Isolation and Genome-Based Characterization of Biocontrol Potential of Bacillus siamensis YB-1631 against Wheat Crown Rot Caused by Fusarium pseudograminearum
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Strain YB-1631 and In Vitro Antifungal Assays
2.2. Biocontrol of FCR and Growth Promotion of Wheat by YB-1631
2.3. Wheat Disease Resistance Enzyme Activities
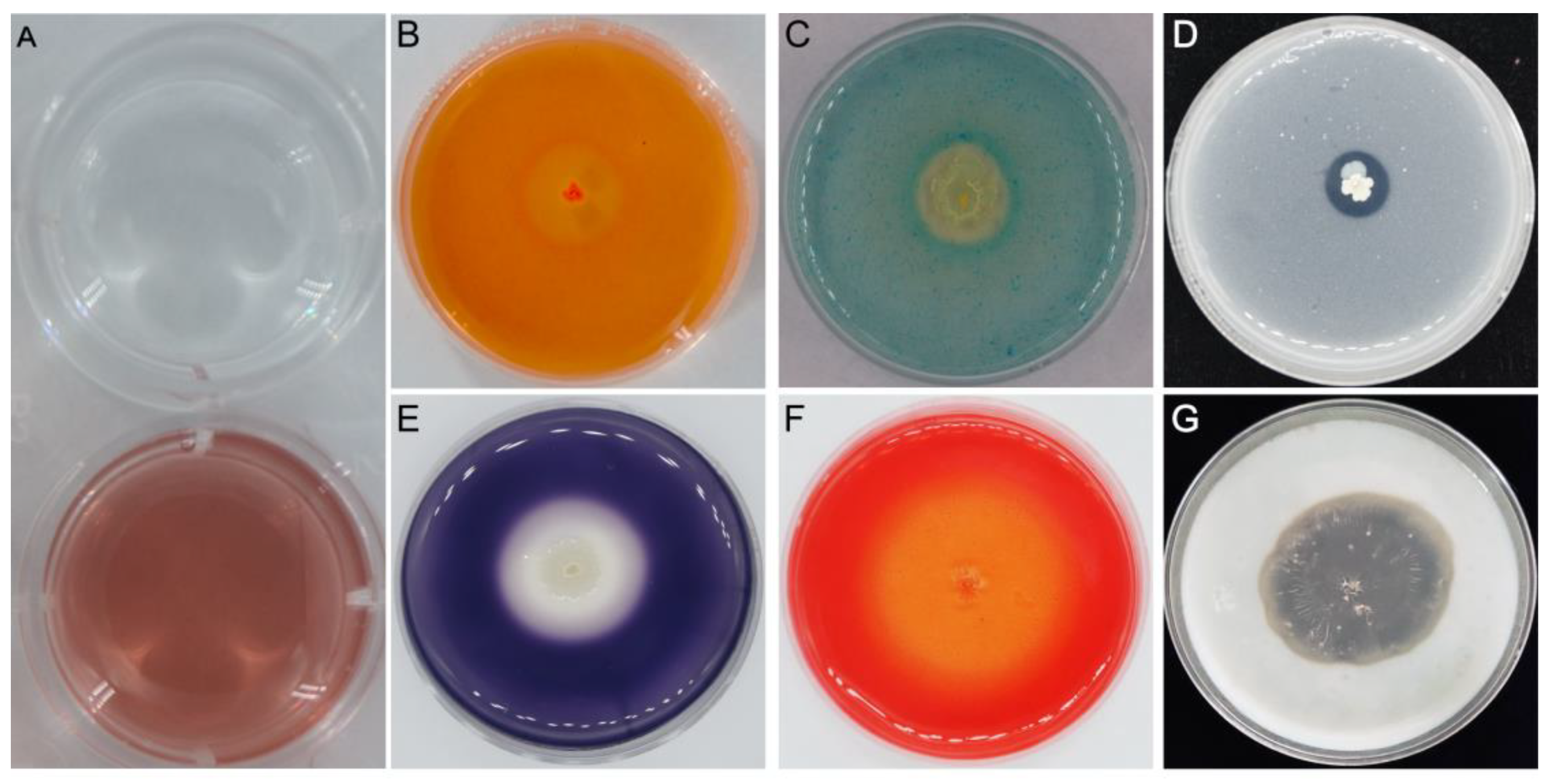
2.4. Plant Growth Promotion and Biocontrol Traits of YB-1631
2.5. Identification of YB-1631
2.6. Genome Sequencing, Assembly, and Annotation
2.7. Statistical Analysis
3. Results
3.1. Isolation and In Vitro Antifungal Activity of YB-1631
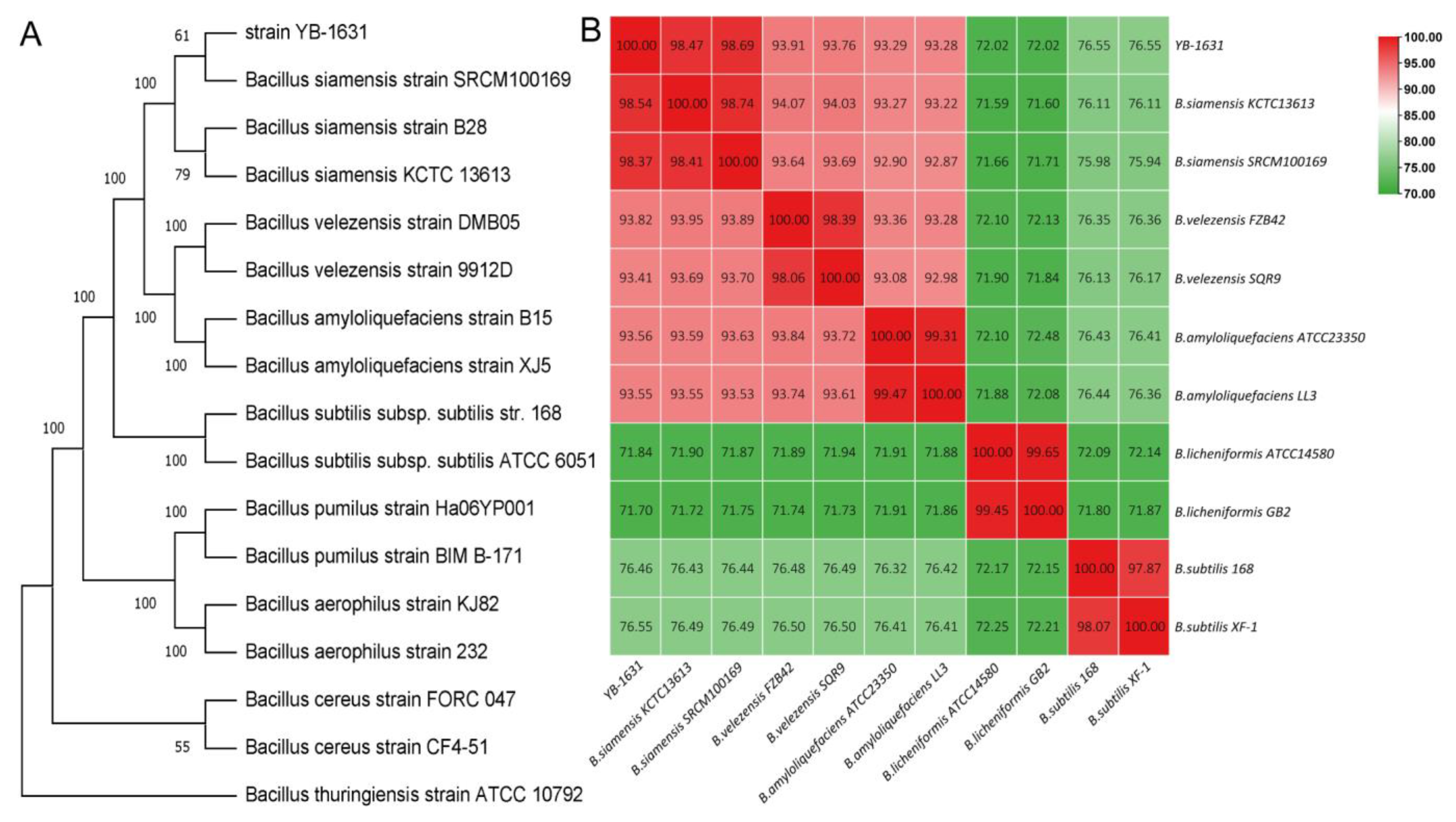
3.2. Identification of YB-1631
3.3. B. siamensis YB-1631 In Vivo Biocontrol and Plant Growth Promotion Activities
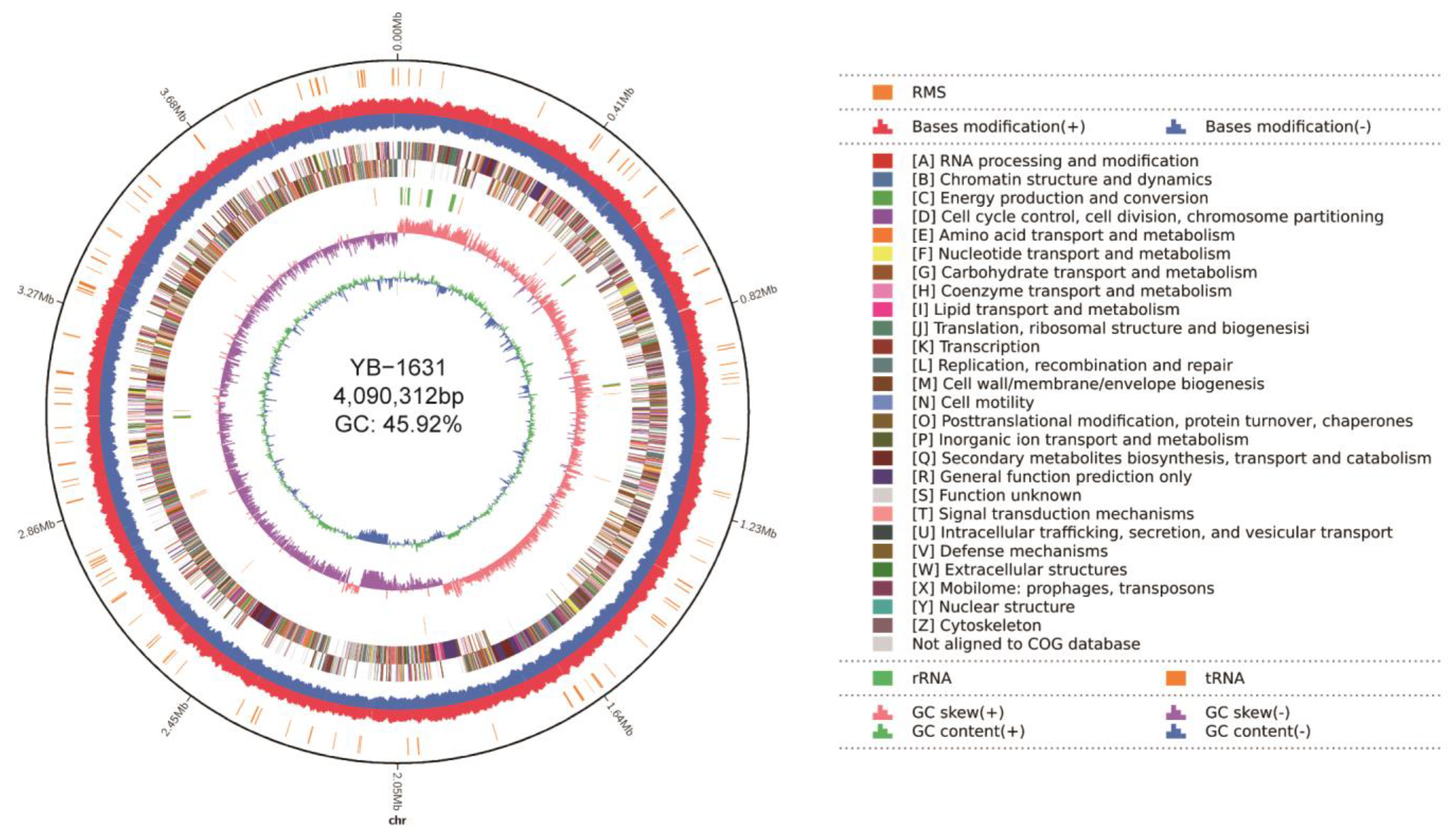
3.4. Genome Assembly and Annotation of B. siamensis YB-1631
3.5. Genes and Activities Potentially Related to FCR Biocontrol, Plant Growth Promotion, and the Interaction of B. siamensis YB-1631 with Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kazan, K.; Gardiner, D.M. Fusarium crown rot caused by Fusarium pseudograminearum in cereal crops: Recent progress and future prospects. Mol. Plant Pathol. 2018, 19, 1547–1562. [Google Scholar] [CrossRef] [PubMed]
- Minati, M.H.; Mohammed-ameen, M.K. Interaction between Fusarium Head Blight and Crown Rot Disease Incidence and Environmental Factors and Soil Physiochemical Analysis on Wheat in the South of Iraq, Basra Province. Karbala Int. J. Mod. Sci. 2019, 5, 6. [Google Scholar] [CrossRef]
- Pecoraro, F.; Giannini, M.; Beccari, G.; Covarelli, L.; Filippini, G.; Pisi, A.; Nipoti, P.; Prodi, A. Comparative studies about fungal colonization and deoxynivalenol translocation in barley plants inoculated at the base with Fusarium graminearum, Fusarium culmorum and Fusarium pseudograminearum. Agric. Food Sci. 2018, 27, 74–83. [Google Scholar] [CrossRef]
- Compant, S.; Duffy, B.; Nowak, J.; Clement, C.; Barka, E.A. Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 2005, 71, 4951–4959. [Google Scholar] [CrossRef] [PubMed]
- Fira, D.; Dimkic, I.; Beric, T.; Lozo, J.; Stankovic, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef]
- Sumpavapol, P.; Tongyonk, L.; Tanasupawat, S.; Chokesajjawatee, N.; Luxananil, P.; Visessanguan, W. Bacillus siamensis sp. nov., isolated from salted crab (poo-khem) in Thailand. Int. J. Syst. Evol. Microbiol. 2010, 60, 2364–2370. [Google Scholar] [CrossRef]
- Xu, B.H.; Lu, Y.Q.; Ye, Z.W.; Zheng, Q.W.; Wei, T.; Lin, J.F.; Guo, L.Q. Genomics-guided discovery and structure identification of cyclic lipopeptides from the Bacillus siamensis JFL15. PLoS ONE 2018, 13, e0202893. [Google Scholar] [CrossRef]
- Xie, Z.; Li, M.; Wang, D.; Wang, F.; Shen, H.; Sun, G.; Feng, C.; Wang, X.; Chen, D.; Sun, X. Biocontrol efficacy of Bacillus siamensis LZ88 against brown spot disease of tobacco caused by Alternaria alternata. Biol. Control. 2021, 154, 104508. [Google Scholar] [CrossRef]
- Gorai, P.S.; Ghosh, R.; Mandal, S.; Ghosh, S.; Chatterjee, S.; Gond, S.K.; Mandal, N.C. Bacillus siamensis CNE6- a multifaceted plant growth promoting endophyte of Cicer arietinum L. having broad spectrum antifungal activities and host colonizing potential. Microbiol. Res. 2021, 252, 126859. [Google Scholar] [CrossRef]
- Awan, S.A.; Ilyas, N.; Khan, I.; Raza, M.A.; Rehman, A.U.; Rizwan, M.; Rastogi, A.; Tariq, R.; Brestic, M. Bacillus siamensis Reduces Cadmium Accumulation and Improves Growth and Antioxidant Defense System in Two Wheat (Triticum aestivum L.) Varieties. Plants 2020, 9, 878. [Google Scholar] [CrossRef]
