Establishment of a CRISPR/Cas9-Mediated Efficient Knockout System of Trichoderma hamatum T21 and Pigment Synthesis PKS Gene Knockout
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and Culture Conditions
2.2. Extraction of DNA and RNA
2.3. Genome Assembly and Annotation
2.4. Strain Identification
2.5. Antibiotic Resistance Screening
2.6. Evolutionary Analyses of Thpyr4 and Thpks1 Genes
2.7. qRT-PCR Analysis
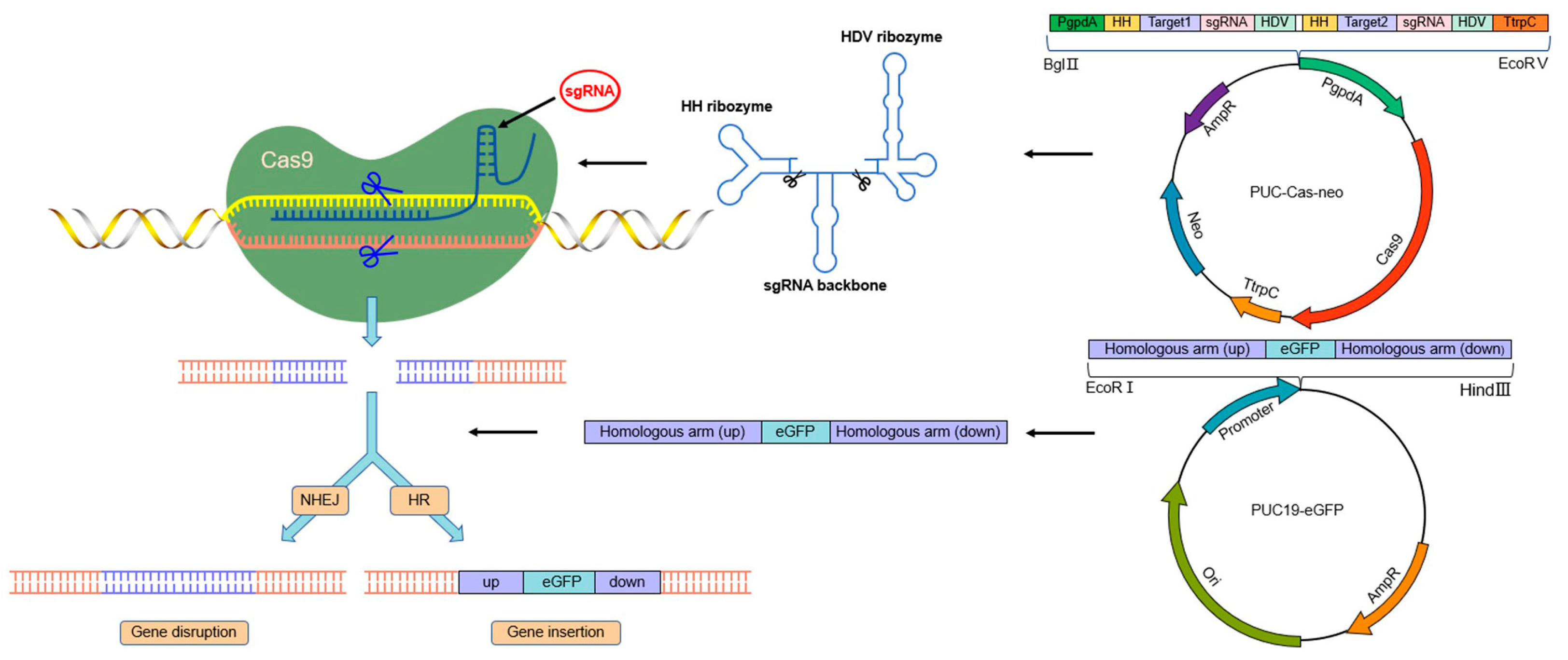
2.8. Construction of CRISPR/Cas9 Plasmid
2.9. Construction of Homologous Recombinant Plasmid
2.10. Preparation of Protoplasts
2.11. Co-Transformation of gRNA with Donor DNA
2.12. Identification of Transformant
2.13. RT-PCR Identification for Cas9 Expression
2.14. Knockout Efficiency Statistics
3. Results
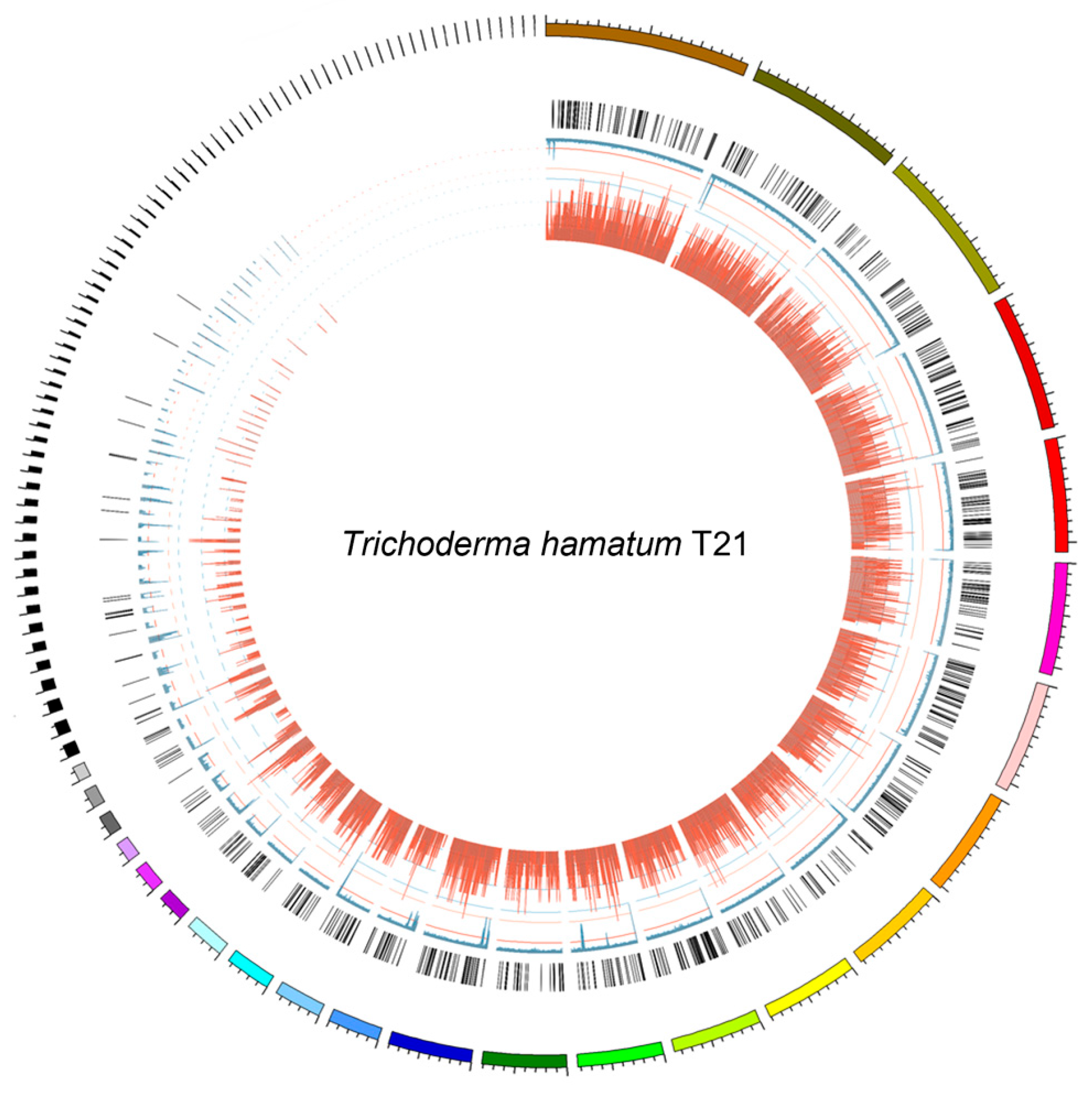
3.1. Strain Identification and Genome Annotation
3.2. Screening of Resistance Concentration of T. hamatum T21 to 5-FOA and G418
3.3. Thpks1 Candidate Genes and qRT-PCR Analysis
3.4. Thpyr4 and Thpks1 Gene Knockout Vector Construction
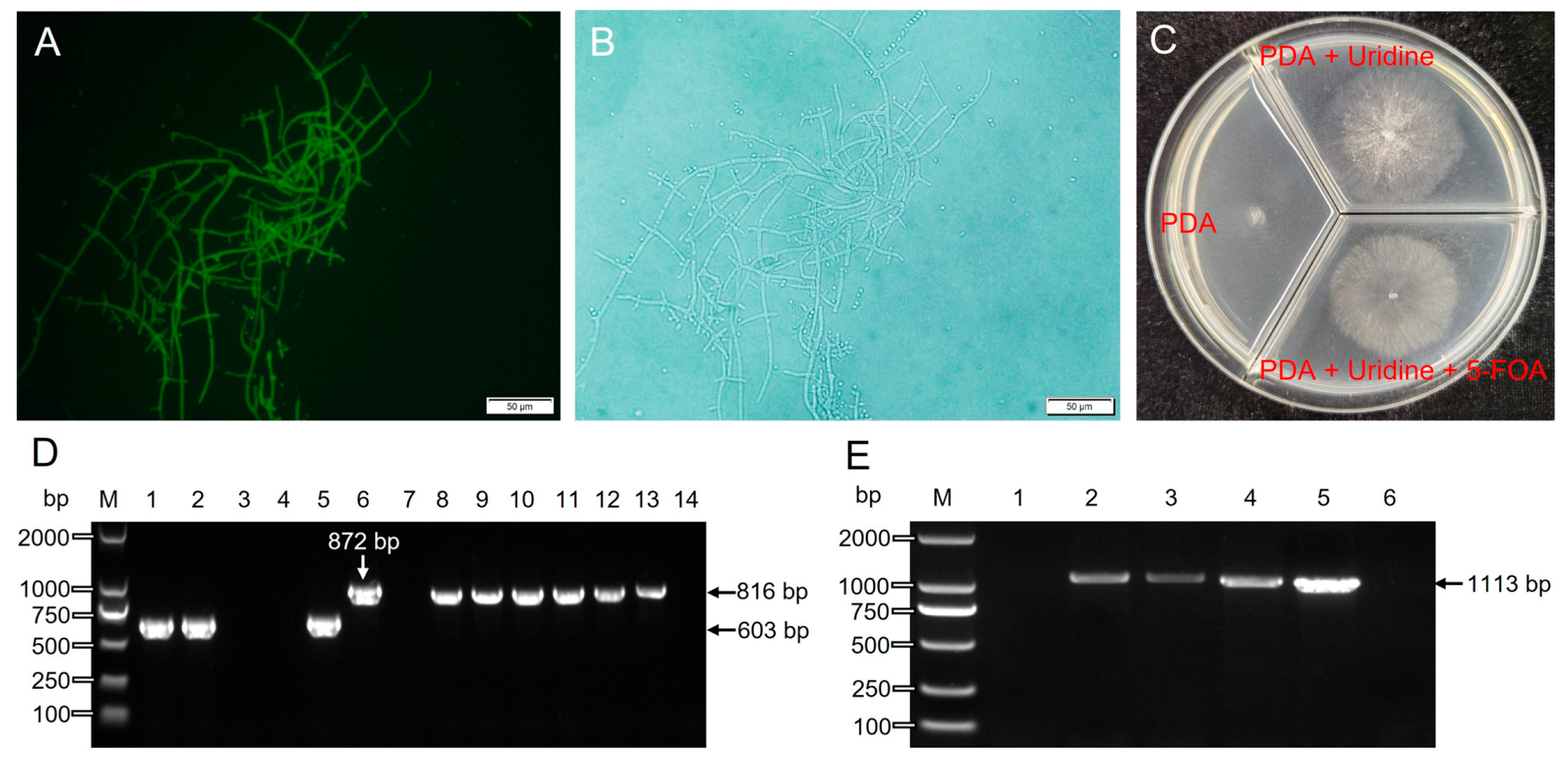
3.5. Thpyr4 Knockout and Identification
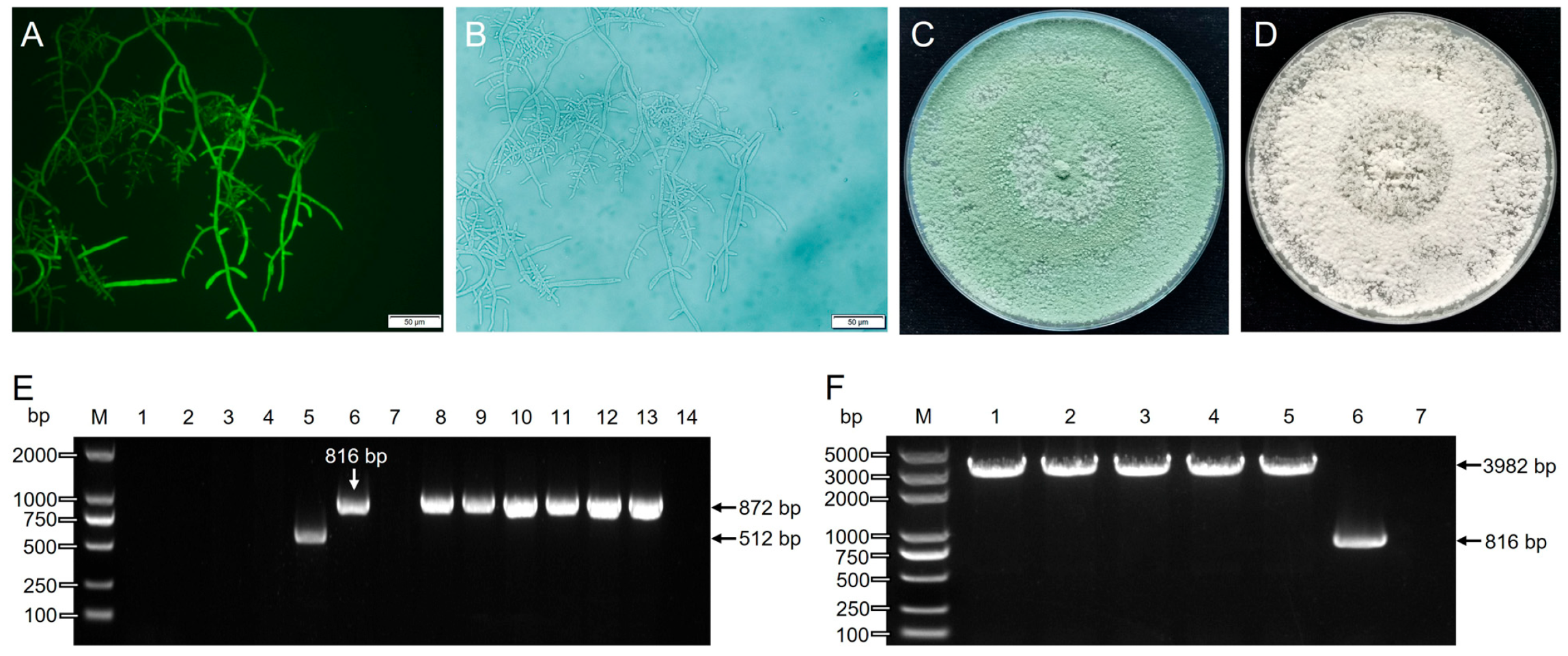
3.6. Thpks1 Knockout and Identification
3.7. Knockout Efficiency Statistics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, R.; Ji, S.; Wang, Z.; Zhang, H.; Wang, Y.; Liu, Z. Trichoderma asperellum xylanases promote growth and induce resistance in poplar. Microbiol. Res. 2021, 248, 126767. [Google Scholar] [CrossRef]
- Kohl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef]
- Romero-Contreras, Y.J.; Ramirez-Valdespino, C.A.; Guzman-Guzman, P.; Macias-Segoviano, J.I.; Villagomez-Castro, J.C.; Olmedo-Monfil, V. Tal6 from Trichoderma atroviride is a LysM effector involved in mycoparasitism and plant association. Front. Microbiol. 2019, 10, 2231. [Google Scholar] [CrossRef]
- Swain, H.; Adak, T.; Mukherjee, A.K.; Mukherjee, P.K.; Bhattacharyya, P.; Behera, S.; Bagchi, T.B.; Patro, R.; Shasmita; Khandual, A.; et al. Novel Trichoderma strains isolated from tree barks as potential biocontrol agents and biofertilizers for direct seeded rice. Microbiol. Res. 2018, 214, 83–90. [Google Scholar] [CrossRef]
