Disruption of the Schizosaccharomyces japonicus lig4 Disturbs Several Cellular Processes and Leads to a Pleiotropic Phenotype
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Identification of the lig4 Putative Orthologue of S. japonicus
3.2. Isolation of the lig4 Mutant Strain
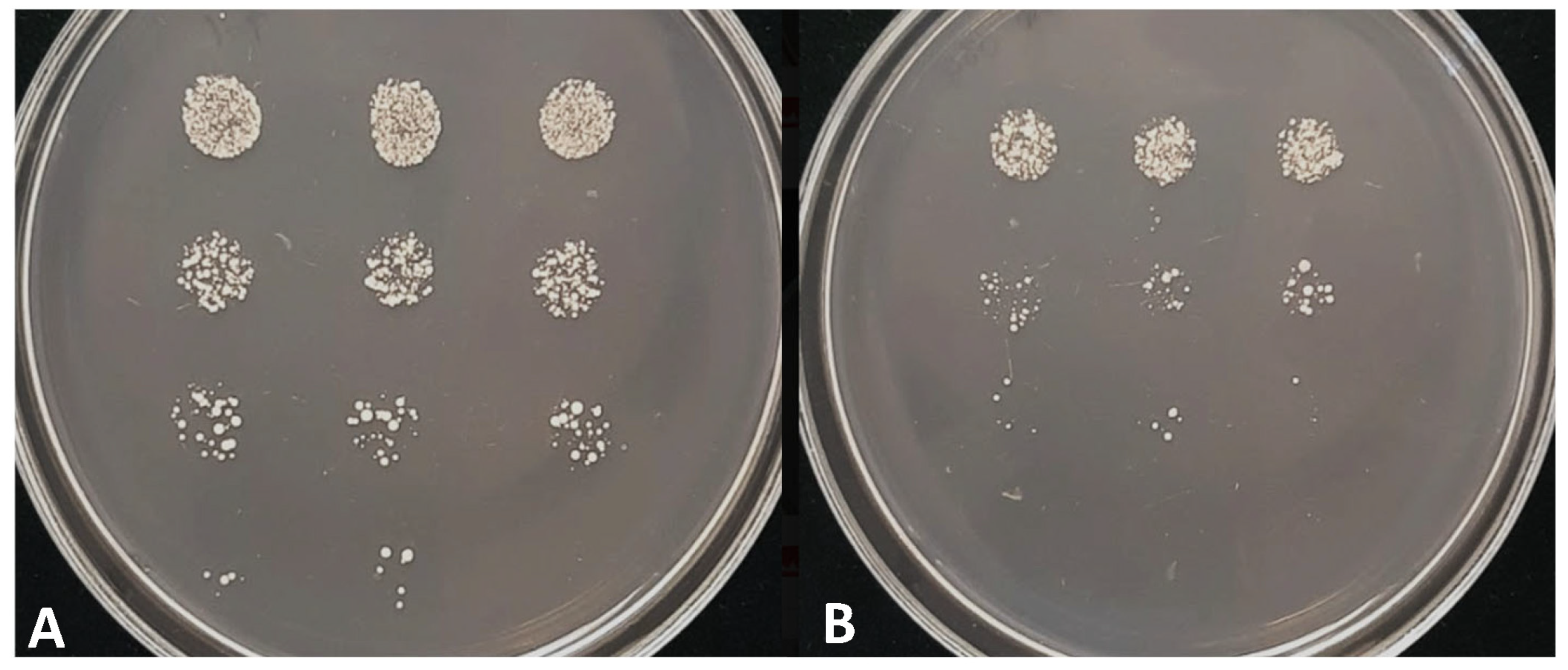
3.3. Phenotypic Analyses of the lig4 Mutant Cells Revealed That They Have Increased Sporulation Capacity on Complete Medium, Decreased Hyphal Growth, and Tend to Produce Chains of Cells in the Presence of Caffeine
3.4. The Study of Stress Response Revealed Increased Chronological Aging and Sensitivity to Heat Shock, UV Light, and Caffeine
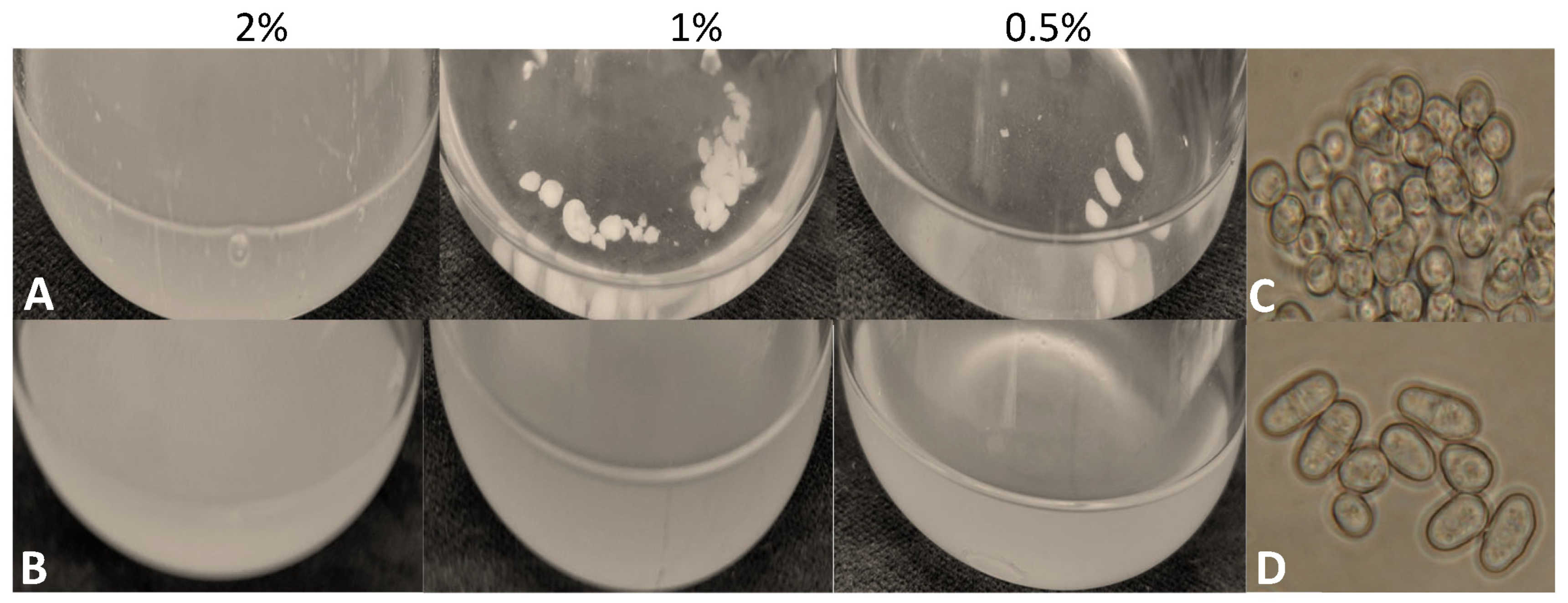
3.5. Higher Flocculation Capacity
3.6. Transcriptional Profiling Confirmed the Phenotypic Changes of the Mutant Strain
3.7. Gene Targeting Was Higher in the lig4 Mutant Strain
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Teo, S.H.; Jackson, S.P. Identification of Saccharomyces cerevisiae DNA ligase IV: Involvement in DNA double-strand break repair. EMBO J. 1997, 16, 4788–4795. [Google Scholar] [CrossRef] [PubMed]
- Schär, P.; Herrmann, G.; Daly, G.; Lindahl, T. A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev. 1997, 11, 1912–1924. [Google Scholar] [CrossRef] [PubMed]
- Gorski, M.M.; Eeken, J.C.J.; de Jong, A.W.M.; Klink, I.; Loos, M.; Romeijn, R.J.; van Veen, B.L.; Mullenders, L.H.; Ferro, W.; Pastink, A. The Drosophila melanogaster DNA Ligase IV Gene Plays a Crucial Role in the Repair of Radiation-Induced DNA Double-Strand Breaks and Acts Synergistically with Rad54. Genetics 2003, 165, 1929–1941. [Google Scholar] [CrossRef]
- Mizutani, O.; Kudo, Y.; Saito, A.; Matsuura, T.; Inoue, H.; Abe, K.; Gomi, K. A defect of LigD (human Lig4 homolog) for nonhomologous end joining significantly improves the efficiency of gene-targeting in Aspergillus oryzae. Fungal Genet. Biol. 2008, 45, 878–889. [Google Scholar] [CrossRef] [PubMed]
- Kito, H.; Fujikawa, T.; Moriwaki, A.; Tomono, A.; Izawa, M.; Kamakura, T.; Ohashi, M.; Sato, H.; Abe, K.; Nishimura, M. MgLig4, a homolog of Neurospora crassa Mus-53 (DNA ligase IV), is involved in, but not essential for, non-homologous end-joining events in Magnaporthe grisea. Fungal Genet. Biol. 2008, 45, 1543–1551. [Google Scholar] [CrossRef]
- Wilson, T.E.; Grawunder, U.; Lieber, M.R. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 1997, 388, 495–498. [Google Scholar] [CrossRef]