- Pastor-Bueis, R.; Mulas, R.; Gómez, X.; González-Andrés, F. Innovative liquid formulation of digestates for producing a biofertilizer based on Bacillus siamensis: Field testing on sweet pepper. J. Plant Nutr. Soil Sci. 2017, 180, 748–758. [Google Scholar] [CrossRef]
- Islam, A.; Kabir, M.S.; Khair, A. Characterization and evaluation of Bacillus siamensis isolate for its growth promoting potential in tomato. Agric./Pol’nohospodárstvo 2019, 65, 42–50. [Google Scholar] [CrossRef]
- Shin, D.J.; Yoo, S.J.; Hong, J.K.; Weon, H.Y.; Song, J.; Sang, M.K. Effect of Bacillus aryabhattai H26-2 and B. siamensis H30-3 on Growth Promotion and Alleviation of Heat and Drought Stresses in Chinese Cabbage. Plant Pathol. J. 2019, 35, 178–187. [Google Scholar] [CrossRef]
- Hussain, T.; Khan, A.A. Determining the antifungal activity and characterization of Bacillus siamensis AMU03 against Macrophomina phaseolina (Tassi) Goid. Indian Phytopathol. 2020, 73, 507–516. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, Z.; Zhang, X.; Bai, W.; Zhang, L.; Pei, H.; Zhang, Y. Control effects of Bacillus siamensis G-3 volatile compounds on raspberry post-harvest diseases caused by Botrytis cinerea and Rhizopus stolonifer. Biol. Control. 2020, 141, 104135. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, X.; Xu, H.; Zhang, F.; Zhang, X.; Yan, Y.; He, L.; Liu, J. Isolation of lipopeptide antibiotics from Bacillus siamensis: A potential biocontrol agent for Fusarium graminearum. Can. J. Microbiol. 2022, 68, 403–411. [Google Scholar] [CrossRef]
- Jiang, Z.; Li, R.; Tang, Y.; Cheng, Z.; Qian, M.; Li, W.; Shao, Y. Transcriptome analysis reveals the inducing effect of Bacillus siamensis on disease resistance in post-harvest mango fruit. Foods 2022, 11, 107. [Google Scholar] [CrossRef]
- Poveda, J.; Calvo, J.; Barquero, M.; González-Andrés, F. Activation of sweet pepper defense responses by novel and known biocontrol agents of the genus Bacillus against Botrytis cinerea and Verticillium dahliae. Eur. J. Plant Pathol. 2022, 164, 507–524. [Google Scholar] [CrossRef]
- Pan, H.; Tian, X.; Shao, M.; Xie, Y.; Huang, H.; Hu, J.; Ju, J. Genome mining and metabolic profiling illuminate the chemistry driving diverse biological activities of Bacillus siamensis SCSIO 05746. Appl. Microbiol. Biotechnol. 2019, 103, 4153–4165. [Google Scholar] [CrossRef]
- Xu, W.; Yang, Q.; Xie, X.; Goodwin, P.H.; Deng, X.; Zhang, J.; Sun, R.; Wang, Q.; Xia, M.; Wu, C.; et al. Genomic and Phenotypic Insights into the Potential of Bacillus subtilis YB-15 Isolated from Rhizosphere to Biocontrol against Crown Rot and Promote Growth of Wheat. Biology 2022, 11, 778. [Google Scholar] [CrossRef]
- Wu, L.; Wu, H.; Chen, L.; Yu, X.; Borriss, R.; Gao, X. Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice pathogens. Sci. Rep. 2015, 5, 12975. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.H.; Ye, Z.W.; Zheng, Q.W.; Wei, T.; Lin, J.F.; Guo, L.Q. Isolation and characterization of cyclic lipopeptides with broad-spectrum antimicrobial activity from Bacillus siamensis JFL15. 3 Biotech 2018, 8, 444. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Y.; Kong, W.L.; Liao, Y.C.; Zhu, L.H. Identification of Bacillus velezensis SBB and Its Antifungal Effects against Verticillium dahliae. J. Fungi 2022, 8, 1021. [Google Scholar] [CrossRef] [PubMed]
