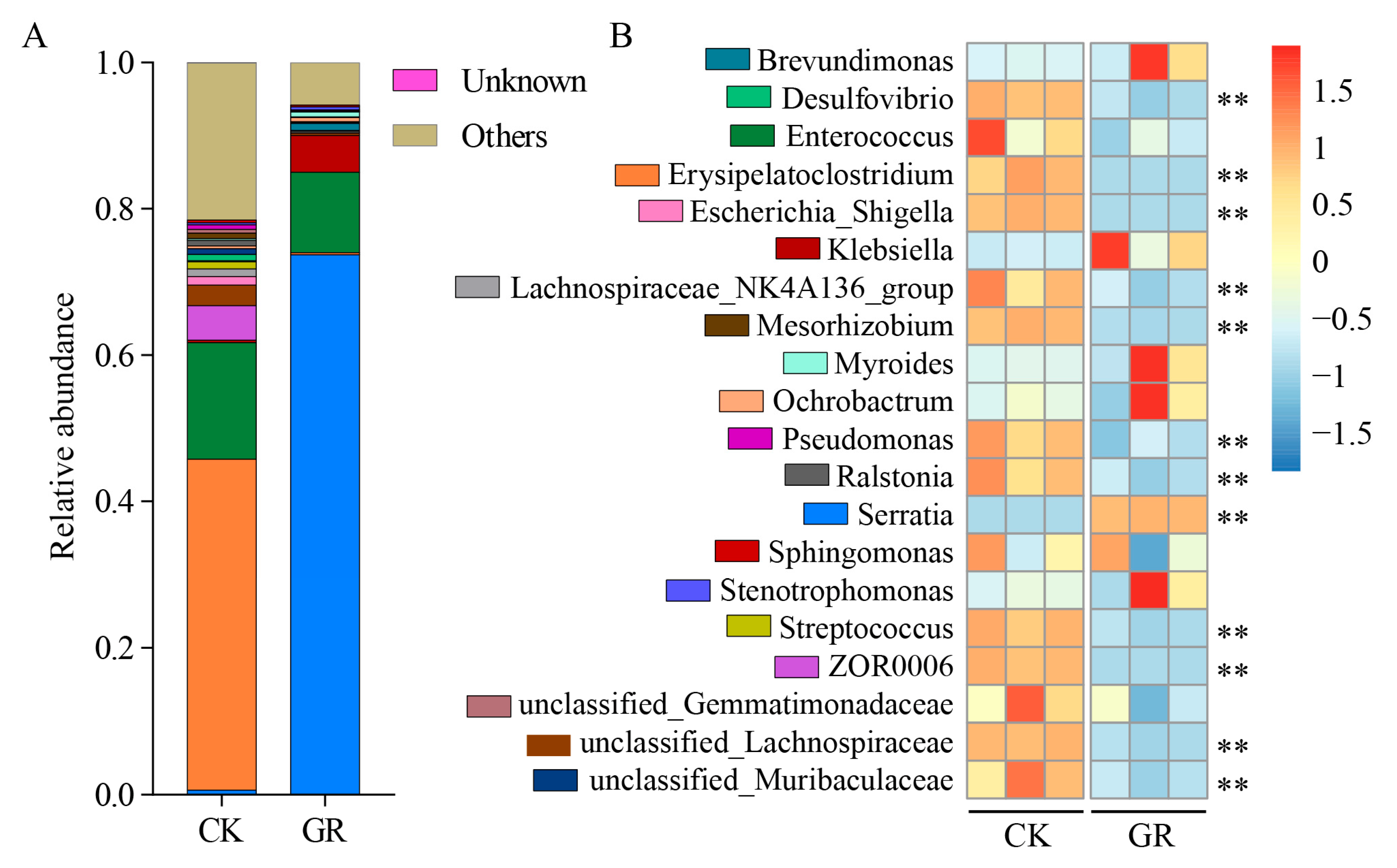
3.4. Analysis of the Abundance and Diversity of the Gut Bacteria at the Genus Level in S. frugiperda
3.5. Prediction of the Microbial Function
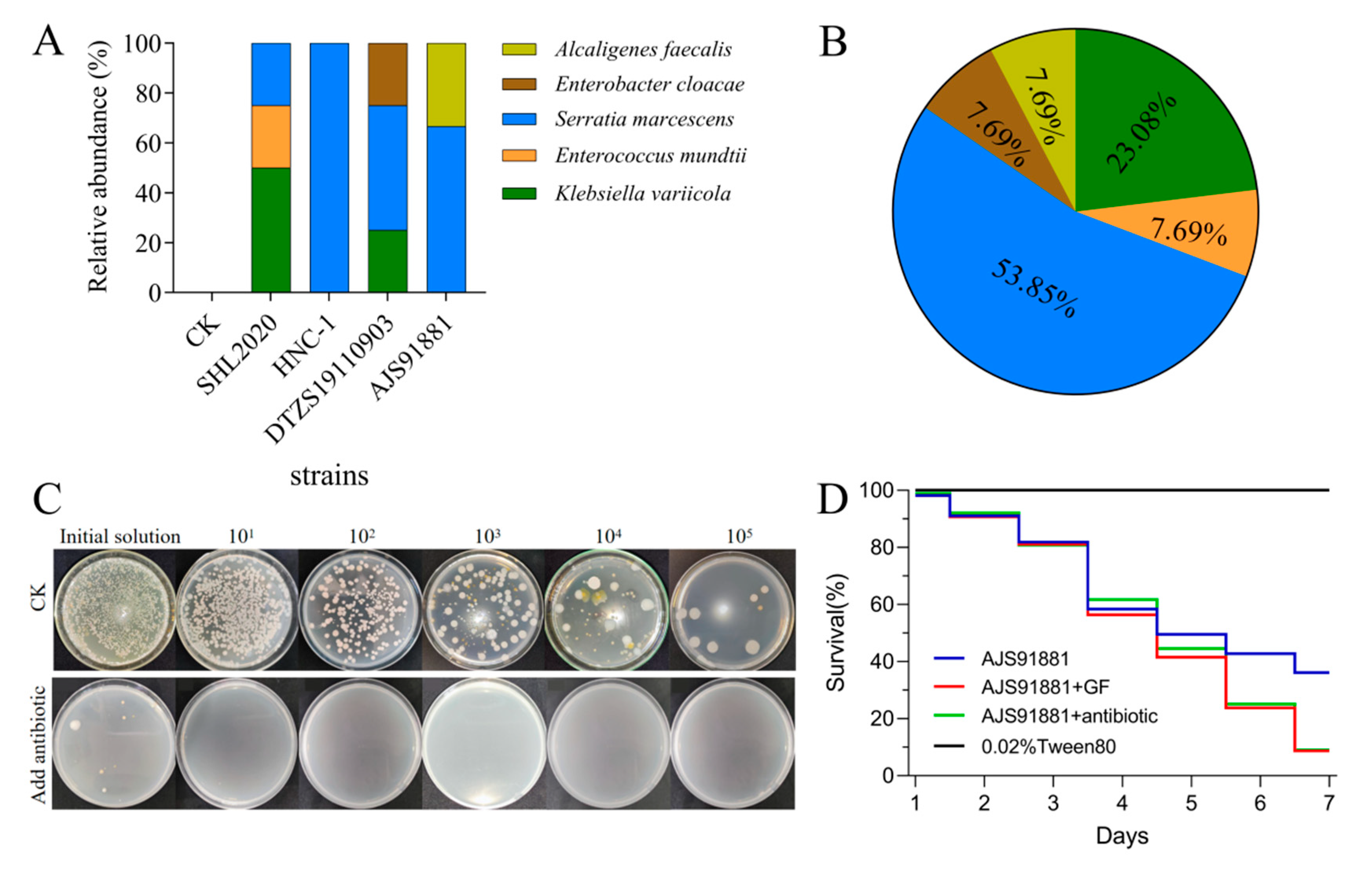
3.6. Infection with B. bassiana Increased the S. marcescens’ Abundance in the Gut of S. frugiperda Larvae
3.7. Intestinal Bacteria Influence the Virulence of Beauveria bassiana to Spodoptera frugiperda
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rioba, N.B.; Stevenson, P.C. Opportunities and Scope for Botanical Extracts and Products for the Management of Fall Armyworm (Spodoptera frugiperda) for Smallholders in Africa. Plants 2020, 9, 207. [Google Scholar] [CrossRef]
- Van den Berg, J.; Brewer, M.J.; Reisig, D.D. A Special Collection: Spodoptera frugiperda (Fall Armyworm): Ecology and Management of its World-Scale Invasion Outside of the Americas. J. Econ. Entomol. 2022, 115, 1725–1728. [Google Scholar] [CrossRef]
- Nagoshi, R.N.; Htain, N.N.; Boughton, D.; Zhang, L.; Xiao, Y.; Nagoshi, B.Y.; Mota-Sanchez, D. Southeastern Asia fall armyworms are closely related to populations in Africa and India, consistent with common origin and recent migration. Sci. Rep. 2020, 10, 1421. [Google Scholar] [CrossRef]
- Beuzelin, J.M.; Larsen, D.J.; Roldan, E.L.; Schwan, R.E. Susceptibility to Chlorantraniliprole in Fall Armyworm (Lepidoptera: Noctuidae) Populations Infesting Sweet Corn in Southern Florida. J. Econ. Entomol. 2022, 115, 224–232. [Google Scholar] [CrossRef]
- Ying, S.H.; Feng, M.G.; Keyhani, N.O. A carbon responsive G-protein coupled receptor modulates broad developmental and genetic networks in the entomopathogenic fungus, Beauveria bassiana. Environ. Microbiol. 2013, 15, 2902–2921. [Google Scholar] [CrossRef]
- Shah, P.A.; Pell, J.K. Entomopathogenic fungi as biological control agents. Appl. Microbiol. Biot. 2003, 61, 413–423. [Google Scholar] [CrossRef]
- Wanchoo, A.; Lewis, M.W.; Keyhani, N.O. Lectin mapping reveals stage-specific display of surface carbohydrates in in vitro and haemolymph-derived cells of the entomopathogenic fungus Beauveria bassiana. Microbiology 2009, 155, 3121–3133. [Google Scholar] [CrossRef]
- Ortiz-Urquiza, A.; Luo, Z.; Keyhani, N.O. Improving mycoinsecticides for insect biological control. Appl. Microbiol. Biot. 2015, 99, 1057–1068. [Google Scholar] [CrossRef]
- Wojda, I. Immunity of the greater wax moth Galleria mellonella. Insect Sci. 2017, 24, 342–357. [Google Scholar] [CrossRef]