- Adachi, N.; Ishino, T.; Ishii, Y.; Takeda, S.; Koyama, H. DNA ligase IV-deficient cells are more resistant to ionizing radiation in the absence of Ku70: Implications for DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 2001, 98, 12109–12113. [Google Scholar] [CrossRef]
- Deshpande, R.A.; Wilson, T.E. Modes of interaction among yeast Nej1, Lif1 and Dnl4 proteins and comparison to human XLF, XRCC4 and Lig4. DNA Repair 2007, 6, 1507–1516. [Google Scholar] [CrossRef]
- Li, J.; Yu, Y.; Suo, F.; Sun, L.L.; Zhao, D.; Du, L.L. Genome-wide Screens for Sensitivity to Ionizing Radiation Identify the Fission Yeast Nonhomologous End Joining Factor Xrc4. G3 Genes Genomes Genet. 2014, 4, 1297–1306. [Google Scholar] [CrossRef]
- Ishibashi, K.; Suzuki, K.; Ando, Y.; Takakura, C.; Inoue, H. Nonhomologous chromosomal integration of foreign DNA is completely dependent on MUS-53 (human Lig4 homolog) in Neurospora. Proc. Natl. Acad. Sci. USA 2006, 103, 14871–14876. [Google Scholar] [CrossRef]
- Altmann, T.; Gennery, A.R. DNA ligase IV syndrome. Orphanet J. Rare Dis. 2016, 11, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Chen, Q.; Wang, Y.; Liu, D.; Hou, J.; Wang, W.; Ying, W.; Hui, X.; Zhou, Q.; Sun, J.; et al. LIG4 syndrome: Clinical and molecular characterization in a Chinese cohort. Orphanet J. Rare Dis. 2020, 15, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Kuhmann, C.; Li, C.; Kloor, M.; Salou, M.; Weigel, C.; Schmidt, C.R.; Ng, L.W.C.; Tsui, W.W.Y.; Leung, S.Y.; Yuen, S.T.; et al. Altered regulation of DNA ligase IV activity by aberrant promoter DNA methylation and gene amplification in colorectal cancer. Hum. Mol. Genet. 2014, 23, 2043–2054. [Google Scholar] [CrossRef] [PubMed]
- Grupp, K.; Roettger, L.; Kluth, M.; Hube-Magg, C.; Simon, R.; Lebok, P.; Minner, S.; Tsourlakis, M.C.; Koop, C.; Graefen, M.; et al. Expression of DNA ligase IV is linked to poor prognosis and characterizes a subset of prostate cancers harboring TMPRSS2:ERG fusion and PTEN deletion. Oncol. Rep. 2015, 34, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Grunebaum, E.; Bates, A.; Roifman, C.M. Omenn syndrome is associated with mutations in DNA ligase IV. J. Allergy Clin. Immunol. 2008, 122, 1219–1220. [Google Scholar] [CrossRef] [PubMed]
- Van der Burg, M.; van Veelen, L.R.; Verkaik, N.S.; Wiegant, W.W.; Hartwig, N.G.; Barendregt, B.H.; Brugmans, L.; Raams, A.; Jaspers, N.G.J.; Zdzienicka, M.Z.; et al. A new type of radiosensitive T--B--NK+ severe combined immunodeficiency caused by a LIG4 mutation. J. Clin. Investig. 2006, 116, 137–145. [Google Scholar] [CrossRef]
