Abstract
In the present study, we report two new asexual fungal species (i.e., Discosia rhododendricola, Neopestalotiopsis rhododendricola (Sporocadaceae) and a new host for a previously described species (i.e., Diaporthe nobilis; Diaporthaceae). All species were isolated from Rhododendron spp. in Kunming, Yunnan Province, China. All taxa are described based on morphology, and phylogenetic relationships were inferred using a multigenic approach (LSU, ITS, RPB2, TEF1 and TUB2). The phylogenetic analyses indicated that D. rhododendronicola sp. nov. is phylogenetically related to D. muscicola, and N. rhododendricola sp. nov is related to N. sonnaratae. Diaporthe nobilis is reported herein as a new host record from Rhododendron sp. for China, and its phylogeny is depicted based on ITS, TEF1 and TUB2 sequence data.
1. Introduction
Rhododendron, a genus of shrub and small to large trees belonging to Ericaceae, is an indicator of health for forest areas [1], commonly found in low-quality acidic soil and sterile conditions. This plant is mainly distributed in India and southeastern Asia, extending from the northwest Himalayas (Arunachal Pradesh) to Bhutan, eastern Tibet, Nepal, north Myanmar, Sikkim, and west central China [2]. Rhododendron flowers are used as food, to produce fermented wine, and to make herbal tea due to their distinctive flavor and color [3,4]. Fungi colonizing Rhododendron include Alternaria alternata, Aspergillus brasiliensis, Chrysomyxa dietelii, C. succinea [5], Diaporthe nobilis [6], Epicoccum nigrum, Mucor hiemalis, Pestalotiopsis sydowiana and Trichoderma koningii [7]. However, given the economic importance of this plant, it is imperative to assess the fungal species associated with it.
Discosia was introduced in Discosiaceae by Maharachchikumbura et al. [8] to accommodate the type genus Discosia and the type species D. artocreas. Senanayake et al. [9,10] introduced Adisciso, Discosia, Discostroma, Immersidiscosia, Sarcostroma and Seimatosporium in the Discosiaceae family. Jaklitsch et al. [11] considered Discosiaceae a synonym of Sporocadaceae based on DNA sequence analyses with strong phylogenetic support. Wijayawardene et al. [12] accepted Discosia species belonging to the family Sporocadaceae. Libert introduced Discosia in 1837, with Discosia strobilina being the lectotype [9,12]. Liu et al. [13] reviewed the generic description of Discosia, an updated morphology, and the phylogenetic relationships based on ITS sequence data [13]. There are 118 epithets of Discosia in Index Fungorum 2022 [14]. Discosia has been identified as an asexual fungus and is characterized by uni- to multilocular conidiomata with muti-layered walls. Conidiogenous cells are monoblastic and phialidic to annellidic. Conidial types are bipolar, polar and subpolar appendages, and usually hyaline to pale brown [9].
The genus Neopestalotiopsis, introduced by Maharachchikumbura et al., (2014) [15], belongs to the family Sporocadaceae (Amphisphaeriales, Sordariomycetes) [8,15,16], with N. protearum being the type species. Neopestalotiopsis species have been reported on saprobes, trees or plant pathogens causing postharvest diseases (fruit rots and leaf blights) [17,18]. The sexual morph of Neopestalotiopsis species remains unknown [15,18,19,20]. Neopestalotiopsis species have a worldwide distribution. This genus has also been reported in caves in China [15,17,18,19,20,21,22]. Studies related to the taxonomy of Neopestalotiopsis included DNA sequence analyses and phylogeny of the ITS, TEF1 and TUB2 [22].
The genus Diaporthe introduced by Nitschke [23] belongs to the families Diaporthaceae, Diaporthales, and Sordariomycetes [8,9]. Diaporthe species have a worldwide distribution [6,24,25,26,27,28]. This genus has been associated with several grapevine diseases in Europe [29] and was detected in Uruguayan deciduous fruit tree (Malus domestica ‘Gala’) wood disease [30]. Studies related to the taxonomy of Diaporthe included DNA sequence analyses and phylogeny of the ITS, TEF1, TUB2 and CAL loci [6,31]. Dissanayake et al. [32] provided phylogenetic relationships of 171 Diaporthe species currently known from culture or direct sequencing, and are linked to their holotype, epitype, isotype or neotype and that can now be recognized with DNA sequence data, essential to species identification [33].
In this study, we introduce the new species D. rhododendricola, N. rhododendricola and a new host record of Diaporthe nobilis, collected from dead leaves of Rhododendron species in China. We further provide descriptions, illustrations, and DNA sequence-based phylogeny to verify identification and placement.
2. Materials and Methods
2.1. Sample Collection, Morphological Observation, and Fungal Isolation
Isolation was performed as described by Senanayake et al. [34]. Dead leaves of Rhododendron spp. were collected from Kunming, Yunnan Province, China and brought to the laboratory in labelled paper envelopes. A light microscope (Nikon ECLIPSE 80i compound microscope, Melville, NY, USA) was used to observe the specimens. Spore mass fruiting bodies were isolated on potato dextrose agar (PDA) plates and incubated at 25 °C.
The isolates were transferred to new PDA plates, incubated at 25 °C, and photographed using a Canon EOS 600D digital camera fitted to the microscope. The Tarosoft (R) Image Frame Work program measured the morphological characteristics. The figures were processed using Adobe Photoshop CS6 Extended version 10.0 (Adobe Systems, San Jose, CA, USA).
