Unleashing the Potential of Bacterial Isolates from Apple Tree Rhizosphere for Biocontrol of Monilinia laxa: A Promising Approach for Combatting Brown Rot Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Antagonistic Bacteria
2.2. Collection, Isolation, and Identification of Fungal Isolates
2.3. Effects of Bacterial Metabolites on the Fungal Pathogen
2.4. In Vitro Dual Culture Bioassay of Bacterial Isolates versus Monilinia laxa
2.5. In Vivo Antagonism Experiment
2.6. Gene Sequencing and Phylogenetic Analysis
2.7. Enzyme Assay
2.7.1. Preparation of Colloidal Chitin
2.7.2. Detection of Chitinolytic Activity on Plates
2.7.3. Determination of Chitinolytic Activity Based on the Colorimetric Method
2.7.4. Detection of Protease Activity on Plates
2.7.5. Determination of Protease Activity Based on the Colorimetric Method
2.8. Statistical Analysis
3. Results
3.1. Isolation and Screening of Chitinase and Protease-Producing Bacterial Strains
3.2. Identification of Fungal Pathogens
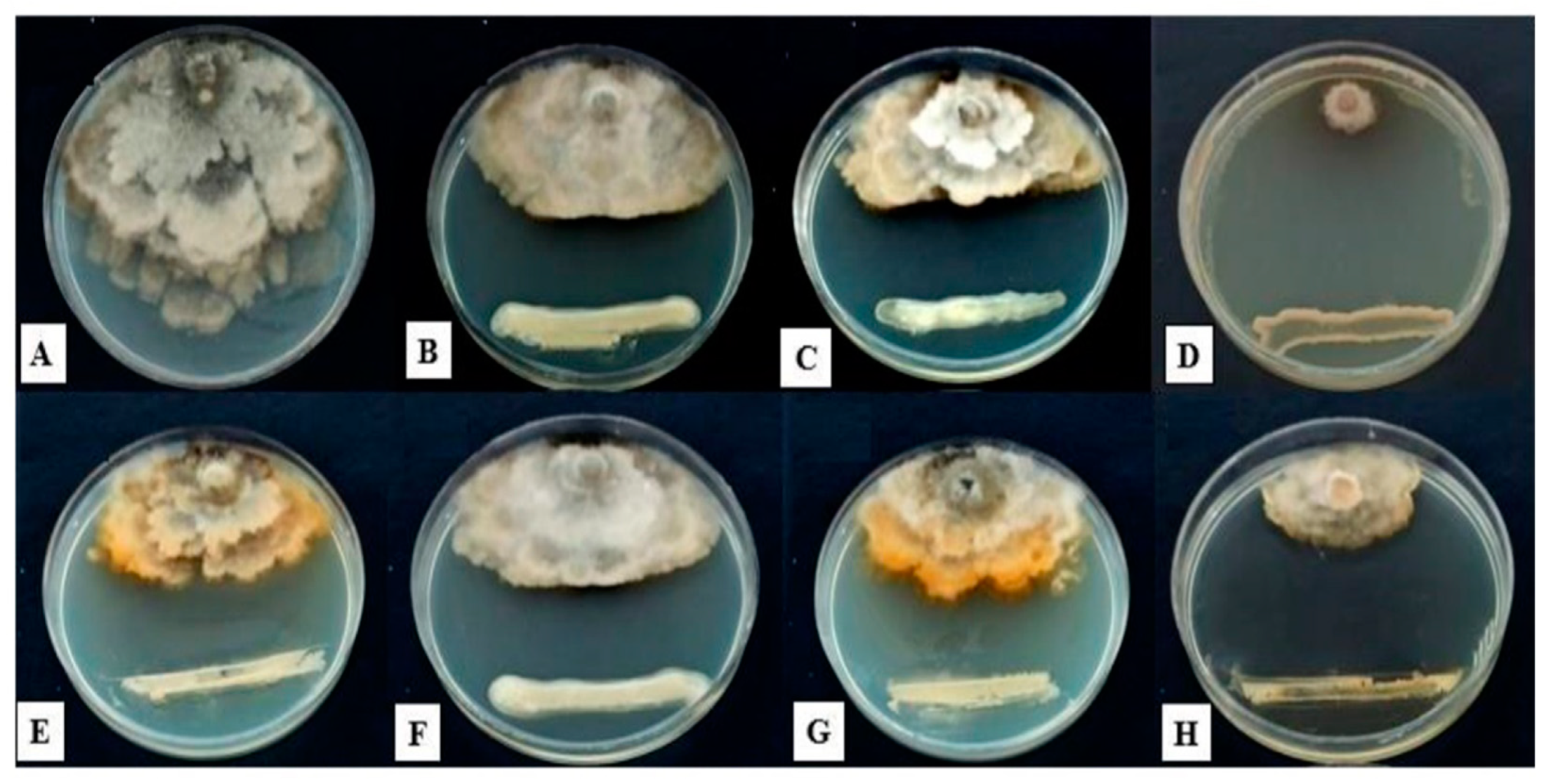
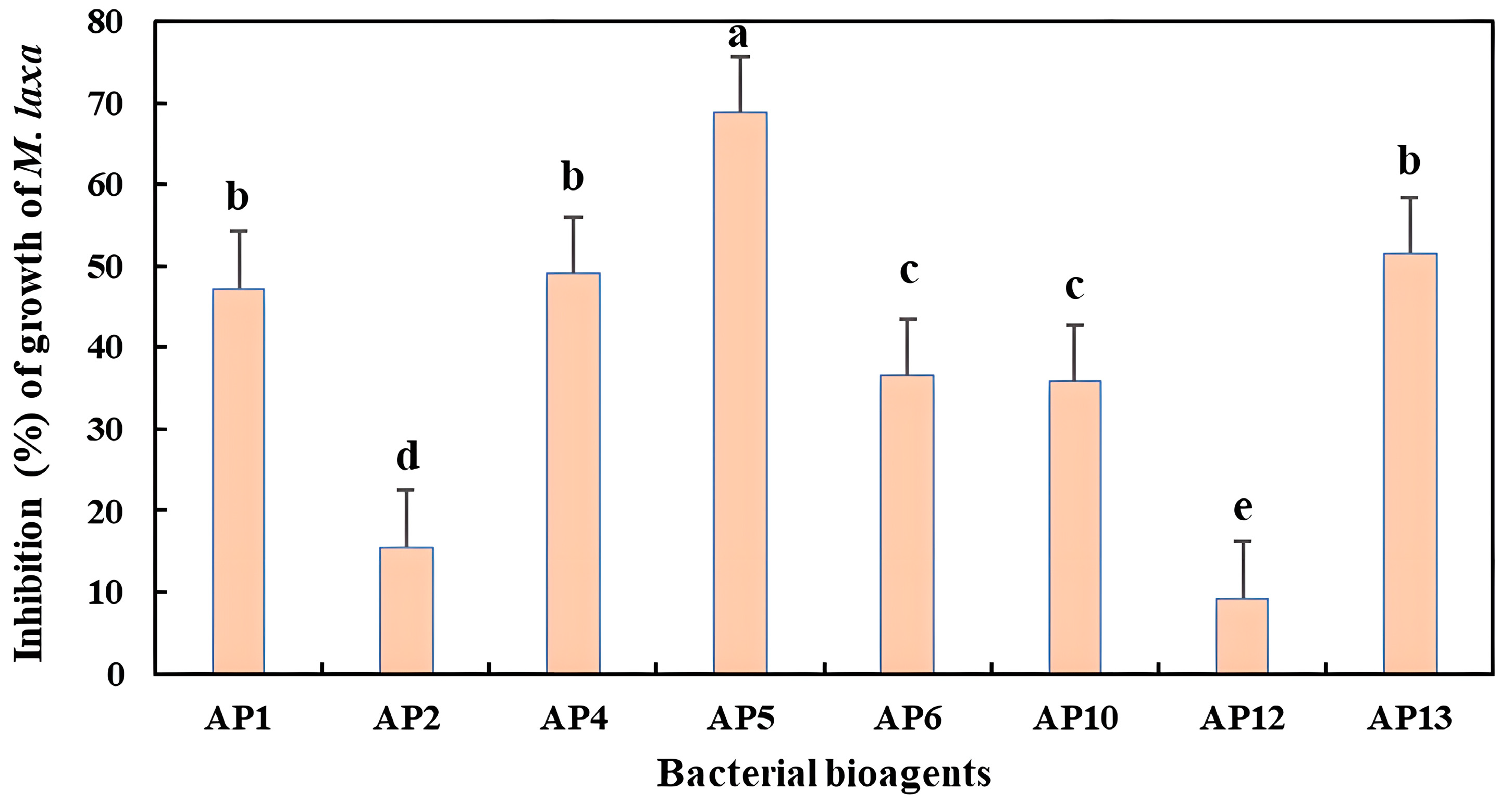
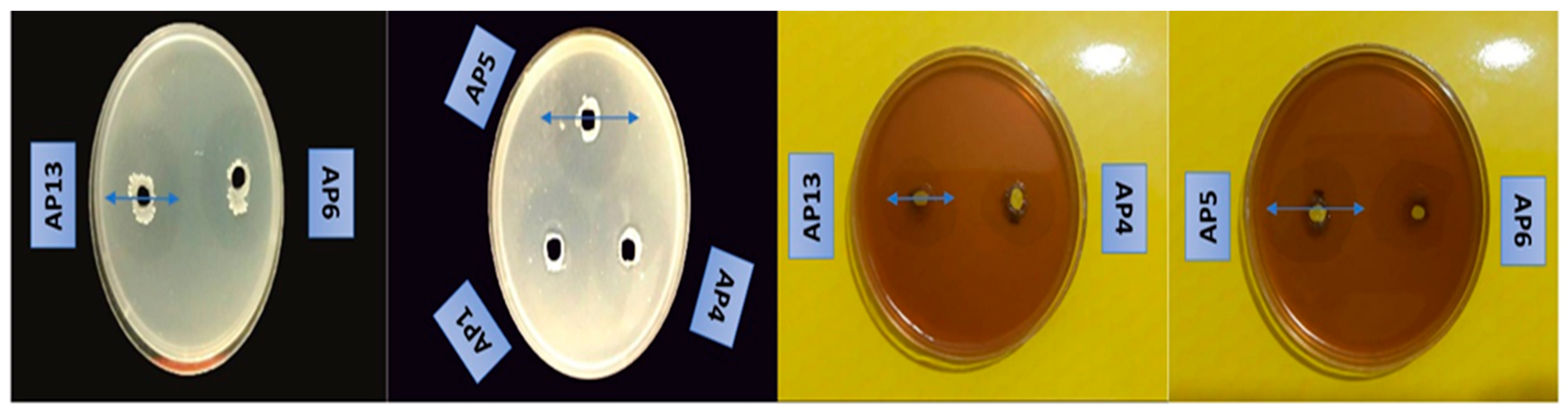
3.3. Antifungal Activity of Isolated Strains
3.4. Extraction of Antifungal Compounds
3.5. Investigation of the Control Effect of Bacterial Isolates on Apple Brown Rot Disease In Vivo
3.6. Morphological and Biochemical Characteristics
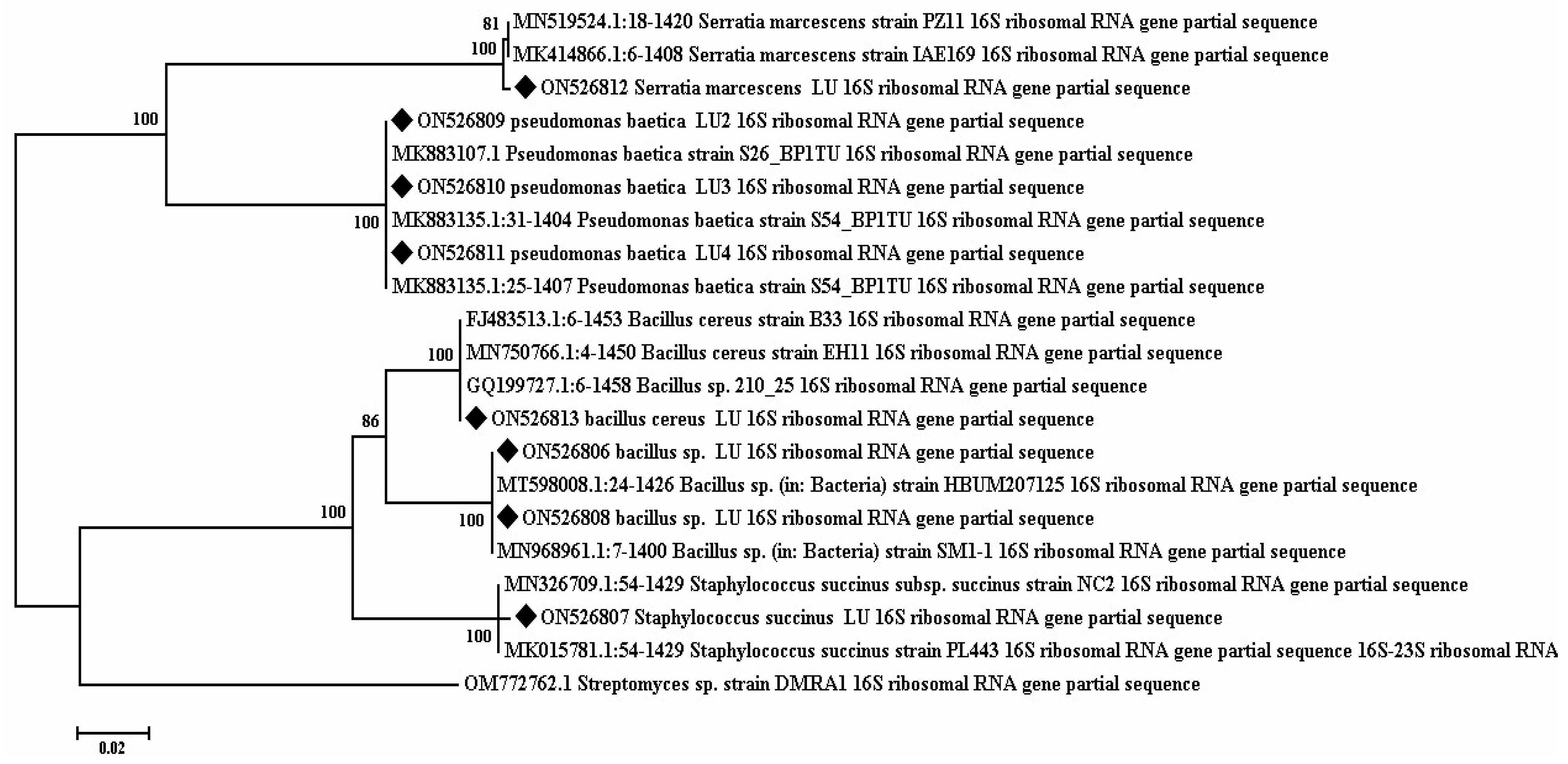
3.7. Phylogenetic Analysis of 16S rRNA Sequences
3.8. Results Regarding Protease and Chitinase Activity
3.9. Chitinase and Protease Activity of the Seven Selected Strains Based on the Colorimetric Method
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ortega, S.F.; López, M.P.B.; Nari, L.; Boonham, N.; Gullino, M.L.; Spadaro, D. Rapid detection of Monilinia fructicola and Monilinia laxa on peaches and nectarines using loopmediated isothermal amplification. Plant Dis. 2019, 103, 2305–2314. [Google Scholar] [CrossRef]