- Joop, G.; Vilcinskas, A. Coevolution of parasitic fungi and insect hosts. Zoology 2016, 119, 350–358. [Google Scholar] [CrossRef]
- Zhang, L.B.; Feng, M.G. Antioxidant enzymes and their contributions to biological control potential of fungal insect pathogens. Appl. Microbiol. Biot. 2018, 102, 4995–5004. [Google Scholar] [CrossRef]
- Chen, B.; Yu, T.; Xie, S.; Du, K.; Liang, X.; Lan, Y.; Sun, C.; Lu, X.; Shao, Y. Comparative shotgun metagenomic data of the silkworm Bombyx mori gut microbiome. Sci. Data 2018, 5, 180285. [Google Scholar] [CrossRef]
- Chen, B.; Teh, B.S.; Sun, C.; Hu, S.; Lu, X.; Boland, W.; Shao, Y. Biodiversity and Activity of the Gut Microbiota across the Life History of the Insect Herbivore Spodoptera littoralis. Sci. Rep. 2016, 6, 29505. [Google Scholar] [CrossRef]
- Martinez-Solis, M.; Collado, M.C.; Herrero, S. Influence of Diet, Sex, and Viral Infections on the Gut Microbiota Composition of Spodoptera exigua Caterpillars. Front. Microbiol. 2020, 11, 753. [Google Scholar] [CrossRef]
- Deguenon, J.M.; Dhammi, A.; Ponnusamy, L.; Travanty, N.V.; Cave, G.; Lawrie, R.; Mott, D.; Reisig, D.; Kurtz, R.; Roe, R.M. Bacterial Microbiota of Field-Collected Helicoverpa zea (Lepidoptera: Noctuidae) from Transgenic Bt and Non-Bt Cotton. Microorganisms 2021, 9, 878. [Google Scholar] [CrossRef]
- Jones, A.G.; Mason, C.J.; Felton, G.W.; Hoover, K. Host plant and population source drive diversity of microbial gut communities in two polyphagous insects. Sci. Rep. 2019, 9, 2792. [Google Scholar] [CrossRef]
- Engel, P.; Moran, N.A. The gut microbiota of insects-diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef]
- Grenier, T.; Leulier, F. How commensal microbes shape the physiology of Drosophila melanogaster. Curr. Opin. Insect Sci. 2020, 41, 92–99. [Google Scholar] [CrossRef]
- Broderick, N.A.; Raffa, K.F.; Handelsman, J. Midgut bacteria required for Bacillus thuringiensis insecticidal activity. Proc. Natl. Acad. Sci. USA 2006, 103, 15196–15199. [Google Scholar] [CrossRef]
- Petersen, L.M.; Tisa, L.S. Friend or foe? A review of the mechanisms that drive Serratia towards diverse lifestyles. Can. J. Microbiol. 2013, 59, 627–640. [Google Scholar] [CrossRef]
- Grimont, P.A.; Grimont, F. The genus Serratia. Annu. Rev. Microbiol. 1978, 32, 221–248. [Google Scholar] [CrossRef]