- Grawunder, U.; Zimmer, D.; Fugmann, S.; Schwarz, K.; Lieber, M.R.D. Ligase IV Is Essential for V(D)J Recombination and DNA Double-Strand Break Repair in Human Precursor Lymphocytes. Mol. Cell 1988, 2, 477–484. [Google Scholar] [CrossRef]
- Roddam, P.L.; Rollinson, S.; O’Driscoll, M.; Jeggo, P.A.; Jack, A.; Morgan, G.J. Genetic variants of NHEJ DNA ligase IV can affect the risk of developing multiple myeloma, a tumour characterized by aberrant class switch recombination. J. Med. Genet. 2002, 39, 900–905. [Google Scholar] [CrossRef]
- Stewart, D.R.; Pemov, A.; Johnston, J.J.; Sapp, J.C.; Yeager, M.; He, J.; Boland, J.F.; Burdett, L.; Brown, C.; Gatti, R.A.; et al. Dubowitz Syndrome Is a Complex Comprised of Multiple, Genetically Distinct and Phenotypically Overlapping Disorders. PLoS ONE 2014, 9, e98686. [Google Scholar] [CrossRef]
- Barnes, D.E.; Stamp, G.; Rosewell, I.; Denzel, A.; Lindahl, T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 1998, 8, 1395–1398. [Google Scholar] [CrossRef]
- Frank, K.M.; JoAnn, M.; Sekiguchi, K.; Seidl, J.; Swat, W.; Rathbun, G.A.; Cheng, H.L.; Davidson, L.; Kangaloo, L.; Alt, F.W. Late embryonic lethality and impaired V (D)J recombination in mice lacking DNA ligase IV. Nature 1998, 396, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Andaluz, E.; Ciudad, A.; Coque, J.R.; Calderone, R.; Larriba, G. Cell cycle regulation of a DNA ligase-encoding gene (CaLIG4) from Candida albicans. Yeast 1999, 15, 1199–1210. [Google Scholar] [CrossRef]
- Andaluz, E.; Calderone, R.; Reyes, G.; Larriba, G. Phenotypic Analysis and Virulence of Candida albicans LIG4 Mutants. Infect. Immun. 2001, 69, 137–147. [Google Scholar] [CrossRef]
- He, Y.; Shao, Y.; Chen, F. Efficient gene targeting in ligase IV-deficient Monascus ruber M7 by perturbing the non-homologous end joining pathway. Fungal Biol. 2014, 118, 846–854. [Google Scholar] [CrossRef]
- Kikukawa, H.; Sakuradani, E.; Ando, A.; Okuda, T.; Ochiai, M.; Shimizu, S.; Ogawa, J. Disruption of lig4 improves gene targeting efficiency in the oleaginous fungus Mortierella alpina. J. Biotechnol. 2015, 208, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Cen, Y.; Fiori, A.; Van Dijck, P. Deletion of the DNA ligase IV gene in Candida glabrata significantly increases gene-targeting efficiency. Eukaryot. Cell 2015, 14, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Watanabe, T.; Aikawa, S.; Nishi, T.; Nishiyama, T.; Nakamura, Y.; Hasunuma, T.; Okubo, Y.; Ishii, J.; Kondo, A. Deletion of DNA ligase IV homolog confers higher gene targeting efficiency on homologous recombination in Komagataella phaffii. FEMS Yeast Res. 2018, 18, foy074. [Google Scholar] [CrossRef] [PubMed]
