Screening and Analysis of Antifungal Strains Bacillus subtilis JF-4 and B. amylum JF-5 for the Biological Control of Fusarium Wilt of Banana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Screening of Antagonists
2.2. Antifungal Effect of Antagonistic Fermentation Supernatant
2.3. Identification of Antagonists
2.4. Scanning Electron Microscopy Detection
2.5. Controlling Effect of Antagonistic Bacteria on Banana Fusarium wilt in Greenhouse
2.6. Effects of Bactericidal Extracts on Pathogen Growth and Metabolism
2.7. Effects of Antagonistic Bacteria on the Related Indices of Pathogens
2.8. Component Analysis of Extracts of JF-4 and JF-5 by Gas Chromatography–Mass Spectrometry
2.9. Genomic DNA Extraction of B. subtilis JF-4
2.10. B. subtilis JF-4 Gene Library
2.11. Sequencing and Assembly of B. subtilis JF-4
2.12. Functional Annotation of B. subtilis JF-4 Genome
2.13. Statistical Analyses
3. Results
3.1. Isolation and Screening of Antagonists
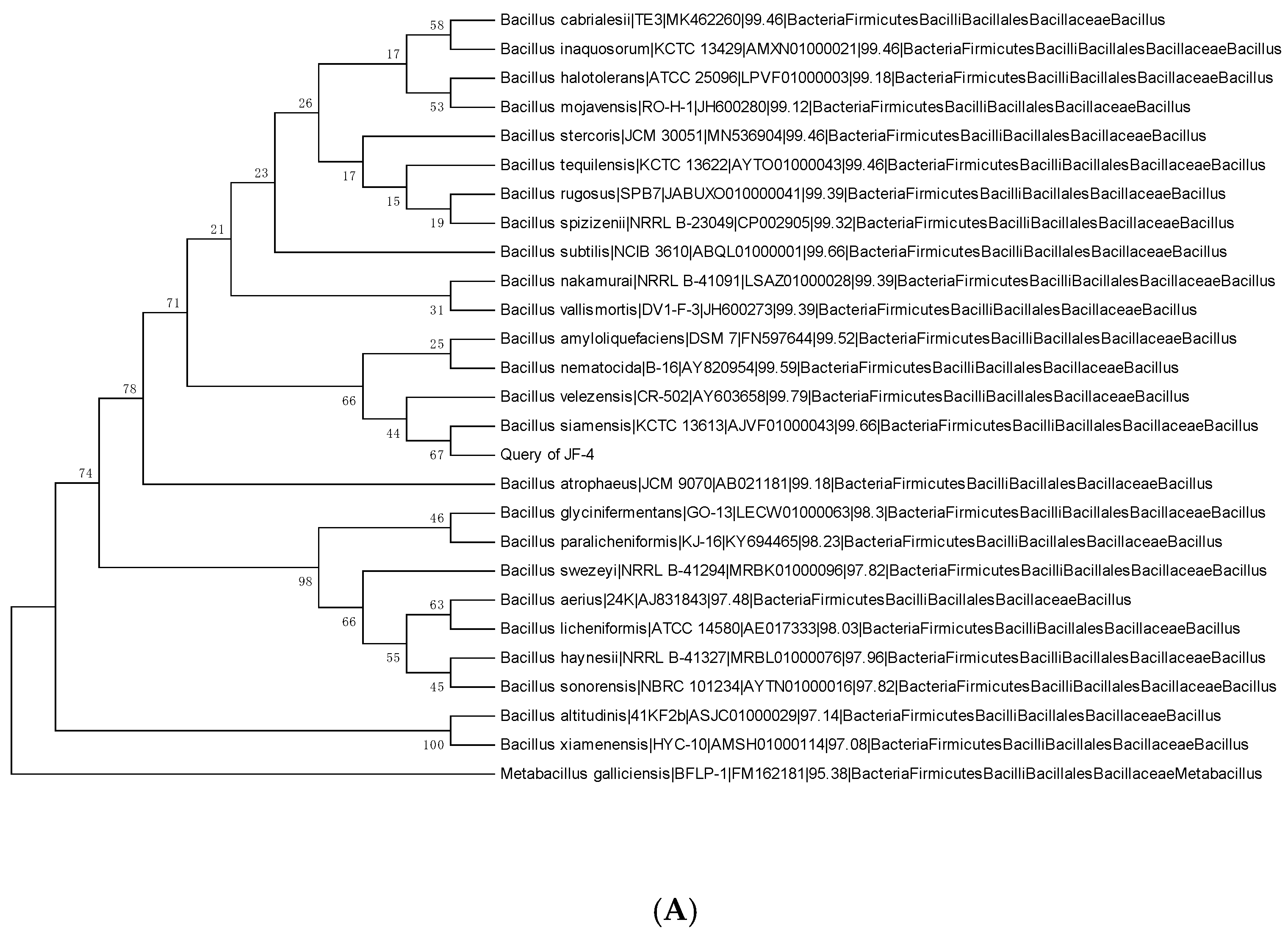
3.2. Physiology and Biochemistry of Antagonists and Identification of the 16S rRNA Gene
3.3. Amplification of Antagonistic-Specific Primers
3.4. Prevention and Control of Antagonistic Bacteria in Greenhouse
3.5. Effect of Antagonistic JF-4 and JF-5 Extracts on the Cell Membrane of Foc TR4
3.5.1. The Effect of CAT Content
3.5.2. Effects of Ergosterol Content in Pathogenic Fungi
3.5.3. Changes in Soluble Protein Content
3.5.4. Effect of the Pectinase Levels
3.6. Analysis of Metabolic Components of Strain JF-4 and JF-5 Extracts Using GC–MS
3.7. Genomic Characterization of B. subtilis JF-4
3.7.1. B. subtilis JF-4 Genome Functional Annotation