- Lu, Z.X.; Tu, G.P.; Zhang, T.; Li, Y.Q.; Wang, X.H.; Chen, J.; Tu, G.P.; Song, W.; Zhang, Q.G. Screening of antagonistic Trichoderma strains and their application for controlling stalk rot in maize. J. Integr. Agr. 2020, 19, 145–152. [Google Scholar] [CrossRef]
- Díaz-Gutiérrez, C.; Arroyave, C.; Llugany, M.; Poschenrieder, C.; Martos, S.; Peláez, C. Trichoderma asperellum as a preventive and curative agent to control Fusarium wilt in Stevia rebaudiana. Biol. Control. 2021, 155, 104537. [Google Scholar] [CrossRef]
- Ngo, M.T.; Nguyen, M.V.; Han, J.W.; Park, M.S.; Kim, H.; Choi, G.J. In vitro and in vivo antifungal activity of sorbicillinoids produced by Trichoderma longibrachiatum. J. Fungi. 2021, 7, 428. [Google Scholar] [CrossRef]
- Fan, H.; Yao, M.; Wang, H.; Zhao, D.; Zhu, X.; Wang, Y.; Liu, X.; Duan, Y.; Chen, L. Isolation and effect of Trichoderma citrinoviride Snef1910 for the biological control of root-knot nematode, Meloidogyne incognita. BMC Microbiol. 2020, 20, 299. [Google Scholar] [CrossRef]
- Martinez-Medina, A.; Van Wees, S.C.M.; Pieterse, C.M.J. Airborne signals from Trichoderma fungi stimulate iron uptake responses in roots resulting in priming of jasmonic acid-dependent defences in shoots of Arabidopsis thaliana and Solanum lycopersicum. Plant Cell Environ. 2017, 40, 2691–2705. [Google Scholar] [CrossRef]
- Contina, J.B.; Dandurand, L.M.; Knudsen, G.R. Use of GFP-tagged Trichoderma harzianum as a tool to study the biological control of the potato cyst nematode Globodera pallida. Appl. Soil Ecol. 2017, 115, 31–37. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, Z.; Zhang, Y.; Wang, Y.; Liu, Z. Biocontrol and growth-promoting effect of Trichoderma asperellum TaspHu1 isolate from Juglans mandshurica rhizosphere soil. Microbiol. Res. 2021, 242, 126596. [Google Scholar] [CrossRef]