- Garcia-Benitez, C.; Casals, C.; Usall, J.; Sánchez-Ramos, I.; Melgarejo, P.; De Cal, A. Impact of Postharvest Handling on Preharvest Latent Infections Caused by Monilinia spp. in Nectarines. J. Fungi 2020, 6, 266. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.S.; Tian, P.; Navarrete-Tindall, N.; Bartelette, W.S. Occurrence of brown rot of wild plum caused by Monilinia fructicola in Missouri. Plant Health Prog. 2023, 24, 112–113. [Google Scholar] [CrossRef]
- Guven, H.; Everhart, S.E.; De Miccolis Angelini, R.M.; Ozkilinc, H. Genetic diversity assessments of brown rot pathogen Monilinia fructicola based on the six simple sequence repeat loci. J. Plant Dis. Prot. 2021, 128, 1459–1465. [Google Scholar] [CrossRef]
- Zaki, O.; Weekers, F.; Thonart, P.; Tesch, E.; Kuenemann, P.; Jacques, P. Limiting factors of mycopesticide development. Biol. Control 2020, 144, 104220. [Google Scholar] [CrossRef]
- World Health Organization. WHO Estimates of the Global Burden of Foodborne Diseases; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Lahlali, R.; Mchachti, O.; Radouane, N.; Ezrari, S.; Belabes, Z.; Khayi, S.; Mentag, R.; Tahiri, A.; Barka, E. The Potential of Novel Bacterial Isolates from Natural Soil for the Control of Brown Rot Disease (Monilinia fructigena) on Apple Fruits. Agronomy 2020, 10, 1814. [Google Scholar] [CrossRef]
- Ahkami, A.H.; White, R.A., III; Handakumbura, P.P.; Jansson, C. Rhizosphere engineering: Enhancing sustainable plant ecosystem productivity. Rhizosphere 2017, 3, 233–243. [Google Scholar] [CrossRef]
- Daniel, R. The metagenomics of soil. Nat. Rev. Microbiol. 2005, 3, 470–478. [Google Scholar] [CrossRef]
- Filonov, A.; Akhmetov, L.; Puntus, I.; Solyanikova, I. Removal of oil spills in temperate and cold climates of Russia: Experience in the creation and use of biopreparations based on effective microbial consortia. In Biodegradation, Pollutants and Bioremediation Principles, 1st ed.; Bidoia, E.D., Montagnolli, R.N., Eds.; Taylor & Francis CRC Press: Boca Raton, FL, USA, 2021; pp. 137–159. [Google Scholar]
- Velten, S.; Leventon, J.; Jager, N.; Newig, J. What Is Sustainable Agriculture? A Systematic Review. Sustainability 2015, 7, 7833–7865. [Google Scholar] [CrossRef]
- Lahlali, R.; Ezrari, S.; Radouane, N.; Kenfaoui, J.; Esmaeel, Q.; El Hamss, H.; Belabess, Z.; Barka, E.A. Biological Control of Plant Pathogens: A Global Perspective. Microorganisms 2022, 10, 596. [Google Scholar] [CrossRef]