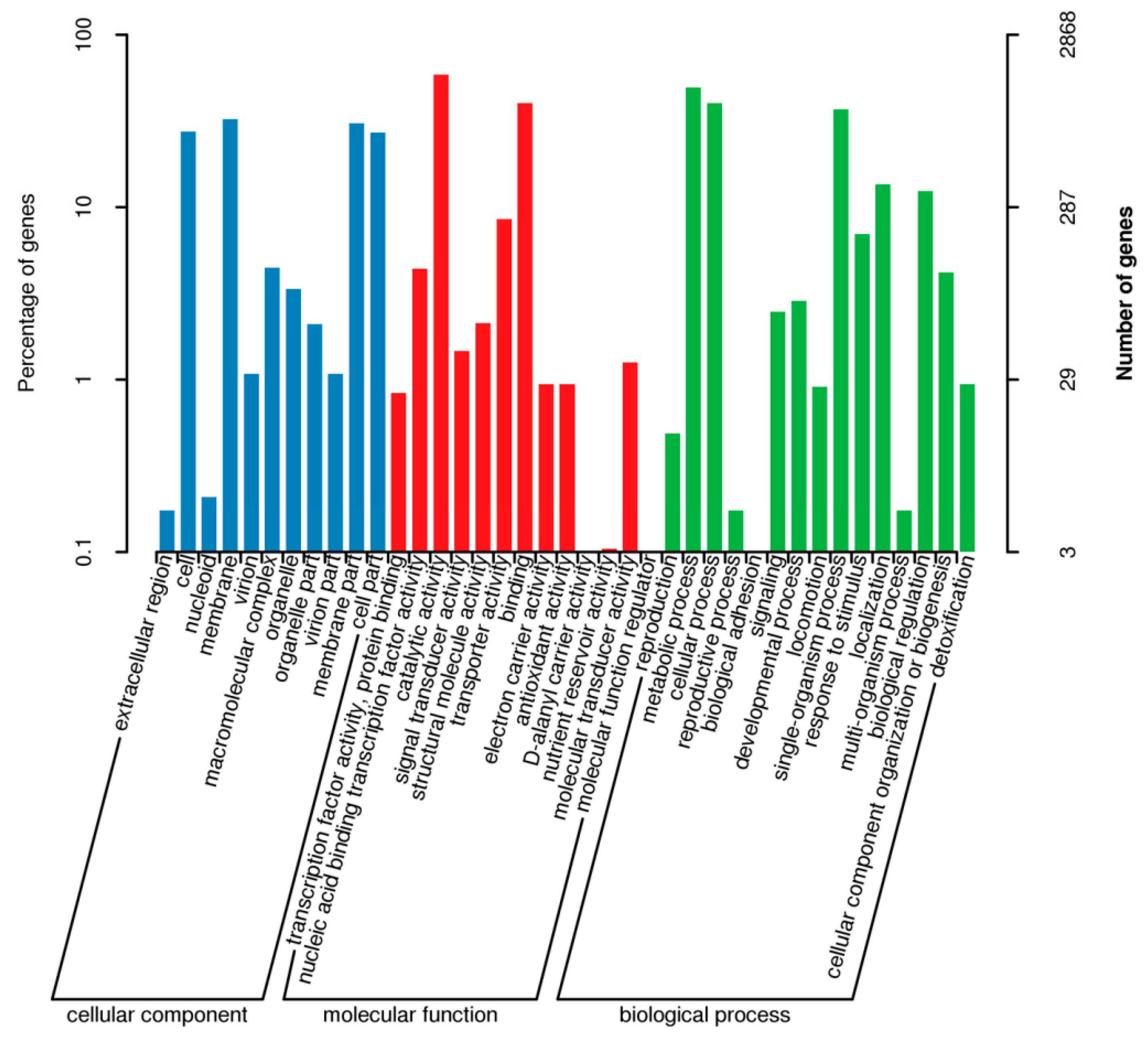
3.7.2. GO Database Functional Annotation
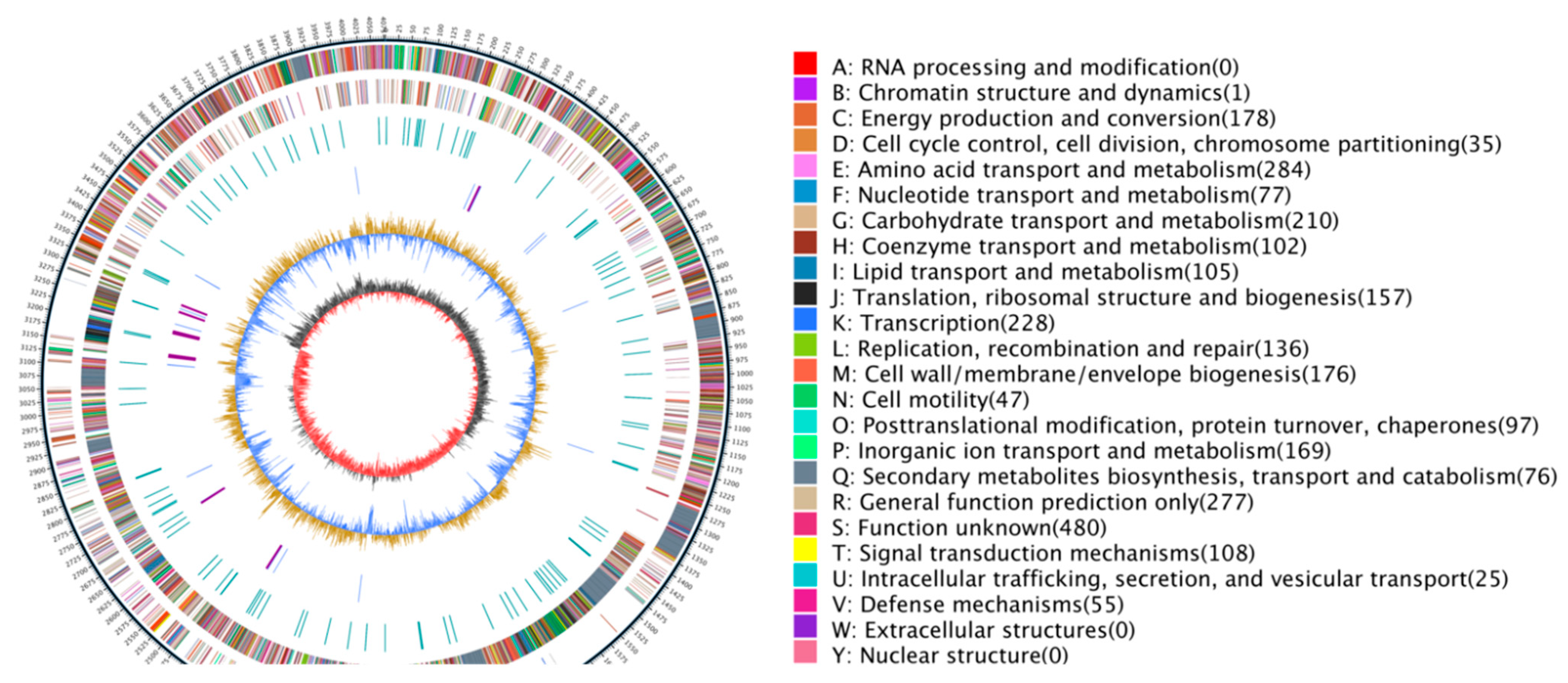
3.7.3. COG Database Functional Notes
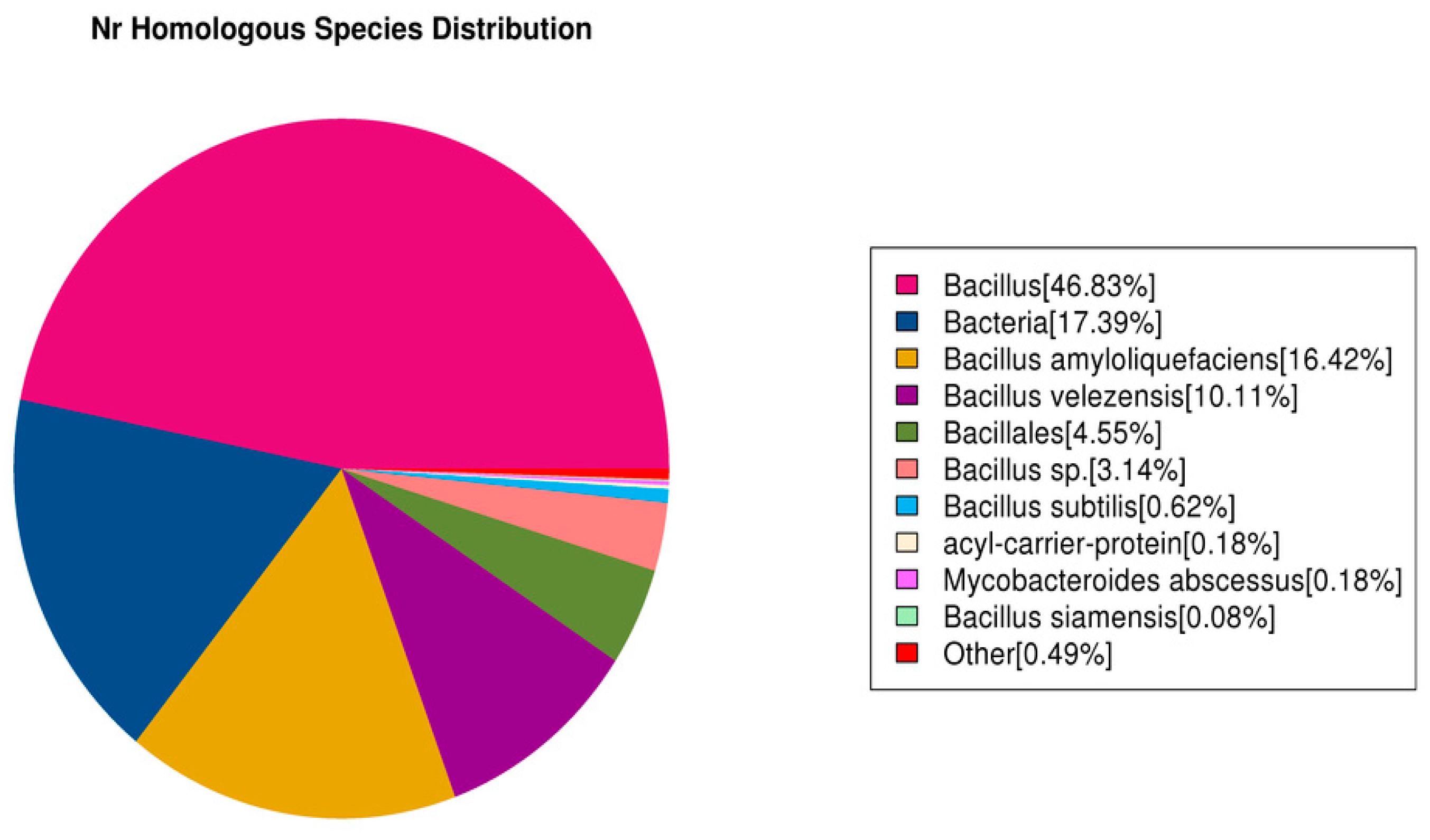
3.7.4. Nr Database Functional Notes
3.7.5. Expression of Key Genes of Secondary Metabolites of B. subtilis JF-4
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Awla, H.K.; Kadir, J.; Othman, R.; Rashid, T.S.; Wong, M.Y. Bioactive compounds produced by Streptomyces sp. isolate UPMRS4 and antifungal activity against Pyricularia oryzae. Am. J. Plant Sci. 2016, 7, 1077–1085. [Google Scholar] [CrossRef]
- Abidin, Z.A.Z.; Malek, N.A.; Zainuddin, Z.; Chowdhury, A.J.K. Selective isolation and antagonistic activity of actinomycetes from mangrove forest of Pahang, Malaysia. Front. Life Sci. 2016, 9, 24–31. [Google Scholar] [CrossRef]
- Alijani, Z.; Amini, J.; Ashengroph, M.; Bahramnejad, B. Antifungal activity of volatile compounds produced by Staphylococcus sciuri strain MarR44 and its potential for the biocontrol of Colletotrichum nymphaeae, causal agent strawberry anthracnose. Int. J. Food Microbiol. 2019, 307, 108276. [Google Scholar] [CrossRef] [PubMed]
- Ambrico, A.; Trupo, M. Efficacy of cell free supernatant from Bacillus subtilis ET-1, an Iturin A producer strain, on biocontrol of green and gray mold. Postharvest Biol. Technol. 2017, 134, 5–10. [Google Scholar] [CrossRef]
- Bagy, H.M.K.; Hassan, E.A.; Nafady, N.A.; Dawood, M.F. Efficacy of arbuscular mycorrhizal fungi and endophytic strain Epicoccum nigrum ASU11 as biocontrol agents against blackleg disease of potato caused by bacterial strain Pectobacterium carotovora subsp. atrosepticum PHY7. Biol. Control 2019, 134, 103–113. [Google Scholar] [CrossRef]