- Steele, M.I.; Motta, E.; Gattu, T.; Martinez, D.; Moran, N.A. The Gut Microbiota Protects Bees from Invasion by a Bacterial Pathogen. Microbiol. Spectr. 2021, 9, e39421. [Google Scholar] [CrossRef]
- Andrejko, M.; Mizerska-Dudka, M.; Jakubowicz, T. Antibacterial activity in vivo and in vitro in the hemolymph of Galleria mellonella infected with Pseudomonas aeruginosa. Comp. Biochem. Phys. B 2009, 152, 118–123. [Google Scholar] [CrossRef]
- Lee, K.S.; Yun, E.Y.; Goo, T.W. Antimicrobial Activity of an Extract of Hermetia illucens Larvae Immunized with Lactobacillus casei against Salmonella Species. Insects 2020, 11, 704. [Google Scholar] [CrossRef]
- Jupatanakul, N.; Pengon, J.; Selisana, S.; Choksawangkarn, W.; Jaito, N.; Saeung, A.; Bunyong, R.; Posayapisit, N.; Thammatinna, K.; Kalpongnukul, N.; et al. Serratia marcescens secretes proteases and chitinases with larvicidal activity against Anopheles dirus. Acta Trop. 2020, 212, 105686. [Google Scholar] [CrossRef]
- Lai, S.; Tremblay, J.; Deziel, E. Swarming motility: A multicellular behaviour conferring antimicrobial resistance. Environ. Microbiol. 2009, 11, 126–136. [Google Scholar] [CrossRef]
- Thong-On, A.; Suzuki, K.; Noda, S.; Inoue, J.; Kajiwara, S.; Ohkuma, M. Isolation and characterization of anaerobic bacteria for symbiotic recycling of uric acid nitrogen in the gut of various termites. Microbes Environ. 2012, 27, 186–192. [Google Scholar] [CrossRef]
- Cirimotich, C.M.; Dong, Y.; Clayton, A.M.; Sandiford, S.L.; Souza-Neto, J.A.; Mulenga, M.; Dimopoulos, G. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 2011, 332, 855–858. [Google Scholar] [CrossRef]
- Cirimotich, C.M.; Ramirez, J.L.; Dimopoulos, G. Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe 2011, 10, 307–310. [Google Scholar] [CrossRef]
- Ceja-Navarro, J.A.; Vega, F.E.; Karaoz, U.; Hao, Z.; Jenkins, S.; Lim, H.C.; Kosina, P.; Infante, F.; Northen, T.R.; Brodie, E.L. Gut microbiota mediate caffeine detoxification in the primary insect pest of coffee. Nat. Commun. 2015, 6, 7618. [Google Scholar] [CrossRef]
- Kalappa, D.M.; Subramani, P.A.; Basavanna, S.K.; Ghosh, S.K.; Sundaramurthy, V.; Uragayala, S.; Tiwari, S.; Anvikar, A.R.; Valecha, N. Influence of midgut microbiota in Anopheles stephensi on Plasmodium berghei infections. Malaria J. 2018, 17, 385. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Lu, X.J.; Wu, Q. Gut Microbiota and Acute Central Nervous System Injury: A New Target for Therapeutic Intervention. Front. Immunol. 2021, 12, 800796. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Peng, Y.; Di, T.; Du, G.; Chen, B. Virulence of Metarhizium rileyi Is Determined by Its Growth and Antioxidant Stress and the Protective and Detoxifying Enzymes of Spodoptera frugiperda. Insects 2023, 14, 260. [Google Scholar] [CrossRef]