- Yadav, A.N.; Verma, P.; Kumar, S.; Kumar, V.; Kumar, M.; Sugitha, T.C.K.; Singh, B.P.; Saxena, A.K.; Dhaliwal, H.S. Actinobacteria from rhizosphere: Molecular diver sity, distributions and potential biotechnological applications. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2018; pp. 13–41. [Google Scholar] [CrossRef]
- Souza, R.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 2015, 38, 401–419. [Google Scholar] [CrossRef]
- Jabeen, F.; Qazi, J.I. Isolation of chitinase yielding Bacillus cereus JF68 from soil employing an edible crab shell chitin. J. Sci. Ind. Res. 2014, 73, 771–776. [Google Scholar]
- Rathore, A.S.; Gupta, R.D. Chitinases from bacteria to human: Properties, applications and future perspectives. Enzym. Res. 2015, 2015, 791907. [Google Scholar] [CrossRef]
- Someya, N.; Nakajima, M.; Hirayae, K.; Hibi, T.; Akutsu, K. Synergistic antifungal activity of chitinolytic enzymes and prodigiosin produced by biocontrol bacterium, Serratia marcescens strain B2 against gray mold pathogen, Botrytis cinerea. J. Gen. Plant Pathol. 2001, 67, 312–317. [Google Scholar] [CrossRef]
- Beier, S.; Bertilsson, S. Bacterial chitin degradation—Mechanisms and ecophysiological strategies. Front. Microbiol. 2013, 4, 149. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Rahim, I.R.; Abo-Elyousr, K.A.M. Using of endophytic Saccharomycopsis fibuligera and thyme oil for management of gray mold rot of guava fruits. Biol. Control. 2017, 110, 124–131. [Google Scholar] [CrossRef]
- Thakur, M.; Sohal, B.S. Role of elicitors in inducing resistance in plants against pathogen infection: A review. Int. Sch. Res. Not. 2013, 2013, 762412. [Google Scholar] [CrossRef]
- Amin, M.; Rakhisi, Z.; Ahmady, A.Z. Isolation and identification of Bacillus species from soil and evaluation of their antibacterial properties. Avicenna Clin. Microb. Infec. 2015, 2, e23233. [Google Scholar] [CrossRef]
- Van Leeuwen, G.C.M.; Baayen, R.P.; Holb, I.J.; Jeger, M.J. Distinction of the Asiatic brown rot fungus Monilia polystroma spp. nov. from M. fructigena. Mycol. Res. 2002, 106, 444–451. [Google Scholar] [CrossRef]