- Bauer, J.S.; Hauck, N.; Christof, L.; Mehnaz, S.; Gust, B.; Gross, H. The Systematic Investigation of the Quorum Sensing System of the Biocontrol Strain Pseudomonas chlororaphis subsp. aurantiaca PB-St2 Unveils aurI to Be a Biosynthetic Origin for 3-Oxo-Homoserine Lactones. PLoS ONE 2016, 11, e0167002. [Google Scholar] [CrossRef] [PubMed]
- Ben Abdallah, D.; Tounsi, S.; Gharsallah, H.; Hammami, A.; Frikha-Gargouri, O. Lipopeptides from Bacillus amyloliquefaciens strain 32a as promising biocontrol compounds against the plant pathogen Agrobacterium tumefaciens. Environ. Sci. Pollut. Res. 2018, 25, 36518–36529. [Google Scholar] [CrossRef]
- DeFilippi, S.; Groulx, E.; Megalla, M.; Mohamed, R.; Avis, T.J. Fungal Competitors Affect Production of Antimicrobial Lipopeptides in Bacillus subtilis Strain B9-5. J. Chem. Ecol. 2018, 44, 374–383. [Google Scholar] [CrossRef]
- Geng, S.; Kong, X.; Song, G.; Jia, M.; Guan, J.; Wang, F.; Qin, Z.; Wu, L.; Lan, X.; Li, A.; et al. Dna methylation dynamics during the interaction of wheat progenitor Aegilops tauschii with the obligate biotrophic fungus Blumeria graminis f sp tritici. New Phytol. 2019, 221, 1023–1035. [Google Scholar] [CrossRef]
- Nigris, S.; Baldan, E.; Tondello, A.; Zanella, F.; Vitulo, N.; Favaro, G.; Guidolin, V.; Bordin, N.; Telatin, A.; Barizza, E.; et al. Biocontrol traits of Bacillus licheniformis GL174, a culturable endophyte of Vitis vinifera cv. Glera. BMC Microbiol. 2018, 18, 133. [Google Scholar] [CrossRef]