- Meng, X.; Miao, Y.; Liu, Q.; Ma, L.; Guo, K.; Liu, D.; Ran, W.; Shen, Q. TgSWO from Trichoderma guizhouense NJAU4742 promotes growth in cucumber plants by modifying the root morphology and the cell wall architecture. Microb. Cell Fact. 2019, 18, 148. [Google Scholar] [CrossRef]
- Liu, H.J.; Duan, W.D.; Liu, C.; Meng, L.X.; Li, H.X.; Li, R.; Shen, Q.R. Spore production in the solid-state fermentation of stevia residue by Trichoderma guizhouense and its effects on corn growth. J. Integr. Agr. 2021, 20, 1147–1156. [Google Scholar] [CrossRef]
- Adnan, M.; Islam, W.; Shabbir, A.; Khan, K.A.; Ghramh, H.A.; Huang, Z.; Chen, H.Y.H.; Lu, G.D. Plant defense against fungal pathogens by antagonistic fungi with Trichoderma in focus. Microb. Pathog. 2019, 129, 7–18. [Google Scholar] [CrossRef]
- Guzman-Guzman, P.; Porras-Troncoso, M.D.; Olmedo-Monfil, V.; Herrera-Estrella, A. Trichoderma species: Versatile plant symbionts. Phytopathology 2019, 109, 6–16. [Google Scholar] [CrossRef]
- Vinale, F.; Sivasithamparam, K. Beneficial effects of Trichoderma secondary metabolites on crops. Phytother. Res. 2020, 34, 2835–2842. [Google Scholar] [CrossRef]
- Martinez-Medina, A.; Fernandez, I.; Lok, G.B.; Pozo, M.J.; Pieterse, C.M.; Van Wees, S.C. Shifting from priming of salicylic acid- to jasmonic acid-regulated defences by Trichoderma protects tomato against the root knot nematode Meloidogyne incognita. New Phytol. 2017, 213, 1363–1377. [Google Scholar] [CrossRef]
- Studholme, D.J.; Harris, B.; Le Cocq, K.; Winsbury, R.; Perera, V.; Ryder, L.; Ward, J.L.; Beale, M.H.; Thornton, C.R.; Grant, M. Investigating the beneficial traits of Trichoderma hamatum GD12 for sustainable agriculture-insights from genomics. Front. Plant Sci. 2013, 4, 258. [Google Scholar] [CrossRef]
- Bao, Z.; Xiao, H.; Liang, J.; Zhang, L.; Xiong, X.; Sun, N.; Si, T.; Zhao, H. Homology-integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae. ACS Synth. Biol. 2015, 4, 585–594. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, B.; Duan, C.; Sun, B.; Yang, J.; Yang, S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microb. 2015, 81, 2506–2514. [Google Scholar]