- Carlone, G.M.; Valadez, M.J.; Pickett, M.J. Methods for distinguishing gram-positive from gram-negative bacteria. J. Clin. Microbiol. 1982, 16, 1157–1159. [Google Scholar] [CrossRef]
- Shields, P.; Cathcart, L. Oxidase Test Protocol; American Society for Microbiology: Washington, DC, USA, 2010; pp. 1–9. [Google Scholar]
- Reiner, K. Catalase Test Protocol; American society for microbiology: Washington, DC, USA, 2010; pp. 1–6. [Google Scholar]
- MacWilliams, M.P. Citrate Test Protocol; American Society for Microbiology: Washington, DC, USA, 2009; pp. 1–7. [Google Scholar]
- Naidu, M.A.; Saranraj, P. Bacterial amylase: A review. Int. J. Pharm. Biol. Arch. 2013, 4, 274–287. [Google Scholar]
- Gupta, P.; Samant, K.; Sahu, A. Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int. J. Microbiol. 2012, 2012, 578925. [Google Scholar] [PubMed]
- Sharmin, S.; Hossain, M.T.; Anwar, M.N. Isolation and characterization of a protease producing bacteria Bacillus amovivorus and optimization of some factors of culture conditions for protease production. J. Biol. Sci. 2005, 5, 358–362. [Google Scholar]
- Janda, J.M.; Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [PubMed]
- Meliah, S.; Sulistiyani, T.R.; Lisdiyanti, P.; Kanti, A.; Sudiana, I.M.; Kobayashi, M. Antifungal Activity of Endophytic Bacteria Associated with Sweet Sorghum (Sorghum bicolor). J. Math. Fundam. Sci. 2021, 53, 16–30. [Google Scholar] [CrossRef]
- Alam, J.; Mathur, A. Evaluation of antifungal potential of chitin and chitin—Based derivatives against pathogenic fungal strains. Biolife 2014, 2, 1354–1358. [Google Scholar]
- Sadfi, N.; Cherif, M.; Fliss, I.; Boudabbous, A.; Antoun, H. Evaluation of bacterial isolates from salty soils Bacillus thuringiensis strains for the biocontrol of Fusarium dry rot of potato tubers. J. Plant Pathol. 2001, 83, 101–118. [Google Scholar]
- Dhanasekaran, D.; Thajuddin, N.; Panneerselvam, A. Applications of Actinobacterial Fungicides in Agriculture and Medicine. In Fungicides for Plant and Animal Diseases; Khanasekaran, D., Ed.; INTECH: London, UK, 2012; pp. 29–54. [Google Scholar] [CrossRef]