- Moura, R.D.; de Castro, L.A.M.; Culik, M.P.; Fernandes, A.A.R.; Fernandes, P.M.B.; Ventura, J.A. Culture medium for improved production of conidia for identification and systematic studies of Fusarium pathogens. J. Microbiol. Methods 2020, 173, 105915. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, W.; Goodwin, P.H.; Lin, Q.; Xia, M.; Xu, W.; Sun, R.; Liang, J.; Wu, C.; Li, H.; et al. Response of Fusarium pseudograminearum to Biocontrol Agent Bacillus velezensis YB-185 by Phenotypic and Transcriptome Analysis. J. Fungi 2022, 8, 763. [Google Scholar] [CrossRef]
- Zhang, D.; Yu, S.; Yang, Y.; Zhang, J.; Zhao, D.; Pan, Y.; Fan, S.; Yang, Z.; Zhu, J. Antifungal Effects of Volatiles Produced by Bacillus subtilis Against Alternaria solani in Potato. Front. Microbiol. 2020, 11, 1196. [Google Scholar] [CrossRef]
- Han, C.; Yang, P. Studies on the molecular mechanisms of seed germination. Proteomics 2015, 15, 1671–1679. [Google Scholar] [CrossRef]
- Smiley, R.W.; Gourlie, J.A.; Easley, S.A.; Patterson, L.M. Pathogenicity of Fungi Associated with the Wheat Crown Rot Complex in Oregon and Washington. Plant Dis. 2005, 89, 949–957. [Google Scholar] [CrossRef]
- Zhou, H.; He, X.; Wang, S.; Ma, Q.; Sun, B.; Ding, S.; Chen, L.; Zhang, M.; Li, H. Diversity of the Fusarium pathogens associated with crown rot in the Huanghuai wheat-growing region of China. Environ. Microbiol. 2019, 21, 2740–2754. [Google Scholar] [CrossRef]
- Teather, R.M.; Wood, P.J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 1982, 43, 777–780. [Google Scholar] [CrossRef]
- Dhouib, H.; Zouari, I.; Ben Abdallah, D.; Belbahri, L.; Taktak, W.; Triki, M.A.; Tounsi, S. Potential of a novel endophytic Bacillus velezensis in tomato growth promotion and protection against Verticillium wilt disease. Biol. Control. 2019, 139, 104092. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, L.; Goodwin, P.H.; Xia, M.; Zhang, J.; Wang, Q.; Liang, J.; Sun, R.; Wu, C.; Yang, L. Isolation, Identification, and Complete Genome Assembly of an Endophytic Bacillus velezensis YB-130, Potential Biocontrol Agent Against Fusarium graminearum. Front. Microbiol. 2020, 11, 598285. [Google Scholar] [CrossRef]
- Yamamoto, S.; Harayama, S. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl. Environ. Microbiol. 2015, 61, 1104–1109. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Richter, M.; Rossello-Mora, R.; Oliver Glockner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Ciufo, S.; Kannan, S.; Sharma, S.; Badretdin, A.; Clark, K.; Turner, S.; Brover, S.; Schoch, C.L.; Kimchi, A.; DiCuccio, M. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018, 68, 2386–2392. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Ramette, A.; Tiedje, J.M. The bacterial species definition in the genomic era. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006, 361, 1929–1940. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Eisenreich, A.; Schneider, K.; Heinemeyer, I.; Morgenstern, B.; Voss, B.; Hess, W.R.; Reva, O.; et al. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 2007, 25, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, S.; Yu, B. Poly-γ-glutamic acid: Recent achievements, diverse applications and future perspectives. Trends Food Sci. Technol. 2022, 119, 1–12. [Google Scholar] [CrossRef]
- Saxena, A.K.; Kumar, M.; Chakdar, H.; Anuroopa, N.; Bagyaraj, D.J. Bacillus species in soil as a natural resource for plant health and nutrition. J. Appl. Microbiol. 2020, 128, 1583–1594. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.-H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef]