- Pinkas, D.; Fišer, R.; Kozlík, P.; Dolejšová, T.; Hryzáková, K.; Konopásek, I.; Mikušová, G. Bacillus subtilis cardiolipin protects its own membrane against surfactin-induced permeabilization. Biochim. Biophys. Acta (BBA) Biomembr. 2020, 1862, 183405. [Google Scholar] [CrossRef] [PubMed]
- Hashem, A.; Tabassum, B.; Fathi Abd Allah, E. Bacillus subtilis: A plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi J. Biol. Sci. 2019, 26, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.H.; Jiang, Z.D. Phylogenetic analysis of Endophytes from Bitter Melon (Momordica charantia) in Guangdong province. Agric. Sci. 2015, 06, 609–621. [Google Scholar]
- Kavino, M.; Manoranjitham, S.K. In vitro bacterization of banana (Musa spp.) with native endophytic and rhi-zospheric bacterial isolates: Novel ways to combat Fusarium wilt. Eur. J. Plant Pathol. 2018, 151, 371–387. [Google Scholar] [CrossRef]
- Kuncharoen, N.; Fukasawa, W.; Mori, M.; Shiomi, K.; Tanasupawat, S. Diversity and Antimicrobial Activity of Endophytic Actinomycetes Isolated from Plant Roots in Thailand. Microbiology 2019, 88, 479–488. [Google Scholar] [CrossRef]
- He, H.; Hao, X.; Zhou, W.; Shi, N.; Feng, J.; Han, L. Identification of antimicrobial metabolites produced by a potential biocontrol Actinomycete strain A217. J. Appl. Microbiol. 2020, 128, 1143–1152. [Google Scholar] [CrossRef]
- Hennessy, R.C.; Glaring, M.A.; Olsson, S.; Stougaard, P. Transcriptomic profiling of microbe–microbe interactions reveals the specific response of the biocontrol strain P. fluorescens In5 to the phytopathogen Rhizoctonia solani. BMC Res. Notes 2017, 10, 376. [Google Scholar] [CrossRef]
- Hopkins, D.L.; Benyamini, Y. Biocontrol of Pierce’s disease of grapevine and citrus greening with a benign strain of Xylella fastidiosa. Phytopathology 2018, 108, 132. [Google Scholar]