- Vancov, T.; Keen, B. Amplification of soil fungal community DNA using the ITS86F and ITS4 primers. FEMS Microbiol. Lett. 2009, 296, 91–96. [Google Scholar] [CrossRef]
- Manter, D.K.; Vivanco, J.M. Use of the ITS primers, ITS1F and ITS4, to characterize fungal abundance and diversity in mixed-template samples by qPCR and length heterogeneity analysis. J. Microbiol. Methods 2007, 71, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Zheng, R.; Cheng, L.; Peng, J.; Li, Q.; Yang, F.; Yang, D.; Xia, Y.; Tang, Q. Comparative analysis of gut microbiota and immune genes linked with the immune system of wild and captive Spodoptera frugiperda (Lepidoptera: Noctuidae). Dev. Comp. Immunol. 2023, 138, 104530. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Li, S.; Wu, J.; Huo, Y.; Zhao, X.; Xue, L. Profiling multiple heavy metal contamination and bacterial communities surrounding an iron tailing pond in Northwest China. Sci. Total Environ. 2021, 752, 141827. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Hill, T.C.; Walsh, K.A.; Harris, J.A.; Moffett, B.F. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 2003, 43, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xiao, G.; Du, G.; Chang, L.; Yang, Y.; Ye, J.; Chen, B. Glutamicibacter halophytocola-mediated host fitness of potato tuber moth on Solanaceae crops. Pest. Manag. Sci. 2022, 78, 3920–3930. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.H.; Feng, M.G. Novel blastospore-based transformation system for integration of phosphinothricin resistance and green fluorescence protein genes into Beauveria bassiana. Appl. Microbiol. Biot. 2006, 72, 206–210. [Google Scholar] [CrossRef]
- Mason, C.J.; Peiffer, M.; Felton, G.W.; Hoover, K. Host-Specific larval lepidopteran mortality to pathogenic Serratia mediated by poor diet. J. Invertebr. Pathol. 2022, 194, 107818. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Urquiza, A.; Keyhani, N.O. Action on the Surface: Entomopathogenic Fungi versus the Insect Cuticle. Insects 2013, 4, 357–374. [Google Scholar] [CrossRef]