- Krappmann, S. Gene targeting in filamentous fungi: The benefits of impaired repair. Fungal Biol. Rev. 2007, 21, 25–29. [Google Scholar] [CrossRef]
- Cen, Y.; Timmermans, B.; Souffriau, B.; Thevelein, J.M.; Van Dijck, P. Comparison of genome engineering using the CRISPR-Cas9 system in C. glabrata wild-type and lig4 strains. Fungal Genet. Biol. 2017, 107, 44–50. [Google Scholar] [CrossRef]
- Klar, A.J.S. Schizosaccharomyces japonicus Yeast Poised to Become a Favorite Experimental Organism for Eukaryotic Research. G3 Genes Genomes Genet. 2013, 3, 1869–1873. [Google Scholar] [CrossRef]
- Papp, L.; Sipiczki, M.; Miklós, M. Expression pattern and phenotypic characterization of the mutant strain reveals target genes and processes regulated by pka1 in the dimorphic fission yeast Schizosaccharomyces japonicus. Curr. Genet. 2017, 63, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Yukawa, M.; Maki, M.T. Schizosaccharomyces japonicus nov. spec. Bul. Sci. Fak. Terkultura Kjusu Imp. Univ. 1931, 4, 224–242. [Google Scholar]
- Sipiczki, M.; Ferenczy, L. Protoplast fusion of Schizosaccharomyces pombe auxotrophic mutants of identical mating-type. Mol. Gen. Genet. 1977, 151, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Mitchison, J.M. Physiological and cytological methods for Schizosaccharomyces pombe. Methods Cell Physiol. 1970, 4, 131–165. [Google Scholar]
- Aoki, K.; Nakajima, R.; Furuya, K.; Niki, H. Novel episomal vectors and a highly efficient transformation procedure for the fission yeast Schizosaccharomyces japonicus. Yeast 2010, 27, 1049–1060. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Rutherford, K.M.; Harris, M.A.; Oliferenko, S.; Wood, V. JaponicusDB: Rapid deployment of a model organism database for an emerging model species Genetics. Genetics 2022, 220, iyab223. [Google Scholar] [CrossRef]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Anisimova, M.; Gascuel, O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Acs-Szabo, L.; Papp, L.A.; Sipiczki, M.; Miklos, I. Genome Comparisons of the Fission Yeasts Reveal Ancient Collinear Loci Maintained by Natural Selection. J. Fungi 2021, 7, 864. [Google Scholar] [CrossRef] [PubMed]
- Moser, B.A.; Brondello, J.M.; Baber-Furnary, B.; Russell, P. Mechanism of Caffeine-Induced Checkpoint Override in Fission Yeast. Mol. Cell Biol. 2000, 20, 4288–4294. [Google Scholar] [CrossRef] [PubMed]