The specimens were deposited at the Herbarium of Mae Fah Luang University (Herb. MFLU) and Herbarium of Kungming Institute of Botany (KUN), Chinese Academy of Science, Kunming, China. Living cultures were deposited at the Culture Collection of Mae Fah Luang University (MFLUCC), Chiang Rai, Thailand and the Culture Collection of Kungming Institute of Botany (KUN), Chinese Academy of Science, Kunming, China. Faces of Fungi and Index Fungorum data are also provided [14,35]. New species were established based on guidelines provided by Jeewon and Hyde [36].
2.2. DNA Extraction, PCR Amplification, and Sequencing
Fungal cultures were grown on PDA at 25 °C for 2–4 weeks. The Biospin Fungus Genomic DNA Extraction Kit-BioFlux (BioFlux®, Hangzhou, China) was used to extract DNA from the mycelium. PCR amplification was performed using primer pairs, ITS4/ITS5 for the internal transcribed spacer region of ribosomal DNA [37], LR0R/LR5 for large subunit nuclear ribosomal DNA [38], EF-728F/EF-986R for translation elongation factor 1-alpha gene [39], fRPB2-5f/fRPB2-7cR for the second largest subunit of RNA polymerase [40] and Bt2a/Bt2b for beta-tubulin [41]. The PCR conditions were based on the methodology as described by Chaiwan et al. [42].
2.3. Phylogenetic Analyses
The sequence alignment and phylogenetic analyses were performed as outlined by Dissanayake et al. [43] and Chaiwan et al. [42,44,45]. Phylogenetic analyses were performed using a combined Discosia dataset of ITS, LSU, RPB2, TEF1 and TUB2 sequence data and a combined Neopestalotiopsis and Diaporthe dataset of ITS, TEF1 and TUB2 sequence data. Taxa used in the analyses were obtained through recent publications [16,28,46]. The phylogenetic analyses were carried out using maximum parsimony (MP), maximum likelihood (ML) and Bayesian posterior probabilities (BYPP). PAUP v4.0b10 was used to conduct the parsimony analysis to obtain the phylogenetic trees [47]. Trees were inferred using the heuristic search option with 1000 random sequence additions. Maxtrees were set to 1000, branches of zero length were collapsed and all multiple parsimonious trees were saved. Descriptive tree statistics for parsimony—tree length (TL), consistency index (CI), retention index (RI), relative consistency index (RC) and homoplasy index (HI)—were calculated for trees generated following the Kishino-Hasegawa test (KHT) criteria [48], which was performed in order to determine whether trees were significantly different. Maximum-parsimony bootstrap values equal or greater than 60% are given as the second set of numbers above the nodes.
Maximum likelihood analysis was performed by using RAxML-HPC2, New Orleans, LA on XSEDE (8.2.8) [45,48,49,50]. The search strategy was set to rapid bootstrapping and the analysis was carried out using the GTRGAMMAI model of nucleotide substitution. Maximum likelihood bootstrap values equal to or greater than 60% are given as the first set of numbers above the nodes.
Bayesian inference (BI) analysis was conducted with MrBayes v. 3.1.2 to evaluate the posterior probabilities (BYPP) using Markov chain Monte Carlo sampling [51]. Two parallel runs were conducted using the default settings, but with the following adjustments: six simultaneous Markov chains were run for 2,000,000 generations and trees were sampled every 200 generations. The distribution of log-likelihood scores were examined to determine stationary phase for each search and to decide if extra runs were required to achieve convergence, using the program Tracer 1.4 [52]. The first 10% of generated trees were discarded and the remaining 90% of trees were used to calculate posterior probabilities (PP) of the majority rule consensus tree. The phylogenetic trees were viewed in FigTree v. 1.4 [53] and edited using Microsoft Office Power Point 2007 and Adobe Photoshop CS6 Extended [42].
2.4. Genealogical Concordance Phylogenetic Species Recognition (GCPSR) Analysis
The related species were analyzed using the Genealogical Concordance Phylogenetic Species Recognition model. The pairwise homoplasy index (PHI) [54] is a model test based on the fact that multiple gene phylogenies will be concordant between species and discordant due to recombination and mutations within a species. The data were analyzed by the pairwise homoplasy index (PHI) test [54]. The test was performed in SplitsTree4 [55,56] as described by Quaedvlieg [57] to determine the recombination level within phylogenetically closely related species using a five-locus concatenated dataset to determine the recombination level within phylogenetically closely related species. If the PHI is below the 0.05 threshold (Φw < 0.05), it indicates that there is significant recombination in the dataset. This means that related species in a group and recombination level are not different. If the PHI is above the 0.05 threshold (Φw > 0.05), it indicates that it is not significant, which means the related species in a group level are different. The new species and its closely related species were analyzed using this model. The relationships between closely related species were visualized by constructing a split graph, using both the LogDet transformation and splits decomposition options.
2.5. Discosia, Habitat and Known Distribution Checklist Associated with Rhododendron sp.
An updated checklist of Discosia based on the SMML database (https://nt.ars-grin.gov/fungaldatabases/) (accessed on 10 June 2022) is provided [58]. Those species for which molecular data are available are indicated. The distribution information regarding the type or original descriptions available and the locality from which Discosia have been recorded on Rhododendron spp. is provided, including all the specimens encountered during this study.