- Li, G.; Zheng, X.; Zhu, Y.; Long, Y.; Xia, X. Bacillus symbiont drives alterations in intestinal microbiota and circulating metabolites of lepidopteran host. Environ. Microbiol. 2022, 24, 4049–4064. [Google Scholar] [CrossRef] [PubMed]
- Heise, P.; Liu, Y.; Degenkolb, T.; Vogel, H.; Schaberle, T.F.; Vilcinskas, A. Antibiotic-Producing Beneficial Bacteria in the Gut of the Burying Beetle Nicrophorus vespilloides. Front. Microbiol. 2019, 10, 1178. [Google Scholar] [CrossRef]
- Bai, S.; Yao, Z.; Raza, M.F.; Cai, Z.; Zhang, H. Regulatory mechanisms of microbial homeostasis in insect gut. Insect Sci. 2021, 28, 286–301. [Google Scholar] [CrossRef]
- Chevrette, M.G.; Carlson, C.M.; Ortega, H.E.; Thomas, C.; Ananiev, G.E.; Barns, K.J.; Book, A.J.; Cagnazzo, J.; Carlos, C.; Flanigan, W.; et al. The antimicrobial potential of Streptomyces from insect microbiomes. Nat. Commun. 2019, 10, 516. [Google Scholar] [CrossRef]
- Zhang, C.; Teng, B.; Liu, H.; Wu, C.; Wang, L.; Jin, S. Impact of Beauveria bassiana on antioxidant enzyme activities and metabolomic profiles of Spodoptera frugiperda. J. Invertebr. Pathol. 2023, 198, 107929. [Google Scholar] [CrossRef]
- Ding, J.L.; Peng, Y.J.; Chu, X.L.; Feng, M.G.; Ying, S.H. Autophagy-related gene BbATG11 is indispensable for pexophagy and mitophagy, and contributes to stress response, conidiation and virulence in the insect mycopathogen Beauveria bassiana. Environ. Microbiol. 2018, 20, 3309–3324. [Google Scholar] [CrossRef]
- Lin, H.Y.; Ding, J.L.; Peng, Y.J.; Feng, M.G.; Ying, S.H. Proteomic and Phosphoryproteomic Investigations Reveal that Autophagy-Related Protein 1, a Protein Kinase for Autophagy Initiation, Synchronously Deploys Phosphoregulation on the Ubiquitin-Like Conjugation System in the Mycopathogen Beauveria bassiana. mSystems 2022, 7, e146321. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.J.; Wang, J.J.; Lin, H.Y.; Ding, J.L.; Feng, M.G.; Ying, S.H. HapX, an Indispensable bZIP Transcription Factor for Iron Acquisition, Regulates Infection Initiation by Orchestrating Conidial Oleic Acid Homeostasis and Cytomembrane Functionality in Mycopathogen Beauveria bassiana. mSystems 2020, 5, e00695-20. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.J.; Ding, J.L.; Feng, M.G.; Ying, S.H. Glc8, a regulator of protein phosphatase type 1, mediates oxidation tolerance, asexual development and virulence in Beauveria bassiana, a filamentous entomopathogenic fungus. Curr. Genet. 