- Sipiczki, M.; Takeo, K.; Yamaguchi, M.; Yoshida, S.; Miklos, I. Environmentally controlled dimorphic cycle in fission yeast. Microbiology 1998, 144, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Sipiczki, M.; Takeo, K.; Grallert, A. Growth polarity transitions in dimorphic fission yeast. Microbiology 1998, 144, 3475–3485. [Google Scholar] [CrossRef]
- Furuya, K.; Niki, H. The DNA Damage Checkpoint Regulates a Transition between Yeast and Hyphal Growth in Schizosaccharomyces japonicus. Mol. Cell Biol. 2010, 30, 2909–2917. [Google Scholar] [CrossRef]
- Papp, L.; Sipiczki, M.; Holb, I.J.; Miklós, I. Optimal conditions for mycelial growth of Schizosaccharomyces japonicus cells in liquid medium: It enables the molecular investigation of dimorphism. Yeast 2014, 31, 475–482. [Google Scholar]
- Sipiczki, M.; Grallert, B.; Miklos, I. Mycelial and syncytial growth in Schizosaccharomyces pombe induced by novel septation mutations. J. Cell Sci. 1993, 104, 485–493. [Google Scholar] [CrossRef]
- Grallert, A.; Grallert, B.; Zilahi, E.; Szilagyi, Z.; Sipiczki, M. Eleven novel sep genes of Schizosaccharomyces pombe required for efficient cell separation and sexual differentiation. Yeast 1999, 15, 669–686. [Google Scholar] [CrossRef]
- Miklos, I.; Ludanyi, K.; Sipiczki, M. The pleiotropic cell separation mutation spl1-1 is a nucleotide substitution in the internal promoter of the proline tRNACGG gene of Schizosaccharomyces pombe. Curr. Genet. 2009, 55, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, T.; Otsubo, Y.; Urano, J.; Tamanoi, F.; Yamamoto, M. Loss of the TOR Kinase Tor2 Mimics Nitrogen Starvation and Activates the Sexual Development Pathway in Fission Yeast. Am. Soc. Microbiol. Mol. Cell. Biol. 2007, 27, 3154–3164. [Google Scholar] [CrossRef] [PubMed]
- AÁlvarez, B.; Moreno, S. Fission yeast Tor2 promotes cell growth and represses cell differentiation. J. Cell Sci. 2006, 119, 4475–4485. [Google Scholar] [CrossRef]
- Otsubo, Y.; Nakashima, A.; Yamamoto, M.; Yamashita, A. TORC1-Dependent Phosphorylation Targets in Fission Yeast. Biomolecules 2017, 7, 50. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Hatanaka, M.; Nagao, K.; Nakaseko, Y.; Kanoh, J.; Kokubu, A.; Ebe, M.; Yanagida, M. Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes Cells 2007, 12, 1357–1370. [Google Scholar] [CrossRef] [PubMed]
- Hayles, J.; Wood, V.; Jeffery, L.; Hoe, K.L.; Kim, D.U.; Park, H.O.; Salas-Pino, S.; Heichinger, C.; Nurse, P. A genome-wide resource of cell cycle and cell shape genes of fission yeast. Open Biol. 2013, 3, 130053. [Google Scholar] [CrossRef]
- Egel, R. Physiological Aspects of Conjugation in Fission Yeast. Planta 1971, 98, 89–96. [Google Scholar] [CrossRef]