3. Results
3.1. Phylogenetic Analyses
The combined sequence alignments of Discosia comprised 54 taxa (Table 1), with Immersidiscosia eucalypti MFLU16-1372 and NBRC 104195 as the outgroup taxa. The dataset comprised 4364 characters including alignment gaps (LSU, ITS, RPB2, TEF1 and TUB2 sequence data). The MP analysis for the combined dataset had 430 parsimony-informative, 3522 constant, and 412 parsimony-uninformative characters, and yielded a single most parsimonious tree (TL = 1353, CI = 0.777, RI = 0.764, RC = 0.594; HI = 0.223). The RAxML analysis of the combined dataset yielded a best scoring tree with a final ML optimization likelihood value of −22,013.917605. The matrix had 840 distinct alignment patterns, with 66% undetermined characters or gaps. Bayesian posterior probabilities from Bayesian inference analysis were assessed with a final average standard deviation of split frequencies = 0.009983. The phylogenetic tree in this study showed that our strain (Discosia rhododendricola KUN-HKAS 123205 and MFLU20-0486) is related to D. muscicola with high support value in the phylogenbetic tree (Figure 1). Sequence alignments are deposited in TreeBASE.

Table 1.
Culture collection numbers and GenBank accession numbers for Discosia used in this study. The type species are indicated in bold. The newly generated sequences are indicated in red. Instances where the GenBank Accession No. did not show the molecular data are marked with a dash.
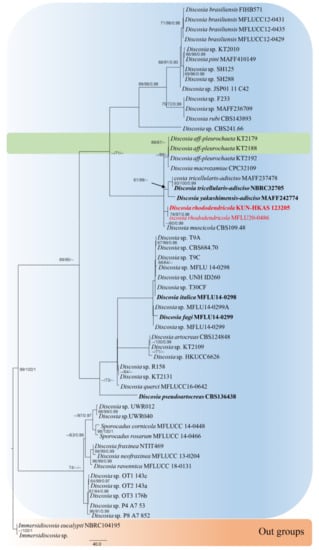
Figure 1.
Phylogram generated from maximum parsimony analysis of LSU, ITS, RPB2, TEF1 and TUB2 gene regions. Bootstrap support values for MP and ML equal to or greater than 60% and Bayesian posterior probabilities (PP) equal to or greater than 0.90 are defined as MP/ML/PP above or below the nodes. Taxonomic novelty is indicated in red. The tree is rooted with Immersidiscosia eucalypti (MFLU 16-1372) and (NBRC 104195).
The combined sequence alignments of Neopestalotiopsis comprised 89 taxa (Table 2), with Monochaetia monochaeta CBS115004 and M. ilexae CBS101009 as the outgroup taxa. The dataset comprised 2634 characters including alignment gaps (ITS, TUB2 and TEF1 sequence data). The MP analysis for the combined dataset had 631 parsimony-informative, 1524 constant, and 479 parsimony-uninformative characters, and yielded a single most parsimonious tree (TL = 2304, CI = 0.679, RI = 0.813, RC = 0.552; HI = 0.321). The RAxML analysis of the combined dataset yielded a best scoring tree with a final ML optimization likelihood value of −24,500.881631. The matrix had 1268 distinct alignment patterns, with 35.77% undetermined characters or gaps. Bayesian posterior probabilities from Bayesian inference analysis were assessed with a standard deviation of split frequencies = 0.024223. The phylogenetic tree in this study showed that N. rhododendricola KUN-HKAS 123204 and MFLU20-0046 belonged to a separate clade, phylogenetically related to N. sonneratae, N. coffeae-arabicae and N. thailandica with 88% MP support (Figure 2). Sequence alignments are deposited in TreeBASE.

Table 2.
Culture collection numbers and GenBank accession numbers for Neopestalotiopsis used in this study. The type species are indicated in bold. The newly generated sequences are indicated in red. Instances where the GenBank Accession No. did not show the molecular data are marked with the dash.
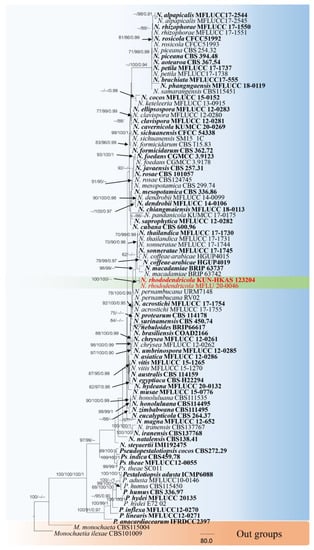
Figure 2.
RAxML tree based on a combined dataset of ITS, TUB2 and TEF1 gene regions. Bootstrap support values for ML and MP equal to or greater than 60% and Bayesian posterior probabilities (PP) equal to or greater than 0.90 are defined as ML/MP/PP above or below the nodes. Our new taxon is indicated in red. The tree was rooted with Monochaetia monochaeta strains (CBS115004) and Monochaetia ilexae strains (CBS101009).
The combined sequence alignments of Diaporthe comprised 56 taxa (Table 3), with Diaporthella corylina CBS 121124 used as the outgroup taxon. The dataset comprised 2350 characters, including alignment gaps (ITS, TEF1 and TUB2 sequence data). After alignment, 641 characters were derived from ITS, 916 from TEF1, and 793 from TUB2. The MP analysis for the combined dataset had 730 parsimony-informative, 1216 constant, and 404 parsimony-uninformative characters, and yielded a single most parsimonious tree (TL = 3968, CI = 0.480, RI = 0.622, RC = 0.298; HI = 0.520). The RAxML analysis of the combined dataset yielded a best scoring tree with a final ML optimization likelihood value of −21,299.667012. The matrix had 1319 distinct alignment patterns, with 37.51% undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.226983, C = 0.316389, G = 0.231894, T = 0.224734; substitution rates AC = 1.153998, AG = 3.111864, AT = 1.039115, CG = 0.869376, CT = 4.271324, GT = 1.000000; gamma distribution shape parameter α = 0.376625. Bayesian posterior probabilities from Bayesian inference analysis were assessed with a standard deviation of split frequencies = 0.009867. The phylogenetic tree in this study showed that D. nobilis KUN-HKAS 123203 grouped with the ex-type strain of D. nobilis, and formed a supported clade with 0.99 PP (Figure 3). Sequence alignments are deposited in TreeBASE.