- Dulbecco, R.; Vogt, M. Plaque formation and isolation of pure lines with poliomyelitis viruses. J. Exp. Med. 1954, 99, 167–182. [Google Scholar] [CrossRef]
- Thi Hong Hanh, T.; Thi Dao, T.; Thi Ngoc An, L.; Thuong Am, L. Isolation selection of chitinase-producing bacteria with antifungal activity against Fusarium oxysporum from Lilium rhizosphere soil. J. Agric. Sci. 2020, 18, 965–975. [Google Scholar]
- Pratika, M.; Ananda, M.; Suwastika, I.N. Protease activity from bacterial isolates of Nepenthes maxima reinw. ex nees. J. Phys. Conf. Ser. 2021, 1763, 012092. [Google Scholar] [CrossRef]
- Musacchi, S.; Serra, S. Apple fruit quality: Overview on pre-harvest factors. Sci. Hortic. 2018, 234, 409–430. [Google Scholar] [CrossRef]
- FAO. FAOSTAT: Food and Agriculture Organization of the United Nations; FAO: Rome, Italy, 2020. [Google Scholar]
- Holb, I. Chapter 6: Brown rot: Causes, detection and control of Monilinia spp. affecting tree fruit. In Integrated Management of Diseases and Insect Pests of Tree Fruit; Burleigh Dodds: Cambridge, UK, 2019; pp. 103–149. [Google Scholar]
- Aiello, D.; Restuccia, C.; Stefani, E.; Vitale, A.; Cirvilleri, G. Postharvest biocontrol ability of Pseudomonas synxantha against Monilinia fructicola and Monilinia fructigena on stone fruit. Postharvest Biol. Technol. 2019, 149, 83–89. [Google Scholar] [CrossRef]
- Bubici, G.; Kaushal, M.; Prigigallo, M.I.; Cabanás, C.G.L.; Mercado-Blanco, J. Biological control agents against Fusarium wilt of banana. Front. Microbiol. 2019, 10, 616. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yin, X.; Wang, Q.; Peng, Y.; Ma, Y.; Liu, P.; Shi, J. Antagonistic activities of volatiles produced by two Bacillus strains against Monilinia fructicola in peach fruit. J. Sci. Food Agric. 2018, 98, 5756–5763. [Google Scholar] [CrossRef] [PubMed]
- Calvo, H.; Marco, P.; Blanco, D.; Oria, R.; Venturini, M.E. Potential of a new strain of Bacillus amyloliquefaciens BUZ-14 as a biocontrol agent of postharvest fruit diseases. Food Microbiol. 2017, 63, 101–110. [Google Scholar] [CrossRef]