- Wang, P.A.; Xiao, H.; Zhong, J.J. CRISPR-Cas9 assisted functional gene editing in the mushroom Ganoderma lucidum. Appl. Microbiol. Biot. 2020, 104, 1661–1671. [Google Scholar] [CrossRef]
- Kuivanen, J.; Wang, Y.J.; Richard, P. Engineering Aspergillus niger for galactaric acid production: Elimination of galactaric acid catabolism by using RNA sequencing and CRISPR/Cas9. Microb. Cell Fact. 2016, 15, 210. [Google Scholar] [CrossRef]
- Pohl, C.; Kiel, J.A.; Driessen, A.J.; Bovenberg, R.A.; Nygard, Y. CRISPR/Cas9 based genome editing of Penicillium chrysogenum. ACS Synth. Biol. 2016, 5, 754–764. [Google Scholar] [CrossRef]
- Chen, J.; Lai, Y.; Wang, L.; Zhai, S.; Zou, G.; Zhou, Z.; Cui, C.; Wang, S. CRISPR/Cas9-mediated efficient genome editing via blastospore-based transformation in entomopathogenic fungus Beauveria bassiana. Sci. Rep. 2017, 8, 45763. [Google Scholar] [CrossRef]
- Liu, R.; Chen, L.; Jiang, Y.; Zhou, Z.; Zou, G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015, 1, 15007. [Google Scholar] [CrossRef]
- Vieira, A.A.; Vianna, G.R.; Carrijo, J.; Aragao, F.J.L.; Vieira, P.M. Generation of Trichoderma harzianum with pyr4 auxotrophic marker by using the CRISPR/Cas9 system. Sci. Rep. 2021, 11, 1085. [Google Scholar] [CrossRef]
- Zhou, J.; Li, S.M. Conversion of viridicatic acid to crustosic acid by cytochrome P450 enzyme-catalysed hydroxylation and spontaneous cyclisation. Appl. Microbiol. Biot. 2021, 105, 9181–9189. [Google Scholar] [CrossRef]
- Koh, C.M.J.; Liu, Y.B.; Moehninsi; Du, M.G.; Ji, L.H. Molecular characterization of KU70 and KU80 homologues and exploitation of a KU70-deficient mutant for improving gene deletion frequency in Rhodosporidium toruloides. BMC Microbiol. 2014, 14, 50. [Google Scholar] [CrossRef]
- Ying, S.H.; Feng, M.G.; Keyhani, N.O. Use of uridine auxotrophy (ura3) for markerless transformation of the mycoinsecticide Beauveria bassiana. Appl. Microbiol. Biot. 2013, 97, 3017–3025. [Google Scholar] [CrossRef]