Table 3.
Culture collection numbers and GenBank accession numbers for Diaporthe used in this study. The type species are indicated in bold. The newly generated sequences are indicated in red. Instances where the GenBank Accession No. did not show the molecular data are marked with the dash.
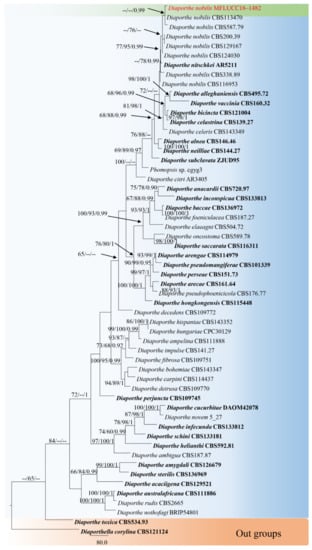
Figure 3.
RAxML tree based on a combined dataset of ITS, TEF1 and TUB2 gene regions. Bootstrap support values for ML and MP equal to or greater than 60% and Bayesian posterior probabilities (PP) equal to or greater than 0.90 are defined as ML/MP/PP above or below the nodes. Our new taxon is indicated in red. The tree was rooted with Diaporthella corylina (CBS 121124).
3.2. Taxonomy
3.2.1. Discosia rhododendricola Chaiwan & K.D. Hyde, sp. Nov. (Figure 4)
MycoBank number: 845145; Facesoffungi number: FoF 09452
Etymology: name reflects the host from which the fungus was isolated.
Holotype: KUN-HKAS 123205

Figure 4.
Discosia rhododendricola (KUN-HKAS 123205, holotype). (a–c) Appearance on host surface; (d) vertical section of conidioma; (e–i) conidiogenous cells and developing conidia; (j–o) conidia from holotype. Scale bars: (b) 500 μm; (c) 200 μm; (d) 100 μm; (g–i) 50 μm; (e,j–o) 20 μm; (f) 10 μm.
Saprobic on dead leaves of Rhododendron sp. Sexual morph: Undetermined. Asexual morph: Conidiomata 200–250 × 30–75 μm, pycnidial, cervular, applanate to disc-like, partly immersed or superficial, black, rounded to irregular in outline, glabrous, unilocular or divided into several locules by tissue conspicuous at the surface. Conidiophores were observed arising from the base, hyaline, filiform to cylindrical, smooth, and reduced to conidiogenous cells. Conidiogenous cells appeared subcylindrical, flask-shaped, hyaline, smooth, phialidic, each producing a single unbranched conidium. Conidia 20–30 × 4–5 μm ( = 25 × 4.5 μm, n = 30), subcylindrical, slightly curved, 3-septate, with slight constrictions at the septa, brown, smooth-walled with unequal cells; bipolar appendages; with a long, tubular base, two median cells subcylindrical, second cell joined to the base, 10–15 μm ( = 12.5 μm) long, the third cell joined to the apex, 11–15 μm ( = 13 μm) long; apical cell subconical with a rounded apex; apical and basal cells each with a subapical, unbranched, filiform, straight appendage; apical appendage, 9–11 μm ( = 10 μm), basal appendage, 20–25 μm ( = 22.5 μm).
Culture characteristics: Colonies grown on PDA were filamentous, raised, filiform margin, reached 4–5 cm in 5 days at 25 °C, brown to black, mycelium superficial, branched, septate, white mycelium with aerial on the surface, and produced black spore mass.
Material examined: CHINA, Kunming Yunnan Province; on dead leaves of Rhododendron sp. (Ericaceae), 28 July 2018, Napalai Chaiwan, KIB009 (KUN-HKAS 123205, holotype; isolate MFLU20-0486; Ex-type living culture KUNCC22-10804, isolate MFLUCC21-0004.
Notes: Discosia rhododendricola is similar to D. macrozamiae CPC 32109 [94] with regards to conidiomata size (D. rhododendricola a: 200–250 μm diam., 30–75 μm high vs. D. macrozamiae CPC 32109: 250 μm diam, 50 μm height). Discosia rhododendricola and D. artocreas (type species) share similar conidiophores lining the inner cavity (0–2-septate, rarely branched at base). There are also similar in conidial characteristics (conidial dimensions between 30 and 32 μm; the second cell joining to the base was 10–15 μm in length ( = 12.5 μm) in D. rhododendricola; 10–11 μm ( = 10.5 μm) in D. macrozamiae CPC 32109; the third cell joining to the apex was 11–15 μm in length ( = 13 μm) in D. rhododendria and 4–5 μm ( = 4.5 μm) in D. macrozamiae CPC 32109. The apical appendage of D. rhododendricola was 9–11 μm in length ( = 10 μm), while in D. macrozamiae (CPC 32109) it was 7–11 μm ( = 9 μm). The basal appendage in D. rhododendricola was 20–25 μm ( = 22.5 μm) in length, and in D. macrozamiae (CPC 32109) 10–16 μm ( = 13 μm).