- McCleary, D.F.; Rine, J. Nutritional Control of Chronological Aging and Heterochromatin in Saccharomyces cerevisiae. Genetics 2017, 205, 1179–1193. [Google Scholar] [CrossRef]
- Roux, A.E.; Quissac, A.; Chartrand, P.; Ferbeyre, G.; Rokeach, L.A. Regulation of chronological aging in Schizosaccharomyces pombe by the protein kinases Pka1 and Sck2. Aging Cell 2006, 5, 345–357. [Google Scholar] [CrossRef]
- Biswas, S.; Van Dijck, P.; Datta, A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 2007, 71, 348–376. [Google Scholar] [CrossRef]
- Papp, L.A.; Acs-Szabo, L.; Batta, G.; Miklos, I. Molecular and comparative genomic analyses reveal evolutionarily conserved and unique features of the Schizosaccharomyces japonicus mycelial growth and the underlying genomic changes. Curr. Genet. 2021, 67, 953–968. [Google Scholar] [CrossRef] [PubMed]
- Otsubo, Y.; Yamashita, A.; Ohno, H.; Yamamoto, M.S. pombe TORC1 activates the ubiquitin-proteasomal degradation of the meiotic regulator Mei2 in cooperation with Pat1 kinase. J. Cell Sci. 2014, 127, 2639–2646. [Google Scholar] [PubMed]
- Wullschleger, S.; Loewith, R.; Hall, M.N.T. Signaling in Growth and Metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Weisman, R. The target of rapamycin (TOR) regulates growth in response to nutritional signals. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.F.; Schreiber, S.L. Target of rapamycin proteins and their kinase activities are required for meiosis. Proc. Natl. Acad. Sci. USA 1997, 94, 3070–3075. [Google Scholar] [CrossRef]
- Laor, D.; Cohen, A.; Kupiec, M.; Weisman, R. TORC1 Regulates Developmental Responses to Nitrogen Stress via Regulation of the GATA Transcription Factor Gaf1. mBio 2015, 6, e00959. [Google Scholar] [CrossRef]
- Rallis, C.; Codlin, S.; Bahler, J. TORC1 signaling inhibition by rapamycin and caffeine affect lifespan, global gene expression, and cell proliferation of fission yeast. Aging Cell 2013, 12, 567–573. [Google Scholar] [CrossRef]
- Powers, R.W.; Kaeberlein, M.; Caldwell, S.D.; Kennedy, B.K.; Fields, S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006, 20, 174–184. [Google Scholar] [CrossRef]
- Alao, J.P.; Legon, L.; Rallis, C. Crosstalk between the mTOR and DNA Damage Response Pathways in Fission Yeast. Cells 2021, 10, 305. [Google Scholar] [CrossRef]
- Cardenas, M.E.; Cutler, N.S.; Lorenz, M.C.; Di Como, C.J.; Heitman, J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999, 13, 3271–3279. [Google Scholar] [CrossRef]