- Suresh, P.; Vellasamy, S.; Almaary, K.S.; Dawoud, T.M.; Elbadawi, Y.B. Fluorescent pseudomonads (FPs) as a potential biocontrol and plant growth promoting agent associated with tomato rhizosphere. J. King Saud Univ. Sci. 2021, 33, 101423. [Google Scholar] [CrossRef]
- Sharma, A.; Kashyap, P.L.; Srivastava, A.K.; Bansal, Y.K.; Kaushik, R. Isolation and characterization of halotolerant bacilli from chickpea (Cicer arietinum L.) rhizosphere for plant growth promotion and biocontrol traits. Eur. J. Plant Pathol. 2019, 153, 787–800. [Google Scholar] [CrossRef]
- Rasool, A.; Mir, M.I.; Zulfajri, M.; Hanafiah, M.M.; Unnisa, S.A.; Mahboob, M. Plant growth promoting and antifungal asset of indigenous rhizobacteria secluded from saffron (Crocus sativus L.) rhizosphere. Microb. Pathog. 2021, 150, 104734. [Google Scholar] [CrossRef]
- Afridi, M.S.; Fakhar, A.; Kumar, A.; Ali, S.; Medeiros, F.H.; Muneer, M.A.; Ali, H.; Saleem, M. Harnessing microbial multitrophic interactions for rhizosphere microbiome engineering. Microbiol. Res. 2022, 265, 127199. [Google Scholar]
- Lahlali, R.; Aksissou, W.; Lyousfi, N.; Ezrari, S.; Blenzar, A.; Tahiri, A.; Ennahli, S.; Hrustic, J.; MacLean, D.; Amiri, S. Biocontrol activity and putative mechanism of Bacillu samyloliquefaciens (SF14andSP10), Alcaligenes faecalis ACBC1, and Pantoea agglomerans ACBP1 against brown rot disease of fruit. Microb. Pathog. 2020, 139, 103914. [Google Scholar] [CrossRef]
- Tan, C.; Kalhoro, M.T.; Faqir, Y.; Ma, J.; Osei, M.D.; Khaliq, G. Climate-Resilient Microbial Biotechnology: A Perspective on Sustainable Agriculture. Sustainability 2022, 14, 5574. [Google Scholar] [CrossRef]
- Choudhary, S.; Singh, A.; Rani, M. Rhizosphere: Hotspot of Soil-Microbes-Plant Interaction. A Mon. Peer Rev. Mag. Agric. Allied Sci. 2022, 1, 80. [Google Scholar]
- Basu, A.; Prasad, P.; Das, S.N.; Kalam, S.; Sayyed, R.Z.; Reddy, M.S.; El Enshasy, H. Plant growth promoting rhizobacteria (PGPR) as green bioinoculants: Recent developments, constraints, and prospects. Sustainability 2021, 13, 1140. [Google Scholar] [CrossRef]