- Léger, G.; Novinscak, A.; Biessy, A.; Lamarre, S.; Filion, M. In Tuber Biocontrol of Potato Late Blight by a Collection of Phenazine-1-Carboxylic Acid-Producing Pseudomonas spp. Microorganisms 2021, 9, 2525. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; Feng, X.; Zhang, J.; He, X. Correction to: A Bjerkandera adust new strain as a potential biocontrol agent against wheat scab. Int. Microbiol. 2022, 25, 831. [Google Scholar] [CrossRef]
- Liu, H.; Zeng, Q.; Yalimaimaiti, N.; Wang, W.; Zhang, R.; Yao, J. Comprehensive genomic analysis of Bacillus velezensis AL7 reveals its biocontrol potential against Verticillium wilt of cotton. Mol. Genet. Genom. 2021, 296, 1287–1298. [Google Scholar] [CrossRef]
- Liu, Y.; Teng, K.; Wang, T.; Dong, E.; Zhang, M.; Tao, Y.; Zhong, J. Antimicrobial Bacillus velezensis HC6: Production of three kinds of lipopeptides and biocontrol potential in maize. J. Appl. Microbiol. 2020, 5, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Moore, G.G.; Lebar, M.D.; Carter-Wientjes, C.H. The role of extrolites secreted by nonaflatoxigenic Aspergillus flavus in biocontrol efficacy. J. Appl. Microbiol. 2019, 126, 1257–1264. [Google Scholar] [CrossRef] [PubMed]
- Moore, G.G.; Lebar, M.D.; Carter-Wientjes, C.H.; Gilbert, M.K. The potential role of fungal volatile organic compounds in Aspergillus flavus biocontrol efficacy. Biol. Control 2021, 160, 104686. [Google Scholar] [CrossRef]
- Pennerman, K.K.; Yin, G.; Bennett, J.W.; Hua, S.-S.T. Aspergillus flavus NRRL 35739, a Poor Biocontrol Agent, May Have Increased Relative Expression of Stress Response Genes. J. Fungi 2019, 5, 53. [Google Scholar] [CrossRef]