- Powers, E.N.; Chan, C.; Doron-Mandel, E.; Allcca, L.L.; Kim, J.K.; Jovanovic, M.; Brar, G.M. Bidirectional promoter activity from expression cassettes can drive off-target repression of neighboring gene translation. eLife 2022, 11, e81086. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.D.; Liu, Y.W.; Bi, H.H.; Yang, X.R.; Han, J.M.; Dong, J.G.; Gu, S.Q. StKU80, a component in the NHEJ repair pathway, is involved in mycelial morphogenesis, conidiation, appressorium development, and oxidative stress reactions in Exserohilum turcicum. J. Integr. Agric. 2021, 20, 147–158. [Google Scholar] [CrossRef]
- Abbasi, S.; Parmar, G.; Kelly, R.D.; Balasuriya, N.; Schild-Poulter, C. The Ku complex: Recent advances and emerging roles outside of non-homologous end-joining. Cell Mol. Life Sci. 2021, 78, 4589–4613. [Google Scholar] [CrossRef] [PubMed]

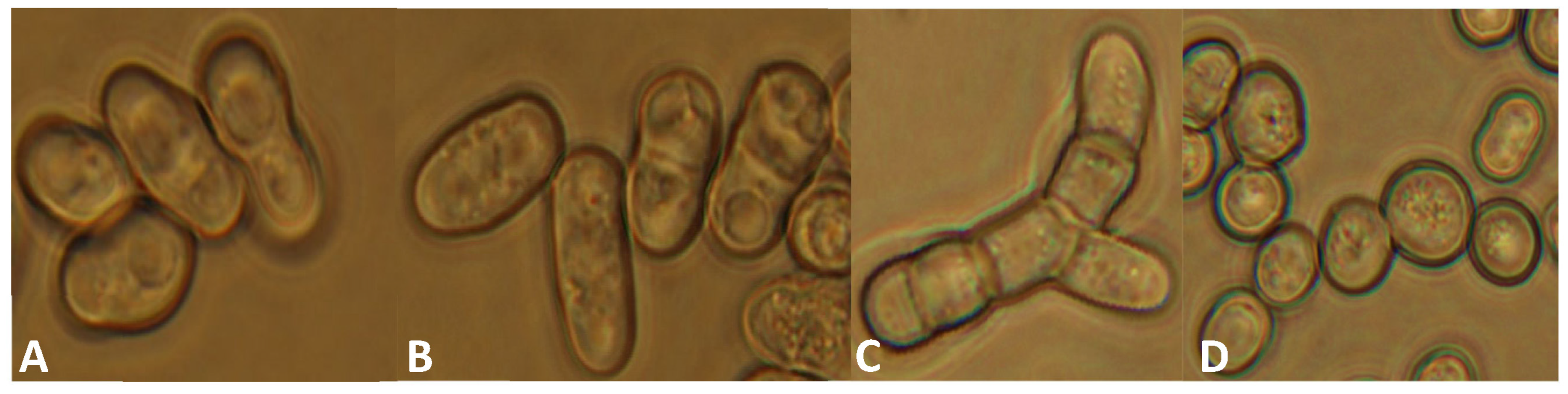
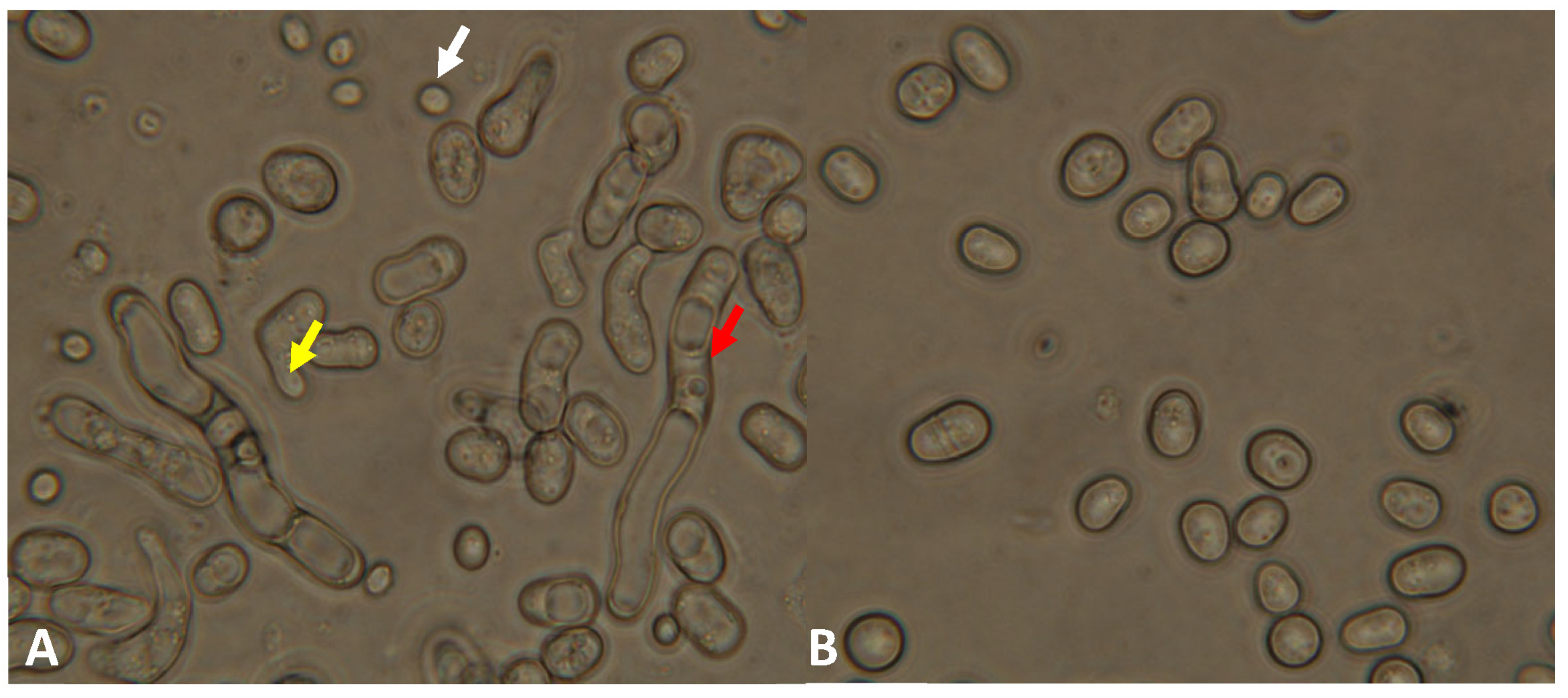
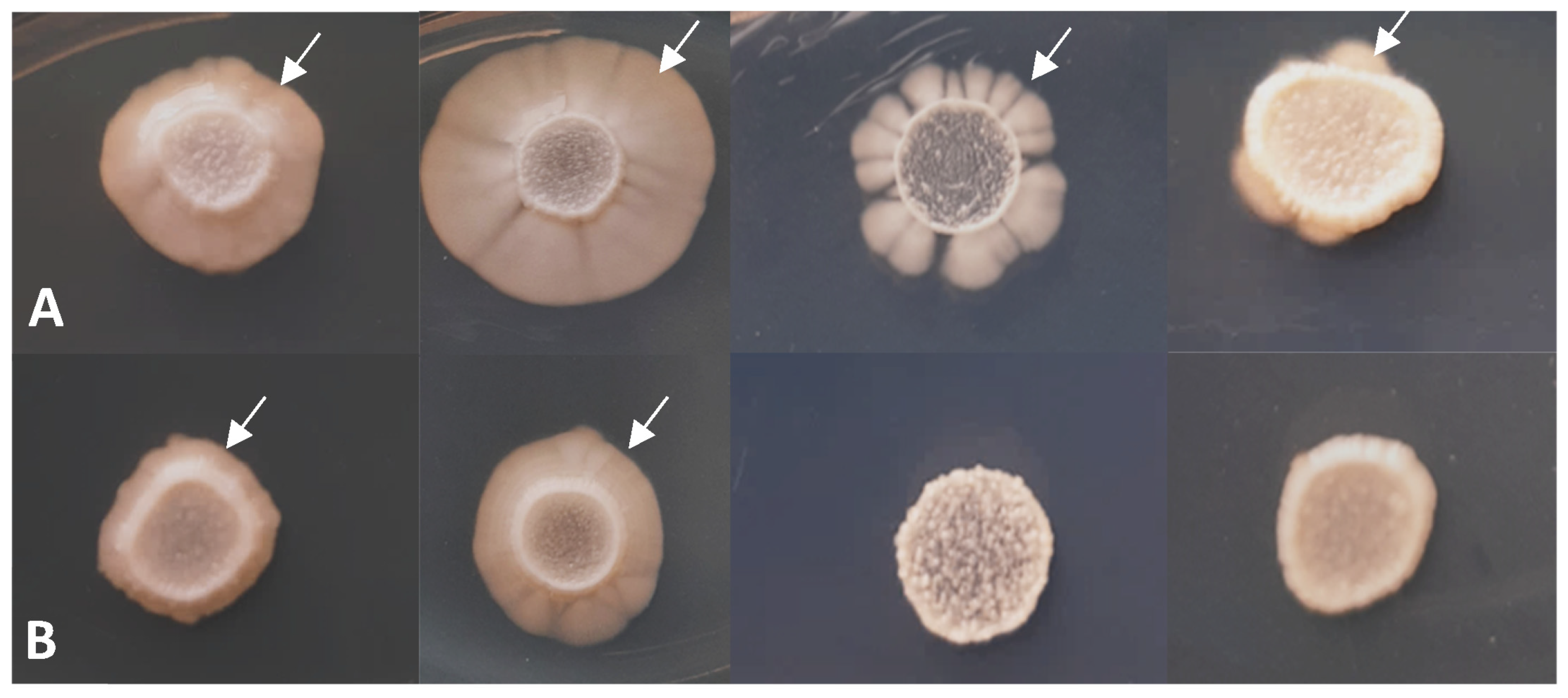
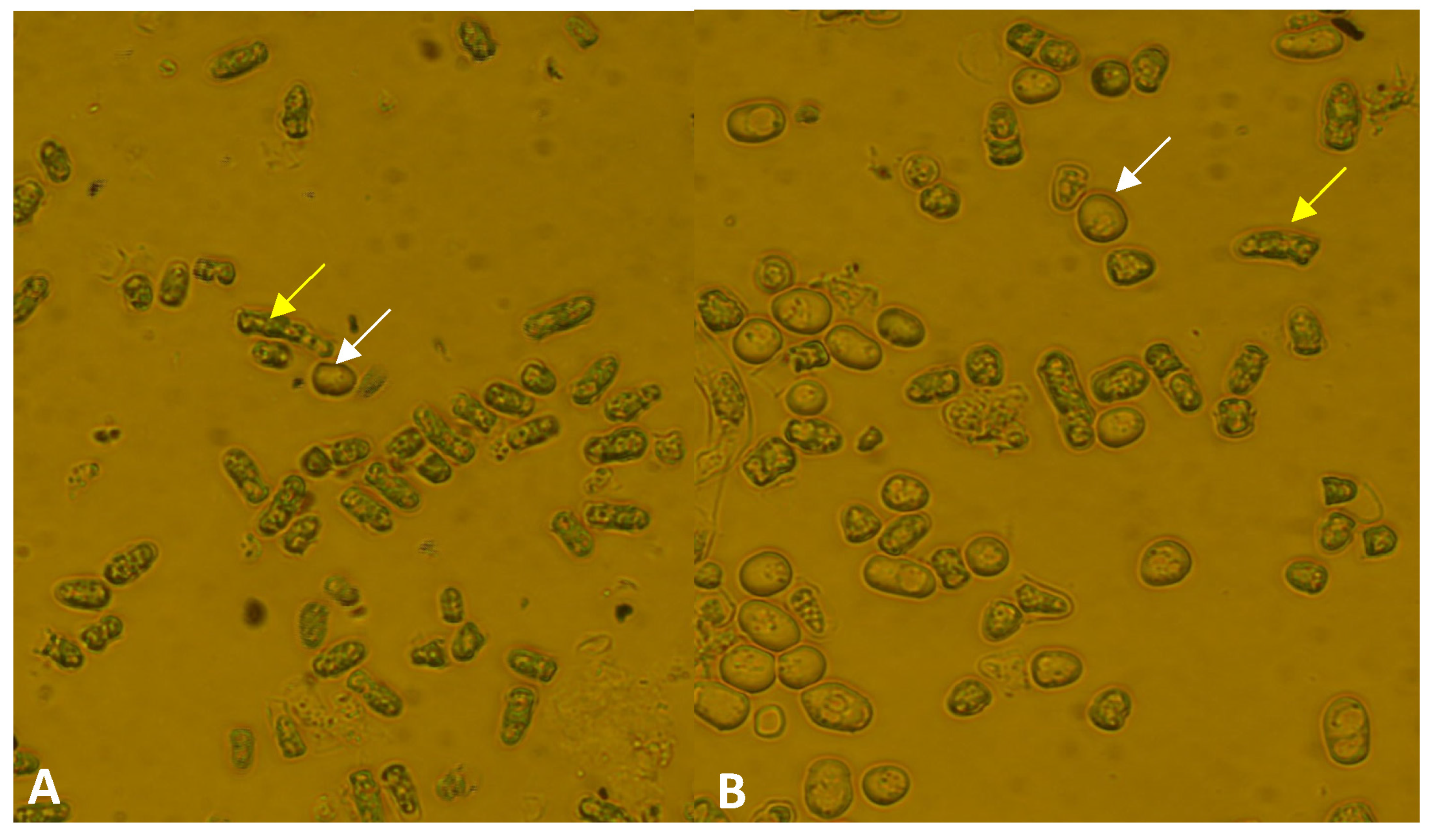
| Strain | Number of Spores and Zygotes (%) | |
|---|---|---|
| Spore | Zygote | |
| Wild-type | 4.3 | 0.6 |
| Mutant | 10.6 | 4.7 |
| Strain | Colony Forming Capacity (%) | One Tailed t-Test p-Values | |
|---|---|---|---|
| Wild-Type | Mutant | ||
| Treatment | |||
| 0 min heat shock | 100 | 100 | 1.00 |
| 5 min heat shock | 99.5 | 78.5 | 0.00521 |
| 10 min heat shock | 93.0 | 45.6 | 1.2796 × 10−6 |
| 0 min UV treatment | 100 | 100 | 1.00 |
| 20 s UV treatment | 63.8 | 42.7 | 0.00223 |
| 40 s UV treatment | 21.0 | 1.6 | 1.8818 × 10−6 |
| 60 s UV treatment | 2.6 | 0.1 | 2.1902 × 10−5 |
| Gene Identifier in S. japonicus | Function in S. japonicus | mRNA Level in the lig4 Mutant Strain | Gene Identifier in S. pombe | Gene Name or Function in S. pombe | mRNA Level in S. pombe tor2-s6 [49] Mutant |
|---|---|---|---|---|---|
| SJAG_05288 | DNA-binding forkhead transcription factor, meiotic Mei4 | + | SPBC32H8.11 | mei4 | + |
| SJAG_00145 | RNA-binding protein involved in meiosis Mei2 | + | SPAC27D7.03 | mei2 [54,55] | + |
| SJAG_03405 | zf-FYVE type zinc finger protein and glutamine sensor Pib2 | − | SPBC9B6.03 | zinc finger protein, Pib2 | + |
| SJAG_04987 | HbrB family protein involved in TOR signaling Bit2 | − | SPAC6B12.03c | bit2 | + |
| SJAG_00100 | meiosis specific cyclin Crs1 | + | SPBC2G2.09 | crs1 | + |
| SJAG_01978 | pheromone p-factor receptor Mam2 | + | SPAC11H11.04 | mam2 | + |
| SJAG_00781 | P-factor pheromone Map2 | + | SPCC1795.06 | map2 | + |
| SJAG_04530 | Rad22 homolog Rti1 | + | SPBC119.14 | rti1 | + |
| SJAG_02652 | linear element-associated protein Hop1 | + | SPBC1718.02 | hop1 | + |
| SJAG_01943 | formin Fus1 | + | SPAC20G4.02 | fus1 | + |