Discosia rhododendricola differs from the type species, D. artocreas, in ascomatal size (D. rhododendricola: 200–250 μm diam., 30–75 μm high; D. artocreas 150–500 μm diam, 60 μm high). The two species share similar conidiophores and conidiogenous cells characteristics. However, D. rhododendricola has hyaline to pale brown conidiogenous cells and conidia, whereas D. artocreas has hyaline conidiogenous cells and conidia. The second cell joining to the base measured 10–15 μm in length ( = 12.5 μm) in D. rhododendricola but 5–9 μm ( = 7.5 μm) in D. artocreas. The third cell joining to the apex was 11–15 μm in length ( = 13 μm) in D. rhododendria and 3–6 μm ( = 4.5 μm) in D. artocreas. The apical appendage of D. rhododendricola was 9–11 μm in length ( = 10 μm), while in D. artocreas it was 6–12 μm ( = 10 μm). The basal appendage in D. rhododendricola was 20–25 μm in length ( = 22.5 μm), while in D. artocreas it was 7–12 μm ( = 10 μm).
The NCBI BLAST search of ITS sequence D. rhododendricola presented 95.32% similarity with Immersdiscosia eucalypti. A comparison of the 542 ITS (+5.8S) nucleotides of D.rhododendricola sp. nov. and I. eucalypti reveals 21 (3.87%) nucleotides differencess. We compared 876 LSU nucleotides of D. rhododendricola with D. muscicola CBS 109.48, and a 0.34% bp difference was observed (a difference of 3 bp in a total 879 bp) (Table 4).

Table 4.
LSU and ITS nucleotides comparisons of Discosia species related to our new taxon.
When analyzing the sequences, D. rhododendricola sp. nov. (KUN-HKAS123205 and MFLU20-0486) were found to be phylogenetically related to D. macrozamiae CPC 32109, D. muscicola CBS 109.48, D. pleurochaeta KT2179, D. pleurochae KT 2188 and KT 2192, while D. tricellularis MAFF237478 and NCBR32705 and D. yakushimensis MAFF 242774 were found to be in a clade. The two isolates of the new taxon (KUN-HKAS123205 and MFLU20-0486) have a high support value in the phylogenetic tree in a distinct clade (Figure 1). The ITS and LSU base pair differences between D. rhododendricola and other related species are shown in Table 4.
Discosia rhododendricola KUN-HKAS 123205 is closely related to the clade consisting of D. muscicola CBS 109.48, D. tricellularis MAFF 237478, NBRC 32705, and D. yakushimensis MAFF 242774 (Figure 5). The results of molecular analyses based on the Genealogical Concordance Phylogenetic Species Recognition (GCPSR) also showed that D. rhododendricola KUN-HKAS 123205 can be distinguished as a separate species by genealogical concordance (PHI = 1.0).
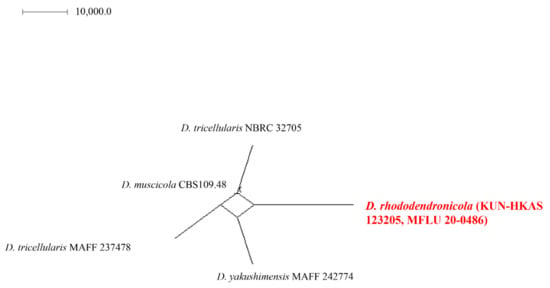
Figure 5.
Results of the pairwise homoplasy index (PHI) test of closely related species using both LogDet transformation and splits decomposition. PHI test results (Φw) <0.05 indicate significant recombination within the dataset. The new taxon is in red bold type.
3.2.2. Neopestalotiopsis Rhododendricola Chaiwan & K.D. Hyde, sp. Nov. (Figure 6)
MycoBank number: 845144; Facesoffungi number: FoF 10475
Etymology: Name reflects the host from which the fungus was isolated.
Holotype: KUN-HKAS 123204

Figure 6.
Neopestalotiopsis rhododendricola (KUN-HKAS 123204). (a) The habitat of the host plant (Rhododendron sp.); (b) Pycnidia with drops of conidial exudate on the leaf surface; (c,d) colonies growing on PDA; (e,f) culture; (g–j) conidiogenous cells and developing conidia; (k–p) conidia. Scale bars: (c) 500 µm; (d) 200 µm; (h) 20 µm; (i–p) 10 µm.
Saprobic on dead leaves of Rhododendron sp. Sexual morph: Undetermined. Asexual morph: Conidiomata (on PDA) 60–80 × 50–75 μm, pycnidial, cervular, applanate to disc-like, partly immersed or superficial, globose to clavate, solitary or confluent, embedded or semi-immersed to erumpent, dark brown, exuding globose, dark brown to black conidial masses, rounded to irregular in outline, glabrous, and unilocular or divided into several locules by tissue cells. Conidiophores are indistinct, arising from the base, hyaline, filiform to cylindrical, smooth, and are often reduced to conidiogenous cells. Conidiogenous cells appeared subcylindrical, flaskshaped, hyaline, smooth, and phialidic, with each producing a single conidium. Conidia 20–30 × 5–7 μm ( = 25 × 6 μm, n = 30), subcylindrical fusoid, ellipsoid, straight to slightly curved, 4-septate, (19–28) × 5–7 μm ( = 23.5 × 6 μm, n = 30), μm; basal cell conic with a truncate base, hyaline, rugose and thin-walled, with constrictions at the septa, hyaline, smooth-walled; with a long, tubular base, two median cells subcylindrical, second cell joined to the base, 10–15 μm ( = 12.5 μm) long, the third cell joined to the apex, 11–15 μm ( = 13 μm) long; apical cell subconical with a rounded apex; apical and basal cells each with a subapical, unbranched, filiform, straight appendage; apical appendage, 9–11 μm ( = 10 μm), basal appendage, 20–25 μm ( = 22.5 μm).