- Chen, Y.; Feng, P.; Shang, Y.; Xu, Y.J.; Wang, C. Biosynthesis of non-melanin pigment by a divergent polyketide synthase in Metarhizium robertsii. Fungal Genet. Biol. 2015, 81, 142–149. [Google Scholar] [CrossRef]
- Zeng, G.; Zhang, P.; Zhang, Q.; Zhao, H.; Li, Z.; Zhang, X.; Wang, C.; Yin, W.B.; Fang, W. Duplication of a Pks gene cluster and subsequent functional diversification facilitate environmental adaptation in Metarhizium species. PLoS Genet. 2018, 14, e1007472. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Yang, H.; Yu, H.; Ma, L.J. Accessory chromosomes in Fusarium oxysporum. Phytopathology 2020, 110, 1488–1496. [Google Scholar] [CrossRef]
- Cai, F.; Druzhinina, I.S. In honor of john bissett: Authoritative guidelines on molecular identification of Trichoderma. Fungal Divers. 2021, 107, 1–69. [Google Scholar] [CrossRef]
- Behiry, S.; Soliman, S.A.; Massoud, M.A.; Abdelbary, M.; Kordy, A.M.; Abdelkhalek, A.; Heflish, A. Trichoderma pubescens elicit induced systemic resistance in tomato challenged by Rhizoctonia solani. J. Fungi. 2023, 9, 167. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jiao, Y.; Li, Y.; Li, Y.; Cao, H.; Mao, Z.; Ling, J.; Yang, Y.; Xie, B. Functional genetic analysis of the leucinostatin biosynthesis transcription regulator lcsL in Purpureocillium lilacinum using CRISPR-Cas9 technology. Appl. Microbiol. Biot. 2019, 103, 6187–6194. [Google Scholar] [CrossRef]
- Labun, K.; Montague, T.G.; Gagnon, J.A.; Thyme, S.B.; Valen, E. CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016, 44, W272–W276. [Google Scholar] [CrossRef]
- Montague, T.G.; Cruz, J.M.; Gagnon, J.A.; Church, G.M.; Valen, E. CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014, 42, W401–W407. [Google Scholar] [CrossRef]
- Bradshaw, R.E.; Jin, H.; Morgan, B.S.; Schwelm, A.; Teddy, O.R.; Young, C.A.; Zhang, S. A polyketide synthase gene required for biosynthesis of the aflatoxin-like toxin, dothistromin. Mycopathologia 2006, 161, 283–294. [Google Scholar] [CrossRef]