- Potshangbam, M.; Sahoo, D.; Verma, P.; Verma, S.; Kalita, M.C.; Devi, S.I. Draft Genome Sequence of Bacillus altitudinis Lc5, a Biocontrol and Plant Growth-Promoting Endophyte Strain Isolated from Indigenous Black Rice of Manipur. Genome Announc. 2018, 6, e00601-18. [Google Scholar] [CrossRef]
- Singh, S.P.; Gaur, R. Evaluation of antagonistic and plant growth promoting activities of chitinolytic endophytic actinomycetes associated with medicinal plants against Sclerotium rolfsii in chickpea. J. Appl. Microbiol. 2016, 121, 506–518. [Google Scholar] [CrossRef]
- Song, S.; Chen, X.; Huang, D.; Xu, Y.; Zeng, H.; Hu, X.; Xu, B.; Jin, Z.; Wang, W. Identification of miRNAs differentially expressed in Fusarium wilt-resistant and susceptible banana varieties. S. Afr. J. Bot. 2016, 106, 244–249. [Google Scholar] [CrossRef]
- Su, L.; Shen, Z.; Ruan, Y.; Tao, C.; Chao, Y.; Li, R.; Shen, Q. Isolation of Antagonistic Endophytes from Banana Roots against Meloidogyne javanica and Their Effects on Soil Nematode Community. Front. Microbiol. 2017, 8, 2070. [Google Scholar] [CrossRef]
- Subedi, N.; Taylor, C.G.; Paul, P.A.; Miller, S.A. Combining partial host resistance with bacterial biocontrol agents improves outcomes for tomatoes infected with Ralstonia pseudosolanacearum. Crop. Prot. 2019, 135, 104776. [Google Scholar] [CrossRef]
- Tran-Dinh, N.; Pitt, J.I.; Markwell, P.J. Use of microsatellite markers to assess the competitive ability of nontoxigenic Aspergillus flavus strains in studies on biocontrol of aflatoxins in maize in Thailand. Biocontrol Sci. Technol. 2018, 28, 215–225. [Google Scholar] [CrossRef]