- Ye, X.; Xu, C.; Xie, T.; Zhang, Y.; Zhao, Y.; Xia, C.; Li, Z.; Huang, Y.; Fan, J.; Cao, H.; et al. Myxobacterial Outer Membrane β-1, 6-Glucanase Induced the Cell Death of Fusarium oxysporum by Destroying the Cell Wall Integrity. Appl. Environ. Microbiol. 2023, 89, e01236-22. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Wilkens, C.; Barrett, K.; Meyer, A.S. Microbial enzymes catalyzing keratin degradation: Classification, structure, function. Biotechnol. Adv. 2020, 44, 107607. [Google Scholar] [CrossRef]
- Rodríguez-Sifuentes, L.; Marszalek, J.E.; Chuck-Hernández, C.; Serna-Saldívar, S.O. Legumes protease inhibitors as biopesticides and their defense mechanisms against biotic factors. Int. J. Mol. Sci. 2020, 21, 3322. [Google Scholar] [CrossRef]
- Kong, W.L.; Li, P.S.; Wu, X.Q.; Wu, T.Y.; Sun, X.R. Forest tree associated bacterial diffusible and volatile organic compounds against various phytopathogenic fungi. Microorganisms 2020, 8, 590. [Google Scholar] [CrossRef]


| Strain No. | BLAST Research Results | Accession | Disease Severity (%) 7 Days | Disease Severity (%) 14 Days |
|---|---|---|---|---|
| AP1 | Pseudomonas baetica | ON526810 | 44.31 d | 31.18 e |
| AP2 | Pseudomonas baetica | ON526809 | 49.03 e | 35.89 f |
| AP4 | Pseudomonas baetica | ON526811 | 24.46 b | 24.66 c |
| AP5 | Serratia marcescens | ON526812 | 00.00 a | 12.34 a |
| AP6 | Bacillus sp. | ON526806 | 34.25 c | 27.98 d |
| AP10 | Bacillus sp. | ON526808 | 52.11 f | 37.90 g |
| AP12 | Staphylococcus succinus | ON526807 | 89.33 h | 61.66 i |
| AP13 | Bacillus cereus | ON526813 | 00.00 a | 14.71 b |
| No. | Strain | Gram | Oxidase | Catalase | Citrate | Amylase | Cellulase | Protease |
|---|---|---|---|---|---|---|---|---|
| 1 | AP1 | − | + | + | + | + | − | + |
| 2 | AP2 | − | + | + | + | − | − | + |
| 3 | AP4 | − | + | + | + | + | + | + |
| 4 | AP5 | − | − | + | + | + | + | + |
| 5 | AP6 | + | + | + | + | + | + | + |
| 6 | AP10 | + | − | + | + | + | − | + |
| 7 | AP12 | + | − | + | + | + | + | + |
| 8 | AP13 | + | − | + | + | + | + | + |
| BLAST Research Results | Bacterial Isolates | Chitinase Activity (k, mm) | Protease Activity (k, mm) |
|---|---|---|---|