- Clemmensen, S.E.; Kromphardt, K.J.K.; Frandsen, R.J.N. Marker-free CRISPR-Cas9 based genetic engineering of the phytopathogenic fungus, Penicillium expansum. Fungal Genet. Biol. 2022, 160, 103689. [Google Scholar] [CrossRef]
- Ferreira, F.V.; Musumeci, M.A. Trichoderma as biological control agent: Scope and prospects to improve efficacy. World J. Microb. Biot. 2021, 37, 90. [Google Scholar] [CrossRef]
- Alfiky, A.; Weisskopf, L. Deciphering Trichoderma-plant-pathogen interactions for better development of biocontrol applications. J. Fungi. 2021, 7, 61. [Google Scholar] [CrossRef]
- Salwan, R.; Sharma, A.; Kaur, R.; Sharma, R.; Sharma, V. The riddles of Trichoderma induced plant immunity. Biol. Control. 2022, 174, 105037. [Google Scholar] [CrossRef]
- Mao, T.; Jiang, X. Changes in microbial community and enzyme activity in soil under continuous pepper cropping in response to Trichoderma hamatum MHT1134 application. Sci. Rep. 2021, 11, 21585. [Google Scholar] [CrossRef]
- Wang, S.; Chen, H.; Tang, X.; Zhang, H.; Chen, W.; Chen, Y.Q. Molecular tools for gene manipulation in filamentous fungi. Appl. Microbiol. Biot. 2017, 101, 8063–8075. [Google Scholar] [CrossRef]
- Yu, L.; Xiao, M.; Zhu, Z.; Wang, Y.; Zhou, Z.; Wang, P.; Zou, G. Efficient genome editing in Claviceps purpurea using a CRISPR/Cas9 ribonucleoprotein method. Synth. Syst. Biotechnol. 2022, 7, 664–670. [Google Scholar] [CrossRef]
- Li, S.; Song, Z.; Liu, C.; Chen, X.L.; Han, H. Biomimetic mineralization-based CRISPR/Cas9 ribonucleoprotein nanoparticles for gene editing. ACS Appl. Mater. Inter. 2019, 11, 47762–47770. [Google Scholar] [CrossRef]
- Nodvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef]
- Paix, A.; Folkmann, A.; Goldman, D.H.; Kulaga, H.; Grzelak, M.J.; Rasoloson, D.; Paidemarry, S.; Green, R.; Reed, R.R.; Seydoux, G. Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc. Natl. Acad. Sci. USA 2017, 114, E10745–E10754. [Google Scholar] [CrossRef]
- Shalem, O.; Sanjana, N.E.; Zhang, F. High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 2015, 16, 299–311. [Google Scholar] [CrossRef]