- Bland, C.; Ramsey, T.L.; Sabree, F.; Lowe, M.; Brown, K.; Kyrpides, N.C.; Hugenholtz, P. CRISPR recognition tool (CRT): A tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinform. 2007, 8, 209. [Google Scholar] [CrossRef]
- Hsiao, W.W.; Ung, K.; Aeschliman, D.; Bryan, J.; Finlay, B.B.; Brinkman, F.S. Evidence of a large novel gene pool associated with prokaryotic genomic islands. PLoS Genet. 2005, 1, e62. [Google Scholar] [CrossRef]
- Akhter, S.; Aziz, R.K.; Edwards, R.A. PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012, 40, e126. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Ali, M.A.; Ren, H.; Ahmed, T.; Luo, J.; An, Q.; Qi, X.; Li, B. Antifungal Effects of Rhizospheric Bacillus Species Against Bayberry Twig Blight Pathogen Pestalotiopsis versicolor. Agronomy 2020, 10, 1811. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, L.; Si, H.; Guo, H.; Liu, J.; Jia, J.; Su, Q.; Wang, Y.; Zang, J.; Xing, J.; et al. Maize stalk rot caused by Fusarium graminearum alters soil microbial composition and is directly inhibited by Bacillus siamensis isolated from rhizosphere soil. Front. Microbiol. 2022, 13, 986401. [Google Scholar] [CrossRef]
- Altimira, F.; Godoy, S.; Arias-Aravena, M.; Araya, B.; Montes, C.; Castro, J.F.; Dardón, E.; Montenegro, E.; Pineda, W.; Viteri, I. Genomic and Experimental Analysis of the Biostimulant and Antagonistic Properties of Phytopathogens of Bacillus safensis and Bacillus siamensis. Microorganisms 2022, 10, 670. [Google Scholar] [CrossRef]
- Khan, M.M.; Kim, Y.K.; Cho, S.S.; Jin, Y.-Y.; Suh, J.-W.; Lee, D.Y.; Yoo, J.C. Response Surface Optimization of Culture Conditions for Cyclic Lipopeptide MS07 from Bacillus siamensis Reveals Diverse Insights Targeting Antimicrobial and Antibiofilm Activity. Processes 2020, 8, 744. [Google Scholar] [CrossRef]
- Thakham, N.; Thaweesak, S.; Teerakulkittipong, N.; Traiosot, N.; Kaikaew, A.; Lirio, G.A.; Jangiam, W. Structural Characterization of Functional Ingredient Levan Synthesized by Bacillus siamensis Isolated from Traditional Fermented Food in Thailand. Int. J. Food Sci. 2020, 2020, 7352484. [Google Scholar] [CrossRef]
- Guesmi, S.; Chouchane, H.; Neifar, M.; Hosni, F.; Cherif, A.; Sghaier, H. Radiation-inducible radioprotective exopolysaccharides of Bacillus siamensis CV5 from irradiated roots of Cistanche violacea to decrease free radical damage produced by ionizing radiation. Int. J. Radiat. Biol. 2019, 95, 1552–1563. [Google Scholar] [CrossRef]
- Hossain, M.T.; Khan, A.; Harun-Or-Rashid, M.; Chung, Y.R. A volatile producing endophytic Bacillus siamensis YC7012 promotes root development independent on auxin or ethylene/jasmonic acid pathway. Plant Soil 2019, 439, 309–324. [Google Scholar] [CrossRef]
- Rungsirivanich, P.; Thongwai, N. Antibacterial Activity and Tannin Tolerance of Bacillus spp. Isolated from Leaves of Miang (Camellia sinensis (L.) Kuntze var. assamica (J.W. Mast.) Kitam.). Int. J. Biosci. Biochem. Bioinform. 2020, 10, 26–33. [Google Scholar] [CrossRef]
- Yang, H.L.; Liu, Z.Y.; Jian, J.T.; Ye, J.D.; Sun, Y.Z. Host-associated Bacillus siamensis and Lactococcus petauri improved growth performance, innate immunity, antioxidant activity and ammonia tolerance in juvenile Japanese seabass (Lateolabrax japonicus). Aquac. Nutr. 2021, 27, 2739–2748. [Google Scholar] [CrossRef]
- Chopra, S.; Kumar, D. Characterization and biodegradation of ibuprofen by Bacillus siamensis strain DSI-1 isolated from wastewater. Rend. Lincei. Sci. Fis. E Nat. 2022, 33, 643–652. [Google Scholar] [CrossRef]