| Pseudomonas baetica | AP1 | 20.11 | 22.31 |
| Pseudomonas baetica | AP2 | 14.63 | 16.54 |
| Pseudomonas baetica | AP4 | 21.02 | 23.11 |
| Serratia marcescens | AP5 | 24.16 | 25.14 |
| Bacillus sp. | AP6 | 20.13 | 21.68 |
| Bacillus sp. | AP10 | 20.05 | 20.89 |
| Staphylococcus succinus | AP12 | 14.36 | 15.02 |
| Bacillus cereus | AP13 | 23.54 | 24.87 |
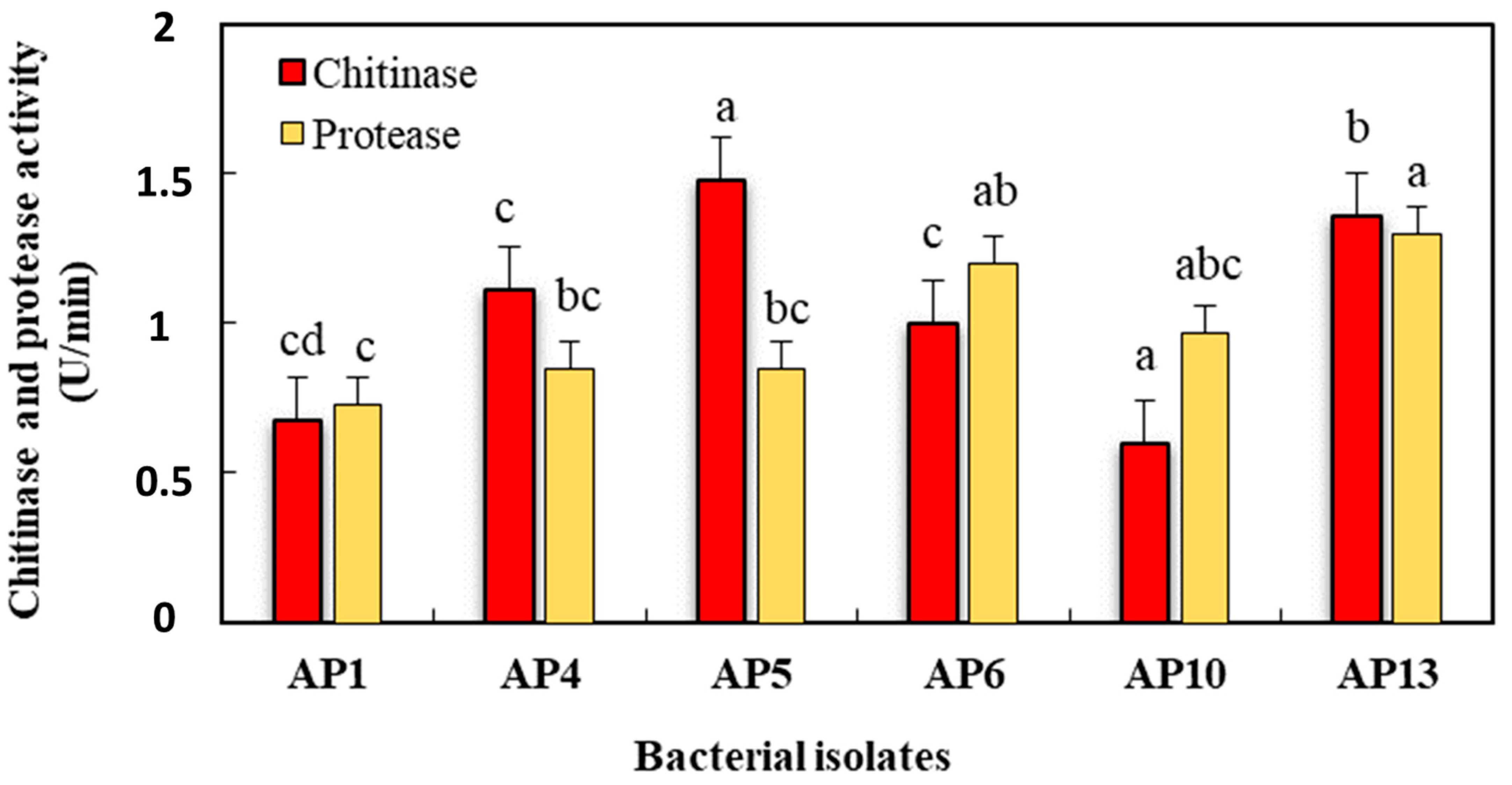
| Strain No. | Chitinase Activity (U/min) | Protease Activity (U/min) |
|---|---|---|
| AP1 | 0.675 cd | 0.725 c |
| AP4 | 1.11 c | 0.85 bc |
| AP5 | 1.475 a | 0.85 bc |
| AP6 | 1 c | 1.2 ab |
| AP10 | 0.6 d | 0.975 abc |
| AP13 | 1.36 b | 1.3 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Derikvand, F.; Bazgir, E.; El Jarroudi, M.; Darvishnia, M.; Mirzaei Najafgholi, H.; Laasli, S.-E.; Lahlali, R. Unleashing the Potential of Bacterial Isolates from Apple Tree Rhizosphere for Biocontrol of Monilinia laxa: A Promising Approach for Combatting Brown Rot Disease. J. Fungi 2023, 9, 828. https://doi.org/10.3390/jof9080828
Derikvand F, Bazgir E, El Jarroudi M, Darvishnia M, Mirzaei Najafgholi H, Laasli S-E, Lahlali R. Unleashing the Potential of Bacterial Isolates from Apple Tree Rhizosphere for Biocontrol of Monilinia laxa: A Promising Approach for Combatting Brown Rot Disease. Journal of Fungi. 2023; 9(8):828. https://doi.org/10.3390/jof9080828
Chicago/Turabian StyleDerikvand, Fatemeh, Eidi Bazgir, Moussa El Jarroudi, Mostafa Darvishnia, Hossein Mirzaei Najafgholi, Salah-Eddine Laasli, and Rachid Lahlali. 2023. "Unleashing the Potential of Bacterial Isolates from Apple Tree Rhizosphere for Biocontrol of Monilinia laxa: A Promising Approach for Combatting Brown Rot Disease" Journal of Fungi 9, no. 8: 828. https://doi.org/10.3390/jof9080828
APA StyleDerikvand, F., Bazgir, E., El Jarroudi, M., Darvishnia, M., Mirzaei Najafgholi, H., Laasli, S.-E., & Lahlali, R. (2023). Unleashing the Potential of Bacterial Isolates from Apple Tree Rhizosphere for Biocontrol of Monilinia laxa: A Promising Approach for Combatting Brown Rot Disease. Journal of Fungi, 9(8), 828. https://doi.org/10.3390/jof9080828