- Shi, T.Q.; Gao, J.; Wang, W.J.; Wang, K.F.; Xu, G.Q.; Huang, H.; Ji, X.J. CRISPR/Cas9-based genome editing in the filamentous fungus Fusarium fujikuroi and its application in strain engineering for gibberellic acid production. ACS Synth. Biol. 2019, 8, 445–454. [Google Scholar] [CrossRef]
- Wei, T.Y.; Wu, Y.J.; Xie, Q.P.; Tang, J.W.; Yu, Z.T.; Yang, S.B.; Chen, S.X. CRISPR/Cas9-based genome editing in the filamentous fungus Glarea lozoyensis and its application in manipulating gloF. ACS Synth. Biol. 2020, 9, 1968–1977. [Google Scholar] [CrossRef]
- Xie, K.; Yang, Y. RNA-guided genome editing in plants using a CRISPR-Cas system. Mol. Plant. 2013, 6, 1975–1983. [Google Scholar] [CrossRef]
- Yamato, T.; Handa, A.; Arazoe, T.; Kuroki, M.; Nozaka, A.; Kamakura, T.; Ohsato, S.; Arie, T.; Kuwata, S. Single crossover-mediated targeted nucleotide substitution and knock-in strategies with CRISPR/Cas9 system in the rice blast fungus. Sci. Rep. 2019, 9, 7427. [Google Scholar] [CrossRef]
- Zhou, H.; Liu, B.; Weeks, D.P.; Spalding, M.H.; Yang, B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014, 42, 10903–10914. [Google Scholar] [CrossRef]
- Katayama, T.; Tanaka, Y.; Okabe, T.; Nakamura, H.; Fujii, W.; Kitamoto, K.; Maruyama, J. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol. Lett. 2016, 38, 637–642. [Google Scholar] [CrossRef]
- Gabriel, R.; Prinz, J.; Jecmenica, M.; Romero-Vazquez, C.; Chou, P.; Harth, S.; Floerl, L.; Curran, L.; Oostlander, A.; Matz, L.; et al. Development of genetic tools for the thermophilic filamentous fungus Thermoascus aurantiacus. Biotechnol. Biofuels. 2020, 13, 167. [Google Scholar] [CrossRef]
- Liu, K.; Sun, B.; You, H.; Tu, J.L.; Yu, X.; Zhao, P.; Xu, J.W. Dual sgRNA-directed gene deletion in basidiomycete Ganoderma lucidum using the CRISPR/Cas9 system. Microb. Biotechnol. 2020, 13, 386–396. [Google Scholar] [CrossRef]
- Liu, G.Y.; Nizet, V. Color me bad: Microbial pigments as virulence factors. Trends Microbiol. 2009, 17, 406–413. [Google Scholar] [CrossRef]