- Fitri, L.; Yasmin, Y.; Fauziah, F.; Septiani, D.A.; Suhartono, S. Characterization of BSL6 isolates isolated from honeybee hive and to determine its antibacterial activity. Biodivers. J. Biol. Divers. 2020, 21. [Google Scholar] [CrossRef]
- Li, S.; Xu, J.; Fu, L.; Xu, G.; Lin, X.; Qiao, J.; Xia, Y. Biocontrol of Wheat Crown Rot Using Bacillus halotolerans QTH8. Pathogens 2022, 11, 595. [Google Scholar] [CrossRef]
- Morales, M.; Munne-Bosch, S. Malondialdehyde: Facts and Artifacts. Plant Physiol. 2019, 180, 1246–1250. [Google Scholar] [CrossRef]
- Choudhary, D.K.; Johri, B.N. Interactions of Bacillus spp. and plants--with special reference to induced systemic resistance (ISR). Microbiol. Res. 2009, 164, 493–513. [Google Scholar] [CrossRef]
- Kildea, S.; Ransbotyn, V.; Khan, M.R.; Fagan, B.; Leonard, G.; Mullins, E.; Doohan, F.M. Bacillus megaterium shows potential for the biocontrol of septoria tritici blotch of wheat. Biol. Control. 2008, 47, 37–45. [Google Scholar] [CrossRef]
- Xie, D.; Cai, X.; Yang, C.; Xie, L.; Qin, G.; Zhang, M.; Huang, Y.; Gong, G.; Chang, X.; Chen, H. Studies on the control effect of Bacillus subtilis on wheat powdery mildew. Pest Manag. Sci. 2021, 77, 4375–4382. [Google Scholar] [CrossRef]
- Chen, L.; Heng, J.; Qin, S.; Bian, K. A comprehensive understanding of the biocontrol potential of Bacillus velezensis LM2303 against Fusarium head blight. PLoS ONE 2018, 13, e0198560. [Google Scholar] [CrossRef]
- Jamal, Q.; Monkhung, S.; Munir, S.; Cho, J.; Moon, J.; Khattak, B.; Malik, M.; Younis, F.; Kim, K. Identification of cyclo (L-Pro-D-Tyr) from Bacillus amyloliquefaciens Y1 exhibiting antifungal activity against Fusarium graminearum to control crown rot in wheat. Appl. Ecol. Environ. Res. 2019, 17, 6299–6314. [Google Scholar] [CrossRef]
- Wang, S.; Sun, L.; Zhang, W.; Chi, F.; Hao, X.; Bian, J.; Li, Y. Bacillus velezensis BM21, a potential and efficient biocontrol agent in control of corn stalk rot caused by Fusarium graminearum. Egypt. J. Biol. Pest Control. 2020, 30, 9. [Google Scholar] [CrossRef]
- You, W.; Ge, C.; Jiang, Z.; Chen, M.; Li, W.; Shao, Y. Screening of a broad-spectrum antagonist-Bacillus siamensis, and its possible mechanisms to control post-harvest disease in tropical fruits. Biol. Control. 2021, 157, 104584. [Google Scholar] [CrossRef]
- Sharma, A.; Kaushik, N.; Sharma, A.; Bajaj, A.; Rasane, M.; Shouche, Y.S.; Marzouk, T.; Djebali, N. Screening of Tomato Seed Bacterial Endophytes for Antifungal Activity Reveals Lipopeptide Producing Bacillus siamensis Strain NKIT9 as a Potential Bio-Control Agent. Front. Microbiol. 2021, 12, 609482. [Google Scholar] [CrossRef]
- Santoso, I.; Fadhilah, Q.G.; Safitri, S.D.; Handayani, S.; Maryanto, A.E.; Yasman, Y. Inhibition of the phytopathogenic fungi Curvularia lunata BM and Ganoderma sp. TB4 by antifungal compounds produced by Bacillus siamensis LDR grown on hanjeli (Coix lacryma-jobi L.) starch. Pharmacia 2022, 69, 203–210. [Google Scholar] [CrossRef]
- Wang, D.; Li, Y.; Yuan, Y.; Chu, D.; Cao, J.; Sun, G.; Ai, Y.; Cui, Z.; Zhang, Y.; Wang, F.; et al. Identification of non-volatile and volatile organic compounds produced by Bacillus siamensis LZ88 and their antifungal activity against Alternaria alternata. Biol. Control. 2022, 169, 104901. [Google Scholar] [CrossRef]
- Park, G.; Nam, J.; Kim, J.; Song, J.; Kim, P.I.; Min, H.J.; Lee, C.W. Structure and Mechanism of Surfactin Peptide from Bacillus velezensis Antagonistic to Fungi Plant Pathogens. Bull. Korean Chem. Soc. 2019, 40, 704–709. [Google Scholar] [CrossRef]