| SJAG_02286 | dynein heavy chain, minus-end directed microtubule motor Dhc1 | + | SPAC1093.06 | dhc1 | + |
| SJAG_04009 | DNA-binding transcription factor Ste11 | + | SPBC32C12.02 | ste11 [54,55] | + |
| SJAG_02237 | XRCC4 nonhomologous end joining factor Xrc4 | + | SPAC6G9.16c | xrc4 | + |
| SJAG_04950 | meu13, Tat binding protein 1(TBP-1)-interacting protein (TBPIP) homolog | + | SPAC222.15 | meu13 | + |
| SJAG_02003 | yippee-like protein | − | SPAPJ691.02 | yippee-like protein | + |
| SJAG_05004 | mug8, DUF1708 family conserved fungal protein, cell division site | + | SPAC32A11.01 | mug8 | + |
| SJAG_03644 | MFS transporter superfamily | + | SPBC1271.09 | tgp1 | + |
| SJAG_02016 | mitochondrial inner membrane anchored oxidoreductase Aif1 | + | SPAC26F1.14c | aif1 | + |
| SJAG_03958 | xylose and arabinose reductase | + | SPAC2F3.05c | xylose and arabinose reductase | + |
| SJAG_00138 | DUF1774 family multi-spanning conserved fungal membrane protein | − | SPAC637.03 | DUF1774 family multi-spanning conserved fungal membrane protein | + |
| SJAG_00377 | Schizosaccharomyces specific protein Meu32 | + | SPAP27G11.08c | meu32 | + |
| SJAG_01869 | NADH/NADPH-dependent indole-3-acetaldehyde reductase, implicated in cellular detoxification | − | SPAC19G12.09 | NADH/NADPH-dependent indole-3-acetaldehyde reductase, implicated in cellular detoxification | + |
| SJAG_00167 | Toc1, Tor complex Tor2 interacting protein 1 | − | SPBP18G5.03 | toc1 [56] | * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acs-Szabo, L.; Papp, L.A.; Takacs, S.; Miklos, I. Disruption of the Schizosaccharomyces japonicus lig4 Disturbs Several Cellular Processes and Leads to a Pleiotropic Phenotype. J. Fungi 2023, 9, 550. https://doi.org/10.3390/jof9050550
Acs-Szabo L, Papp LA, Takacs S, Miklos I. Disruption of the Schizosaccharomyces japonicus lig4 Disturbs Several Cellular Processes and Leads to a Pleiotropic Phenotype. Journal of Fungi. 2023; 9(5):550. https://doi.org/10.3390/jof9050550
Chicago/Turabian StyleAcs-Szabo, Lajos, Laszlo Attila Papp, Szonja Takacs, and Ida Miklos. 2023. "Disruption of the Schizosaccharomyces japonicus lig4 Disturbs Several Cellular Processes and Leads to a Pleiotropic Phenotype" Journal of Fungi 9, no. 5: 550. https://doi.org/10.3390/jof9050550
APA StyleAcs-Szabo, L., Papp, L. A., Takacs, S., & Miklos, I. (2023). Disruption of the Schizosaccharomyces japonicus lig4 Disturbs Several Cellular Processes and Leads to a Pleiotropic Phenotype. Journal of Fungi, 9(5), 550. https://doi.org/10.3390/jof9050550