2019, 65, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.J.; Ding, J.L.; Lin, H.Y.; Feng, M.G.; Ying, S.H. A virulence-related lectin traffics into eisosome and contributes to functionality of cytomembrane and cell-wall in the insect-pathogenic fungus Beauveria bassiana. Fungal Biol. 2021, 125, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.J.; Zhang, H.; Feng, M.G.; Ying, S.H. SterylAcetyl Hydrolase 1 (BbSay1) Links Lipid Homeostasis to Conidiogenesis and Virulence in the Entomopathogenic Fungus Beauveria bassiana. J. Fungi 2022, 8, 292. [Google Scholar] [CrossRef]
- Ding, J.L.; Hou, J.; Feng, M.G.; Ying, S.H. Transcriptomic analyses reveal comprehensive responses of insect hemocytes to mycopathogen Beauveria bassiana, and fungal virulence-related cell wall protein assists pathogen to evade host cellular defense. Virulence 2020, 11, 1352–1365. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Yi, C.; Chen, X.; Liu, C.; Zhang, C.; Chen, Q.; Chen, S.; Liu, H.; Pu, D. The Adaptive Evolution in the Fall Armyworm Spodoptera frugiperda (Lepidoptera: Noctuidae) Revealed by the Diversity of Larval Gut Bacteria. Genes 2023, 14, 321. [Google Scholar] [CrossRef]
- Jang, S.; Kikuchi, Y. Impact of the insect gut microbiota on ecology, evolution, and industry. Curr. Opin. Insect Sci. 2020, 41, 33–39. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Y.; Wen, S.; Wang, G.; Zhang, X.; Di, T.; Du, G.; Chen, B.; Zhang, L. Reconstruction of Gut Bacteria in Spodoptera frugiperda Infected by Beauveria bassiana Affects the Survival of Host Pest. J. Fungi 2023, 9, 906. https://doi.org/10.3390/jof9090906
Peng Y, Wen S, Wang G, Zhang X, Di T, Du G, Chen B, Zhang L. Reconstruction of Gut Bacteria in Spodoptera frugiperda Infected by Beauveria bassiana Affects the Survival of Host Pest. Journal of Fungi. 2023; 9(9):906. https://doi.org/10.3390/jof9090906
Chicago/Turabian StylePeng, Yuejin, Shaohai Wen, Guang Wang, Xu Zhang, Teng Di, Guangzu Du, Bin Chen, and Limin Zhang. 2023. "Reconstruction of Gut Bacteria in Spodoptera frugiperda Infected by Beauveria bassiana Affects the Survival of Host Pest" Journal of Fungi 9, no. 9: 906. https://doi.org/10.3390/jof9090906
APA StylePeng, Y., Wen, S., Wang, G., Zhang, X., Di, T., Du, G., Chen, B., & Zhang, L. (2023). Reconstruction of Gut Bacteria in Spodoptera frugiperda Infected by Beauveria bassiana Affects the Survival of Host Pest. Journal of Fungi, 9(9), 906. https://doi.org/10.3390/jof9090906