Culture characteristics: Colonies grown on PDA, with an undulating edge, reached 4–5 cm in 5 days at 25 °C, mycelium superficial, branched, septate, white mycelium with aerial on the surface, and produced black spore mass.
Material examined: CHINA, Kunming Yunnan Province; on dead leaves of Rhododendron sp. (Ericaceae), 28 July 2018, Napalai Chaiwan, KIB008 (KUN-HKAS 123204, holotype; isolate MFLU20-0046; Ex-type living culture KUNCC22-10802; isolate MFLUCC22-0004).
Notes: Neopestalotiopsis rhododendricola (KUN-HKAS 123204 and MFLU20-0046) were isolated from a Rhododendron sp. in China. In the phylogenetic analyses, N. rhododendricola forms a distinct highly supported lineage sister to N. sonneratae (MFLUCC17-1745T, MFLUCC17-1744), N. coffeae-arabicae (HGUP4019T, HGUP4015), N. thailandica (MFLUCC17-1730T, MFLUCC17-1731) and N. macadamiae (Figure 2). Neopestalotiopsis sonneratae was reported from leaf spots on Sonneronata alba in Thailand [21], Neopestalotiopsis thailandica was reported from leaf spots on Rhizophora mucronata Lam. in Thailand [21], and N. coffeae-arabicae was found on leaves of Coffea arabica in China [75].
Neopestalotiopsis rhododendricola sp. nov. resembles N. thailandica in having similar conidial size [21], but the difference is that N. rhododendricola has two to three tubular appendages on the apical cell, while N. thailandica showed only one to two tubular appendages on the apical cell. Comparison of ITS sequence differences revealed 2 base pairs, comparison of TEF sequence differences revealed 15 base pairs, and comparison of TUB differences revealed 6 base pairs of N. rhododendricola and N. thailandica. Therefore, based on morphology and phylogeny, we justify the description of N. rhododendricola as a new species in the Neopestalotiopsis genus.
3.2.3. Diaporthe nobilis Tanaka & S. Endô, in Endô, J. Pl. Prot. Japan 13: (1927) (Figure 7)
Faces of Fungi number: FoF 02717
Saprobic on dead leaves of Rhododendron sp. Sexual morph: Undetermined. Asexual morph: Conidiomata pycnidial 50–100 × 25–75 μm. ( = 75 × 50 μm, n = 10), globose to stromatic, multilocular, dark brown to black, scattered. Conidiophores were observed arising from the base, hyaline, filiform to cylindrical, smooth, straight. Conidiogenous cells, 35–40 × 1–2 μm ( = 37.5 × 1.5 μm, n = 10), phialidic, cylindrical, terminal and lateral, slightly tapered towards the apex, with visible periclinal, thickening, hyaline, and smooth-walled. Beta conidia 16–20 × 1–2 μm ( = 18 × 1.5 μm, n = 30), hyaline smooth, guttulate, fusoid to ellipsoid, straight, tapered towards both ends, apex sub obtuse, base sub truncate, and aseptate. Alpha conidia not found.
Culture characteristics: Colonies grew on PDA, filamentous, flattened, dense and felty, reaching 5–6 cm in 14 days at 25 °C, white to brown on the surface, mycelium superficial, branched, and septate.
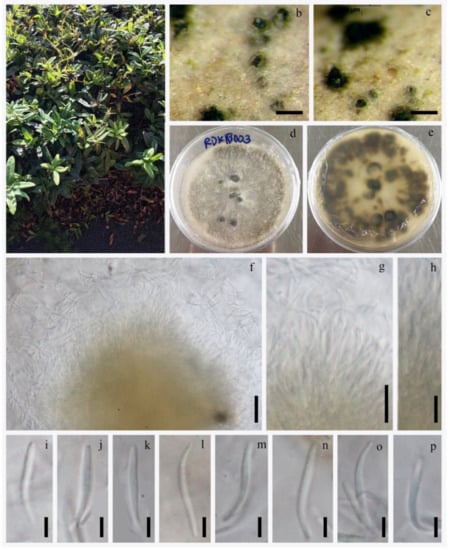
Figure 7.
Diaporthe nobilis (KUN-HKAS 123203). (a) Habitat of host; (b,c) appearance of fungi on host surface; (d,e) culture characters on PDA; (g,h) conidiophore with attached conidium; (i–p) conidia. Scale bars: (b,c) 200 μm; (d–f,i–p) 10 μm.
Material examined: CHINA, Kunming Yunnan Province, on dead leaves of Rhododendron sp. (Ericaceae), 28 July 2018, Napalai Chaiwan, KIB003 (KUN-HKAS 123203, new host record; isolate MFLU20-0485; living culture KUNCC22-10803; isolate MFLUCC 18–1482.