- Kim, P.I.; Bai, H.; Bai, D.; Chae, H.; Chung, S.; Kim, Y.; Park, R.; Chi, Y.T. Purification and characterization of a lipopeptide produced by Bacillus thuringiensis CMB26. J. Appl. Microbiol. 2004, 97, 942–949. [Google Scholar] [CrossRef]
- Wang, J.; Qiu, J.; Yang, X.; Yang, J.; Zhao, S.; Zhou, Q.; Chen, L. Identification of Lipopeptide Iturin A Produced by Bacillus amyloliquefaciens NCPSJ7 and Its Antifungal Activities against Fusarium oxysporum f. sp. niveum. Foods 2022, 11, 2996. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, J.; Wu, F.; Yin, C.; Kim, K.H.; Zhang, Y. Termite Nest Associated Bacillus siamensis YC-9 Mediated Biocontrol of Fusarium oxysporum f. sp. cucumerinum. Front. Microbiol. 2022, 13, 893393. [Google Scholar] [CrossRef]
- Feng, X.; Xu, R.; Zhao, N.; Wang, D.; Cun, M.; Yang, B. Isolation, Identification, and Characterization of Endophytic Bacillus from Walnut (Juglans sigillata) Root and Its Biocontrol Effects on Walnut Anthracnose. Agriculture 2022, 12, 2102. [Google Scholar] [CrossRef]
- Manikandan, A.; Jaivel, N.; Johnson, I.; Krishnamoorthy, R.; Senthilkumar, M.; Raghu, R.; Gopal, N.O.; Mukherjee, P.K.; Anandham, R. Suppression of Macrophomina root rot, Fusarium wilt and growth promotion of some pulses by antagonistic rhizobacteria. Physiol. Mol. Plant Pathol. 2022, 121, 101876. [Google Scholar] [CrossRef]
- Fonseca, R.; Silva, A.; De França, F.; Cardoso, V.; Sérvulo, E. Optimizing carbon/nitrogen ratio for biosurfactant production by a Bacillus subtilis strain. Appl. Biochem. Biotecnol. 2007, 137, 471–486. [Google Scholar] [CrossRef]
- Zhao, X.; Han, Y.; Tan, X.Q.; Wang, J.; Zhou, Z.J. Optimization of antifungal lipopeptide production from Bacillus sp. BH072 by response surface methodology. J. Microbiol. 2014, 52, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.-S.; Mo, M.-H.; Gu, Y.-Q.; Zhou, J.-P.; Zhang, K.-Q. Possible contributions of volatile-producing bacteria to soil fungistasis. Soil Biol. Biochem. 2007, 39, 2371–2379. [Google Scholar] [CrossRef]
- Jeong, H.; Jeong, D.E.; Kim, S.H.; Song, G.C.; Park, S.Y.; Ryu, C.M.; Park, S.H.; Choi, S.K. Draft genome sequence of the plant growth-promoting bacterium Bacillus siamensis KCTC 13613T. J. Bacteriol. 2012, 194, 4148–4149. [Google Scholar] [CrossRef]
- Poudel, P.; Khanal, S.; Panta, O.P.; Dhakal, S.; Shah, R.; Maharjan, R.; Gautam, A.; Khadka, S.; Sapkota, S. Screening and Optimization of Thermo-Tolerant Bacillus Sp. For Amylase Production and Antifungal Activity. J. Inst. Sci. Technol. 2019, 24, 47–56. [Google Scholar] [CrossRef]
- Zhao, M.; Liu, D.; Liang, Z.; Huang, K.; Wu, X. Antagonistic activity of Bacillus subtilis CW14 and its β-glucanase against Aspergillus ochraceus. Food Control. 2021, 131, 108475. [Google Scholar] [CrossRef]
- Tomita, M.; Kikuchi, A.; Kobayashi, M.; Yamaguchi, M.; Ifuku, S.; Yamashoji, S.; Ando, A.; Saito, A. Characterization of antifungal activity of the GH-46 subclass III chitosanase from Bacillus circulans MH-K1. Antonie Van Leeuwenhoek 2013, 104, 737–748. [Google Scholar] [CrossRef]
- Aktuganov, G.E.; Galimzyanova, N.F.; Melent’ev, A.I.; Kuz’mina, L.Y. Extracellular hydrolases of strain Bacillus sp. 739 and their involvement in the lysis of micromycete cell walls. Microbiology 2007, 76, 413–420. [Google Scholar] [CrossRef]
- Chang, W.-T.; Chen, M.-L.; Wang, S.-L. An antifungal chitinase produced by Bacillus subtilis using chitin waste as a carbon source. World J. Microbiol. Biotechnol. 2009, 26, 945–950. [Google Scholar] [CrossRef]