- Avalos, J.; Carmen Limon, M. Biological roles of fungal carotenoids. Curr. Genet. 2015, 61, 309–324. [Google Scholar] [CrossRef]
- Cary, J.W.; Harris-Coward, P.Y.; Ehrlich, K.C.; Di Mavungu, J.D.; Malysheva, S.V.; De Saeger, S.; Dowd, P.F.; Shantappa, S.; Martens, S.L.; Calvo, A.M. Functional characterization of a veA-dependent polyketide synthase gene in Aspergillus flavus necessary for the synthesis of asparasone, a sclerotium-specific pigment. Fungal Genet. Biol. 2014, 64, 25–35. [Google Scholar] [CrossRef]
- Yu, X.; Huo, L.; Liu, H.; Chen, L.F.; Wang, Y.; Zhu, X.D. Melanin is required for the formation of the multi-cellular conidia in the endophytic fungus Pestalotiopsis microspora. Microbiol. Res. 2015, 179, 1–11. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, N.; Li, Z.; Ling, J.; Zhao, J.; Li, Y.; Yang, Y.; Mao, Z.; Xie, B.; Li, H.; Jiao, Y. Establishment of a CRISPR/Cas9-Mediated Efficient Knockout System of Trichoderma hamatum T21 and Pigment Synthesis PKS Gene Knockout. J. Fungi 2023, 9, 595. https://doi.org/10.3390/jof9050595
Luo N, Li Z, Ling J, Zhao J, Li Y, Yang Y, Mao Z, Xie B, Li H, Jiao Y. Establishment of a CRISPR/Cas9-Mediated Efficient Knockout System of Trichoderma hamatum T21 and Pigment Synthesis PKS Gene Knockout. Journal of Fungi. 2023; 9(5):595. https://doi.org/10.3390/jof9050595
Chicago/Turabian StyleLuo, Ning, Zeyu Li, Jian Ling, Jianlong Zhao, Yan Li, Yuhong Yang, Zhenchuan Mao, Bingyan Xie, Huixia Li, and Yang Jiao. 2023. "Establishment of a CRISPR/Cas9-Mediated Efficient Knockout System of Trichoderma hamatum T21 and Pigment Synthesis PKS Gene Knockout" Journal of Fungi 9, no. 5: 595. https://doi.org/10.3390/jof9050595
APA StyleLuo, N., Li, Z., Ling, J., Zhao, J., Li, Y., Yang, Y., Mao, Z., Xie, B., Li, H., & Jiao, Y. (2023). Establishment of a CRISPR/Cas9-Mediated Efficient Knockout System of Trichoderma hamatum T21 and Pigment Synthesis PKS Gene Knockout. Journal of Fungi, 9(5), 595. https://doi.org/10.3390/jof9050595