Notes: Diaporthe nobilis KUN-HKAS 123203 clustered with D. nobilis CBS 587.79 and CBS113470 with high 0.99 PP bootstrap support. Conidiomata from the MFLUCC 18–1482 strain was acervular, semi-immersed, globose to eustromatic, and multilocular, while D. nobilis CBS 587.79 has pycnidia subcuticular, scattered to confluent, and uniloculate. Our strain was observed to share similar morphological characteristics with other Diaporthe nobilis strains in having conidiogenous cells formed at the apex of the conidiophores, cylindric, straight or curved hyaline and smooth-walled. Comparison of ITS, TEF1 and TUB2 sequence data of isolate KUN-HKAS 123203 and D. nobilis CBS113470, revealed 9 bp (1.41%) in 637 ITS (+5.8S) nucleotides, 2 bp (0.40%) in 496 TEF1 nucleotides and 6 bp (0.71%) in 844 TUB2 nucleotides. Therefore, we consider our strain (KUN-HKAS 123203) as D. nobilis and as a new host record from Rhododendron sp. in China.
4. Discussion
Discosia species are distributed on various vascular plants and a wide range of hosts, and occur primarily in their asexual state as endophytes, saprobes and pathogens [20,58]. Host-specificity of species in this genus has not yet been established. Discosia species can be found on Fagus sylvatica (Fagaceae), Gaultheria procumbens (Ericaceae), Platanus orientalis (Platanaceae), Quercus sp. (Fagaceae), Syzygium cumini (Myrtaceae), Smilax rotundifolia (Smilacaceae), and leaves of undetermined plants [60]. Discosia blumencronii Bubák was reported from Rhododendron poniicum [92], while other species can be found on leaves of Beilschmeidia tarairi (Lauraceae), Brachychiton populneus (Malvaceae), Ceanothus fiedleri (Rhamnaceae), Eucalyptus sp. (Myrtaceae), Laurus nobilis (Lauraceae) and Phillyrea latifolia (Oleaceae) [9,60]. Discosia species is distributed in temperate regions, being previously reported in Algeria, Austria, Brazil, France, Germany, India, Italy, New Zealand, Portugal, the USA, Sweden, Tunisia and Turkey [9].
The new taxon, D. rhododendricola, was phylogenetically related to D. muscicola, described by Nicot-Toulouse Morelet (1968), and isolated from Cephalozia bicuspidate (Cephaloziaceae) in France. However, no morphological data are available for comparison [94]. Discosia rhododendricola sp. nov. was isolated from Rhododendron sp. and its morphology was compared. The ascomata and conidia of D. rhododendricola were larger than those of D. artocreas, whereas the sizes of conidiophores, conidiogenous cells and apical appendage were similar.
Discosia rhododendricola is similar to D. macrozamiae (CPC 32109) [62], but the phylogenetic tree showed that our species was closely related to D. muscicola CBS 109.48. However, for D. muscicola CBS 109.48, only rDNA sequence data were available (Figure 1). It should be pointed out that when the ITS DNA sequences of Discosia muscicola were subjected to a blast search, the closest hits were Aspergillus species similar to A. avenaceus. Our novel species have DNA sequence data from three regions (LSU, ITS, and RPB2), but we can only compare the LSU region for D. muscicola CBS 109.48, as there are no sequence data of the protein coding gene available for comparison. Based on the previous study of Wijayawardene et al. [16], 34 genera are recognized in Sporocadaceae. In this study, we introduce Discosia rhododendricola as a new species based on phylogenetic analyses and the pairwise homoplasy index.
Discosia species share similar morphological characters, but most characters are not meaningful in species delineation. In this study, our new species constitutes a different branching pattern in our phylogeny and appears distinct from extant species. A relationship among species based on similar conidial characters does not necessarily correlate with our phylogenetic relationships, and this indicates that morphology has little significance for reliable species identification.
Herein, we introduce a new species, Neopestalotiopsis rhododendricola KUN-HKAS 123204, within the Neopestalotiopsis genus that was separated from the other Neopestalotiopsis clade based on morphological and molecular phylogenetic analyses (Figure 2). Neopestalotiopsis are characterized by their conidia with versicolor median cells, by indistinct conidiogenous cells [15] and the ITS, TUB2 and TEF1 sequences. The newly described species is phylogenetically related to the group of N. sonneratae, N. coffeae-arabicae and N. thailandica in the phylogenetic tree (Figure 2), and the relationship is not strongly supported. Our new species was found on a Rhododendron sp. plant host from China, while N. sonneratae was reported on leaf spots on Sonneronata alba L. [21] and Neopestalotiopsis thailandica was reported on leaf spots of Rhizophora mucronata Lam. Both strains have been reported in Thailand [21], and N. coffeae-arabicae was found on leaves of Coffea arabica in China [75].
Diaporthe species have been reported as plant pathogens, saprobes and endophytes on many plant hosts [23,28,58]. Species of Diaporthe are not host-specific [6,28,40]. Substrates colonized by members of Diaporthe recorded to date are mainly dicotyledons of Ericaceae, Fagaceae, Pinaceae, Rhizophoraceae, Rosaceae and Theaceae. Some species of Diaporthe can be found on more than one host. For example, Diaporthe nobilis was reported on Camellia sinensis (Theaceae), Castanea sativa (Fagaceae), Malus pumila (Rosaceae), Pinus pantepella (Pinaceae), Pyrus pyrifolia (Rosaceae) and Rhododendron sp. (Ericaceae) [6,28,40,58]. Diaporthe is mostly presented in the asexual morph as coelomycetes [23]. Diaporthe nobilis complex [6] has alpha and beta conidia [28]. However, our strain was only found to have beta conidia.