- Xu, W.; Yang, Q.; Yang, F.; Xie, X.; Goodwin, P.H.; Deng, X.; Tian, B.; Yang, L. Evaluation and Genome Analysis of Bacillus subtilis YB-04 as a Potential Biocontrol Agent Against Fusarium Wilt and Growth Promotion Agent of Cucumber. Front. Microbiol. 2022, 13, 885430. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef]
- Wybouw, B.; De Rybel, B. Cytokinin—A Developing Story. Trends Plant Sci. 2019, 24, 177–185. [Google Scholar] [CrossRef]
- Fan, B.; Wang, C.; Song, X.; Ding, X.; Wu, L.; Wu, H.; Gao, X.; Borriss, R. Bacillus velezensis FZB42 in 2018: The gram-positive model strain for plant growth promotion and biocontrol. Front. Microbiol. 2018, 9, 2491. [Google Scholar] [CrossRef]
- Suthar, K.; Patel, R.; Singh, D. Polyamine Modulation by antagonistic bacteria Bacillus subtilis in chickpea during Fusarium oxysporum f. sp. ciceri interaction. Legume Res. Int. J. 2021, 44, 109–114. [Google Scholar] [CrossRef]
- Ryu, C.M.; Farag, M.A.; Paré, P.W.; Kloepper, J.W. Invisible Signals from the Underground: Bacterial Volatiles Elicit Plant GrowthPromotion and Induce Systemic Resistance. Plant Pathol. J. 2005, 21, 7–12. [Google Scholar] [CrossRef]
- Wang, L.; Ning, W.; Mi, D.; Luo, Y.; Guo, J. Poly-γ-glutamic acid productivity of Bacillus subtilis BsE1 has positive function in motility and biocontrol against Fusarium graminearum. J. Microbiol. 2017, 55, 554–560. [Google Scholar] [CrossRef]
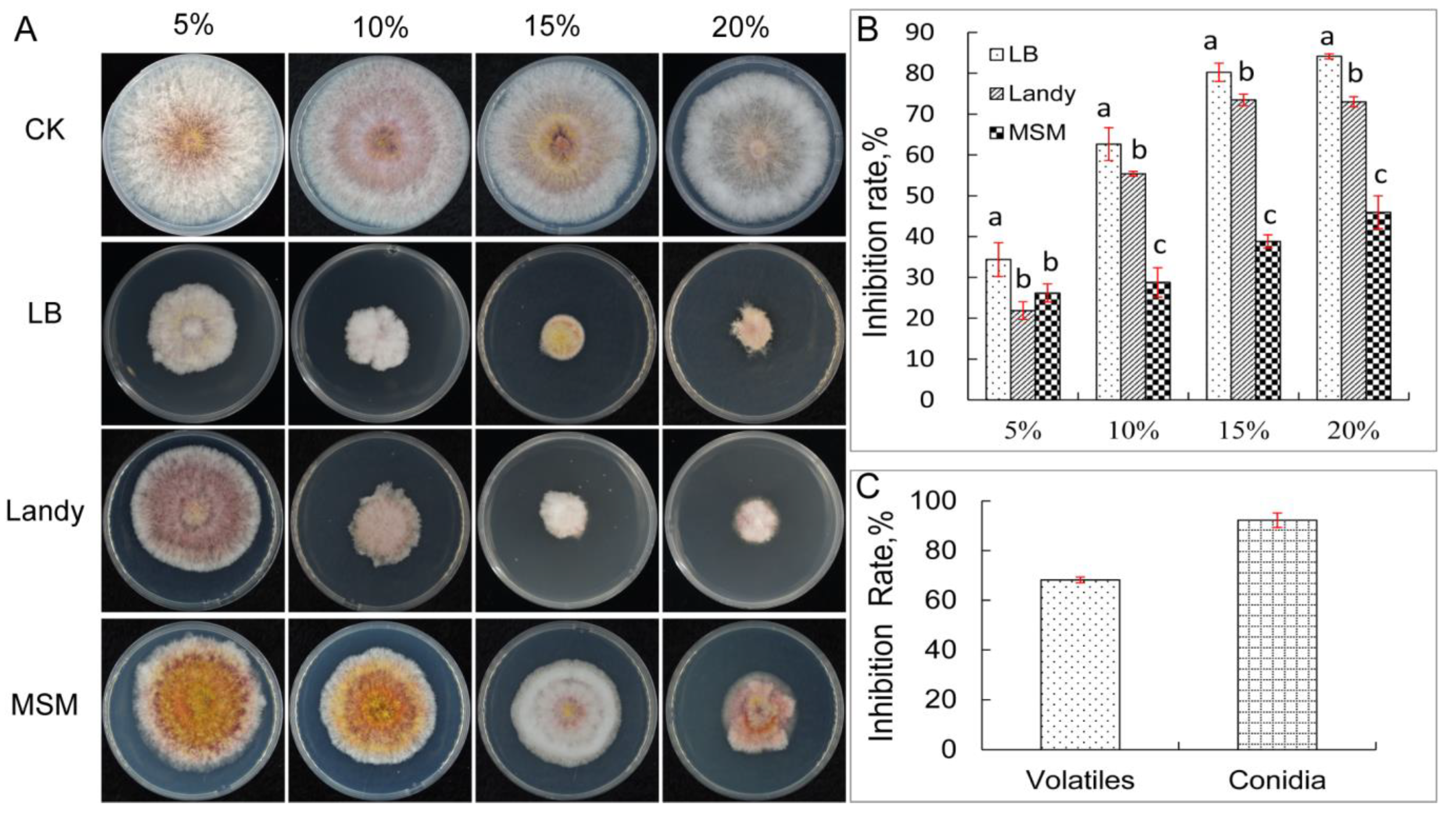
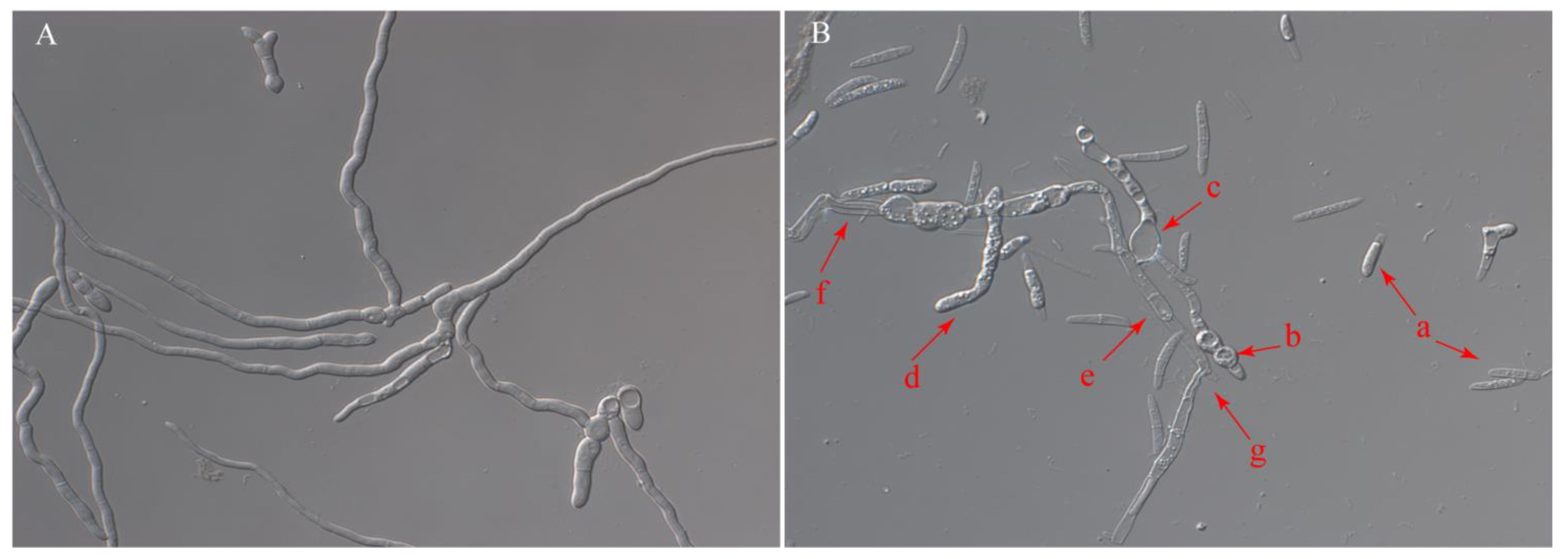
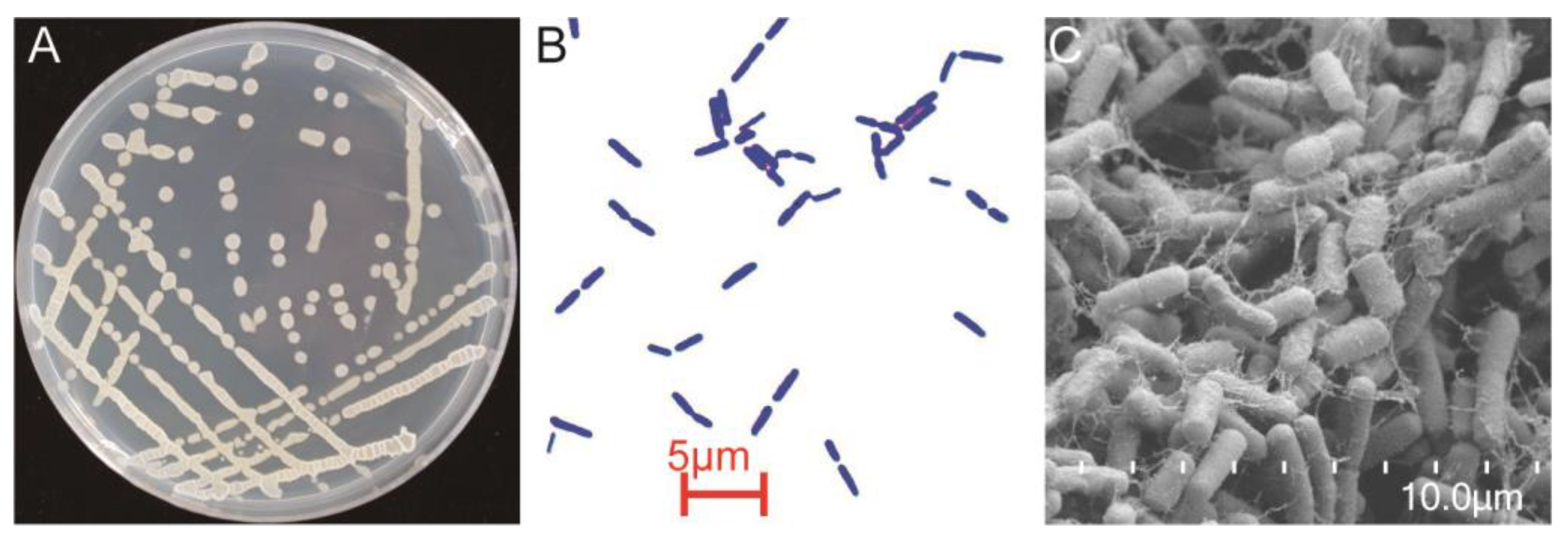

| Strain | Width | Strain | Width |
|---|---|---|---|
| Mq202003-1 | 2.33 ± 0.29 lm | Mq202003-30 | 4.60 ± 0.17 abcde |
| Mq202003-2 | 1.17 ± 0.29 no | Mq202003-31 | 3.40 ± 0.17 hi |
| Mq202003-3 | 4.17 ± 0.29 cdefg | Mq202003-32 | 4.07 ± 0.12 cdefg |
| Mq202003-4 | 2.83 ± 0.29 jk | Mq202003-33 | 3.87 ± 0.12 fgh |
| Mq202003-5 | 0.67 ± 0.29 opq | Mq202003-34 | 2.07 ± 0.12 m |
| Mq202003-8 | 0.40 ± 0.17 pq | Mq202003-36 | 4.40 ± 0.17 bcdef |
| Mq202003-9 | 4.83 ± 0.29 ab | Mq202003-37 | 4.07 ± 0.12 defg |
| Mq202003-10 | 4.67 ± 0.29 abc | Mq202003-38 | 1.23 ± 0.25 no |
| Mq202003-11 | 4.00 ± 1.00 fg | Mq202003-39 | 2.40 ± 0.17 lm |
| Mq202003-12 | 4.17 ± 0.29 cdefg | Mq202003-40 | 3.67 ± 0.29 ghi |
| Mq202003-13 | 0.83 ± 0.29 nop | Mq202003-41 | 3.93 ± 0.12 fg |
| Mq202003-14 | 1.00 ± 0.00 no | Mq202003-42 | 3.13 ± 0.12 ijk |
| Mq202003-15 | 2.50 ± 0.50 lm | Mq202003-43 | 2.33 ± 0.29 lm |
| Mq202003-16 | 5.07 ± 0.12 a | Mq202003-44 | 4.17 ± 0.29 cdefg |
| Mq202003-17 | 4.23 ± 0.25 cdefg | Mq202003-45 | 4.67 ± 0.29 ab |
| Mq202003-18 | 4.17 ± 0.29 cdefg | Mq202003-47 | 4.40 ± 0.17 bcdef |
| Mq202003-19 | 3.17 ± 0.29 ijk | Mq202003-48 | 1.33 ± 0.29 n |
| Mq202003-20 | 2.50 ± 0.50 lm | Mq202003-49 | 3.33 ± 0.29 ij |
| Mq202003-21 | 4.17 ± 0.29 cdefg | Mq202003-50 | 2.17 ± 0.29 lm |
| Mq202003-22 | 0.83 ± 0.29 nop | Mq202003-51 | 2.67 ± 0.29 kl |
| Mq202003-23 | 4.67 ± 0.29 abc | Mq202003-53 | 0.83 ± 0.29 nop |
| Mq202003-24 | 3.17 ± 0.29 ijk | Mq202003-54 | 0.67 ± 0.29 opq |
| Mq202003-25 | 2.50 ± 0.50 lm | Mq202003-56 | 1.33 ± 0.29 n |
| Mq202003-27 | 0.23 ± 0.25 q | Mq202003-57 | 1.40 ± 0.17 n |
| Mq202003-28 | 0.67 ± 0.29 opq | Mq202003-58 | 4.83 ± 0.29 ab |
| Treatment | Root Length (cm) | Plant Height (cm) | Root Fresh Weight (mg) | Total Fresh Weight (mg) | FCR Incidence (FCRI %) | Disease Severity Index (DSI %) | Relative Control Effect (RCE %) |
|---|---|---|---|---|---|---|---|
| CK | 8.89 ± 0.15 b | 25.29 ± 0.58 b | 26.36 ± 1.13 c | 407.40 ± 6.34 c | |||
| YB-1631 | 12.19 ± 0.52 a | 28.90 ± 0.73 a | 31.88 ± 0.58 a | 449.60 ± 10.48 a | |||