- Xiong, H.; Li, Y.; Cai, Y.; Cao, Y.; Wang, Y. Isolation of Bacillus amyloliquefaciens JK6 and identification of its lipopeptides surfactin for suppressing tomato bacterial wilt. RSC Adv. 2015, 13, 35–48. [Google Scholar] [CrossRef]
- Zouari, I.; Jlaiel, L.; Tounsi, S.; Trigui, M. Biocontrol activity of the endophytic Bacillus amyloliquefaciens strain CEIZ-11 against Pythium aphanidermatum and purification of its bioactive compounds. Biol. Control 2016, 100, 54–62. [Google Scholar] [CrossRef]
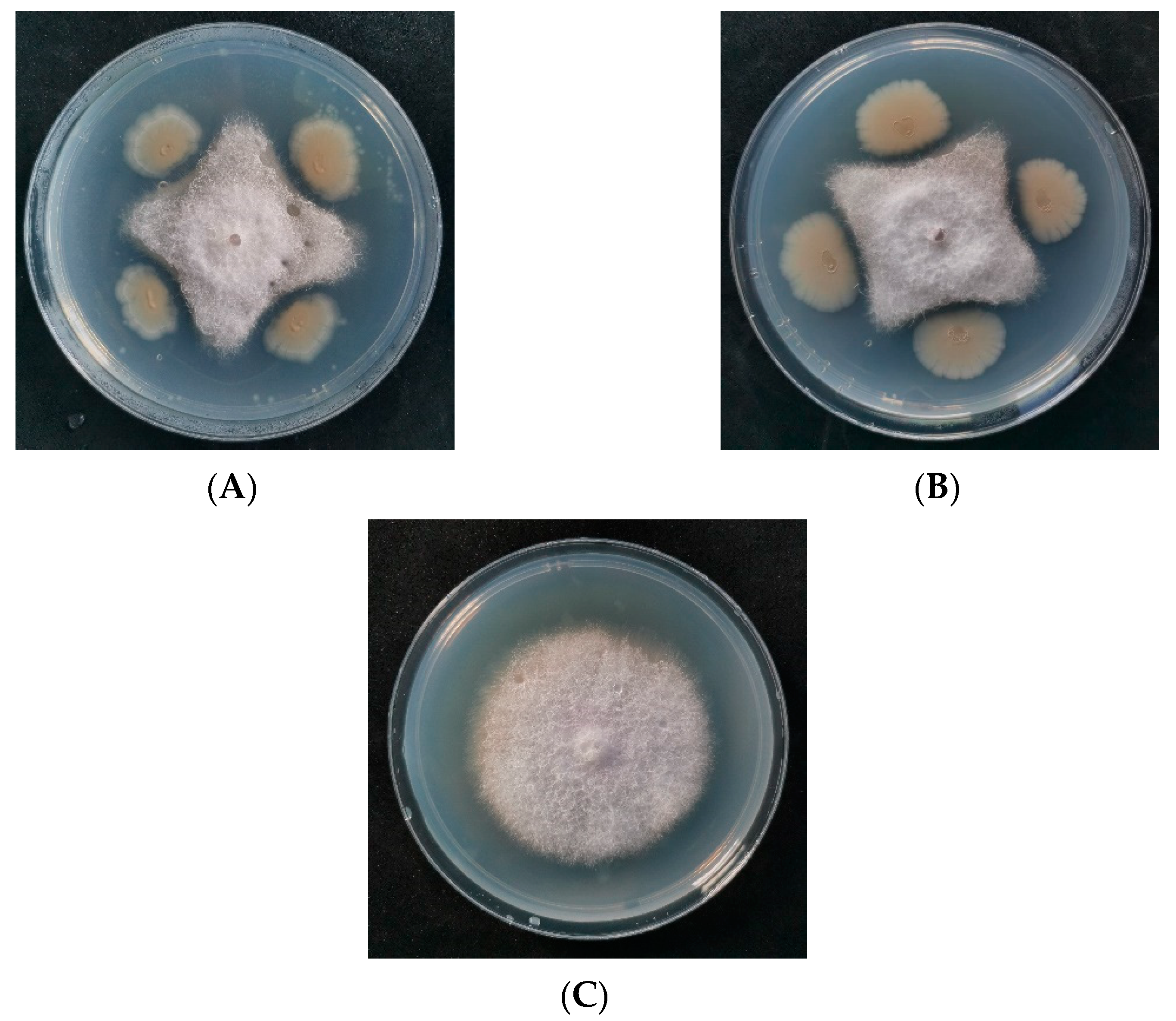





| Characteristics | JF-4 | JF-5 |
|---|---|---|
| Citrate | + | + |
| Hydrogen sulfide | − | − |
| Urease | − | − |
| Lactose | + | + |
| Gelatine | + | + |
| Glucose | + | + |
| Arabinose | + | + |
| Gram stain | + | + |
| Retention Time | Molecular Formula | Molecular Mass | Possible Compound Classification | |
|---|---|---|---|---|
| 3.45 | C6H12O2 | 116 | Methylguanidine | |
| 4.30 | C9H16ClN5 | 229 | Phenylacetamide, Others | |
| 11.38 | C12H20F3NO3 | 283 | 3-Furancarboxylic acid, methyl ester | |
| 15.33 | C5H4N4O3 | 168 | 2,4-bis (1,1-dimethylethyl)-phenol | |
| 2.23 | C14H22O | 206 | 2-hydroxypropanediamine acid dimethyl | |
| 6.71 | C7H10N2O2 | 154 | diethyl phthalate | 0.71 |
| 9.32 | C16H22ClNO | 279 | methyl ester, Hydrocarbons | 0.32 |
| 17.96 | C8H9NO | 135 | 2,4-decadienal | 1.96 |
| 20.43 | C27H54 | 378 | 7-methyl-Z-tetradecene-1-ol acetate | |
| 26.44 | C18H36O2 | 284 | dimethyltetradecan-1-thiol | |
| 30.32 | CH3(CH2)12CH3 | 198 | Tetradecane | |
| 4.89 | C8H8O2 | 136 | 2-phenylacetic acid | |
| 8.42 | C18H36O2 | 284 | hexadecanoic acid | |