| Fp | 6.41 ± 0.19 c | 18.15 ± 1.63 c | 13.80 ± 2.56 d | 225.40 ± 15.87 d | 90.75 ± 3.67 a | 48.69 ± 1.33 a | |
| YB-1631 + Fp | 12.07 ± 0.18 a | 27.98 ± 0.40 b | 29.80 ± 0.84 b | 426.20 ± 5.84 b | 14.50 ± 1.60 b | 8.17 ± 0.93 b | 83.23 ± 1.75 |
| Treatment | PPO (U/g) | CAT (U/g) | PAL (U/g) | SOD (U/g) | POD (U/g) | LOX (U/g) | MDA (nmol/g) |
|---|---|---|---|---|---|---|---|
| CK | 22.38 ± 1.04 d | 143.56 ± 3.11 d | 24.86 ± 2.38 c | 9.61 ± 1.54 c | 15,175.45 ± 249.40 d | 752.40 ± 13.14 d | 21.36 ± 0.79 b |
| YB-1631 | 25.5 ± 0.67 c | 156.03 ± 5.19 c | 34.17 ± 2.38 b | 24.64 ± 1.70 b | 16,674.29 ± 384.66 c | 3261.24 ± 94.44 c | 16.66 ± 2.15 c |
| Fp | 38.06 ± 0.46 b | 238.68 ± 3.48 b | 37.20 ± 1.37 b | 31.66 ± 1.64 a | 34,486.16 ± 544.57 b | 1008.90 ± 22.57 b | 37.88 ± 1.52 a |
| YB-1631 + Fp | 45.89 ± 1.28 a | 567.73 ± 10.22 a | 51.76 ± 0.84 a | 34.53 ± 0.70 a | 42,529.88 ± 651.12 a | 4022.64 ± 77.42 a | 20.93 ± 2.44 c |
| Clusters | Types | From | To | Most Similar Known Clusters | Similarity |
|---|---|---|---|---|---|
| Cluster 1 | NRPS | 337,277 | 401,867 | Surfactin biosynthetic gene cluster from B. velezensis FZB42 | 82% |
| Cluster 2 | NRPS | 1,865,326 | 1,973,480 | Fengycin biosynthetic gene cluster from B. velezensis FZB42 | 93% |
| Cluster 3 | NRPS | 3,131,564 | 3,183,345 | Bacillibactin biosynthetic gene cluster from B. subtilis subsp. subtilis str. 168 | 100% |
| Cluster 4 | TransAT-PKS | 1,709,598 | 1,809,529 | Bacillaene biosynthetic gene cluster from B. velezensis FZB42 | 92% |
| Cluster 5 | TransAT-PKS | 2,383,511 | 2,477,284 | Difficidin biosynthetic gene cluster from B. velezensis FZB42 | 100% |
| Cluster 6 | PKS-like | 1,011,800 | 1,053,044 | Butirosin A biosynthetic gene cluster from B. circulans | 7% |
| Cluster 7 | terpene | 1,137,529 | 1,154,562 | None | |
| Cluster 8 | terpene | 2,002,946 | 2,023,869 | None | |
| Cluster 9 | NRP + Polyketide | 1,882,031 | 1,926,708 | Bacillomycin D biosynthetic gene cluster from B. velezensis FZB42 | 84% |
| Cluster 10 | NRP + Polyketide | 199,769 | 267,925 | Locillomycin biosynthetic gene cluster from B. subtilis | 21% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Q.; Liu, Q.; Goodwin, P.H.; Deng, X.; Xu, W.; Xia, M.; Zhang, J.; Sun, R.; Wu, C.; Wang, Q.; et al. Isolation and Genome-Based Characterization of Biocontrol Potential of Bacillus siamensis YB-1631 against Wheat Crown Rot Caused by Fusarium pseudograminearum. J. Fungi 2023, 9, 547. https://doi.org/10.3390/jof9050547
Dong Q, Liu Q, Goodwin PH, Deng X, Xu W, Xia M, Zhang J, Sun R, Wu C, Wang Q, et al. Isolation and Genome-Based Characterization of Biocontrol Potential of Bacillus siamensis YB-1631 against Wheat Crown Rot Caused by Fusarium pseudograminearum. Journal of Fungi. 2023; 9(5):547. https://doi.org/10.3390/jof9050547
Chicago/Turabian StyleDong, Qianqian, Qingxiang Liu, Paul H. Goodwin, Xiaoxu Deng, Wen Xu, Mingcong Xia, Jie Zhang, Runhong Sun, Chao Wu, Qi Wang, and et al. 2023. "Isolation and Genome-Based Characterization of Biocontrol Potential of Bacillus siamensis YB-1631 against Wheat Crown Rot Caused by Fusarium pseudograminearum" Journal of Fungi 9, no. 5: 547. https://doi.org/10.3390/jof9050547
APA StyleDong, Q., Liu, Q., Goodwin, P. H., Deng, X., Xu, W., Xia, M., Zhang, J., Sun, R., Wu, C., Wang, Q., Wu, K., & Yang, L. (2023). Isolation and Genome-Based Characterization of Biocontrol Potential of Bacillus siamensis YB-1631 against Wheat Crown Rot Caused by Fusarium pseudograminearum. Journal of Fungi, 9(5), 547. https://doi.org/10.3390/jof9050547