| 25.67 | C16H32O2 | 256 | hexadecanoic acid, Acids | |
| 26.01 | C18H36O2 | 284 | 9,12-octadecadienoic acid | |
| 35.11 | C16H32O2 | 256 | oleic acid | |
| 10.48 | C17H34O2 | 270 | pyrazine-1,4-dione | |
| 22.44 | C19H38O2 | 298 | 2-diphthalic acid isooctyl esters | |
| 31.87 | C21H38O2 | 322 | 13-docosadecamide, Terpene | |
| 42.64 | C20H36O2 | 308 | Guaia-10(14),11-diene |
| Genome | Genome Assembly | ||
|---|---|---|---|
| Number of sequences | 117,186 | Scaffold length (bp) | 4,078,417 |
| Number of bases (bp) | 1,486,274,400 | Scaffold number | 1 |
| N50 reads length (bp) | 21,608 | Scaffold N50 (bp) | 4,078,417 |
| N90 reads length (bp) | 5366 | Scaffold N90 (bp) | 4,078,417 |
| Mean reads length (bp) | 12,683 | GC content (%) | 46.34 |
| Mean reads quality | 12.21 | Gaps number | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, Y.; Pang, Z.; Yin, S.; Xiao, W.; Hu, H.; Xie, J. Screening and Analysis of Antifungal Strains Bacillus subtilis JF-4 and B. amylum JF-5 for the Biological Control of Fusarium Wilt of Banana. J. Fungi 2023, 9, 886. https://doi.org/10.3390/jof9090886
Duan Y, Pang Z, Yin S, Xiao W, Hu H, Xie J. Screening and Analysis of Antifungal Strains Bacillus subtilis JF-4 and B. amylum JF-5 for the Biological Control of Fusarium Wilt of Banana. Journal of Fungi. 2023; 9(9):886. https://doi.org/10.3390/jof9090886
Chicago/Turabian StyleDuan, Yajie, Zhencai Pang, Shunli Yin, Weijun Xiao, Huigang Hu, and Jianghui Xie. 2023. "Screening and Analysis of Antifungal Strains Bacillus subtilis JF-4 and B. amylum JF-5 for the Biological Control of Fusarium Wilt of Banana" Journal of Fungi 9, no. 9: 886. https://doi.org/10.3390/jof9090886