Diaporthe have been reported on Rhododendron spp. from Europe (Latvia) [6]. The strain (KUN-HKAS 123203) was isolated from Asia (China), indicating that the species is distributed in different geographical locations on the host; however, there is a need for more collections of microfungi associated with Rhododendron, targeting a wide variety of geographical locations. A checklist for Discosia species associated with Rhododendron is also provided herein.
5. Discosia Species Associated with Rhododendron sp.: Habitat, Known Distribution and Checklist
The above information is based on the USDA Systematic Mycology and Microbiology Laboratory (SMML) database [58], relevant literature, date from this study while current names and fungal classifications used are according to Index Fungorum (2022) [14], and an outline of Ascomycota [16]. Species confirmed with DNA sequence data are marked with an asterisk.
- Discosia artocreas (Tode) Fr., Summa veg. Scand., Sectio Post. (Stockholm): 423 (1849)= Sphaeria artocreas Tode, F. Meckl. 2: 77, 1791; Fries, Syst. Myc. 2: 523, 1823.Habitat: Rhododendron arboretum, R. campylocarpum, R. nudiflorum [95,96], R. catawbiense, R. maximum [97], R. ponticum [98,99] and Rhododendron sp. [95,96,100]Known distribution: Italy [98], Maryland [95,96,97], New York [97], United Kingdom [100], Turkey* [99], Washington [95,96].
- Discosia blumencronii Bubák, in Handel-Mazzetti, Annln K. K. naturh. Hofmus. Wien 23: 106 (1910)Habitat: Rhododendron ponticum (on dead leaves) [101]Known distribution: Turkey [101]
- Discosia himalayensis Died., Annls mycol. 14(3/4): 218 (1916)= Discosia strobilina Lib. ex Sacc., Syll. Fung. (Abellini) 3: 656 (1884)Habitat: Rhododendron arboretum, R. campanulatum (on dead leaves) [101,102,103]Known distribution: India [101,102,103]
- Discosia rhododendri (Speschnew, Monit. Jard. Bot. Tiflis 4: 10 (1906)Habitat: Rhododendron albrechtii (on dead leaves)* [104], R. ponticum* [99], Rhododendron sp. (on leaves) [101]Known distribution: Japan* [104], Turkey* [99]
- Discosia rhododendricola (This study *)Habitat: Rhododendron sp. (on dead leaves) (This study *)Known distribution: China (This study *)
- Discosia sp.Habitat: Rhododendron sp.* [104]Known distribution: Japan* [104]
- Discosia tricellularis (Okane, Nakagiri & Tad. Ito) F. Liu, L. Cai & Crous, in Liu, Bonthond, Groenewald, Cai & Crous, Stud. Mycol. 92: 322 (2018) (2019)Habitat: Rhododendron indicum [105]Known distribution: Japan [105]
- Discosia vagans De Not., Atti Acad. Tor.: 354 (1849)Habitat: Rhododendron arboretum, R. nilagiricum, R. veitchianum* [59,103], R. ponticum* [61]Known distribution: India* [99,105], Scotland* [59]
Author Contributions
N.C., D.P., R.J., R.S.J., N.N., A.M., I.P. and K.D.H. designed the study and were involved in the writing of the paper. N.C. performed the sample collections. N.C. and R.S.J. were involved in phylogenetic analyses. N.N. contributed to paper planning and editing. All authors have read and agreed to the published version of the manuscript.
Funding
We are grateful to the Thailand Research Fund (TRF) grant no PHD60K0147 and Kunming Institute of Botany for partially supporting this work. Shaun Pennycook is thanked for nomenclatural advice. K.D. Hyde would like to thank the Thailand Research Fund projects entitled ‘The future of specialist fungi in a changing climate: baseline data for generalist and specialist fungi associated with ants, Rhododendron species and Dracaena species (No. DBG6080013)’ and ‘Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion (No. RDG6130001)’. K.D. Hyde thanks Chiang Mai University for the award of Visiting Professor. Adam Kaplan is thanked for the English editing of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequences generated in this study were submitted to GenBank.
Acknowledgments
N.C. is grateful to the Thailand Research Fund (TRF) grant no PHD60K0147 and the Centre of Excellence in Fungal Research, Mea Fha Luang University (Thailand). K.D.H. thanks Chiang Mai University for the award of Visiting Professor.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mainra, A.; Badola, H.K.; Mohanty, B. Rhododendrons: Conservation and Sustainable Use International Conference; Government of Sikkim: Gangtok-Sikkim, India, 2010.
- Sekar, K.C.; Srivastava, S.K. Rhododendrons in Indian Himalayan Region: Diversity and Conservation. Am. J. Plant Sci. 2010, 1, 131–137. [Google Scholar] [CrossRef]
- Samant, S.S.; Dhar, U. Diversity, endemism and economic potential of wild edibles plants of Indian Himalaya. Int. J. Sustain. Dev. World Ecol. 1997, 4, 179–191. [Google Scholar] [CrossRef]
- Negi, V.S.; Maikhuri, R.K.; Rawat, L.S.; Chandra, A. Bioprospecting of Rhododendron arboreum for Livelihood Enhancement in Central Himalaya. Indian J. Sci. Technol. 2013, 8, 61–70. [Google Scholar]
- Crane, P.E. Rust fungi on rhododendrons in Asia: Diaphanopellis forrestii gen. et sp. nov., new species of Caeoma, and expanded descriptions of Chrysomyxa dietelii and C. succinea. Mycologia 2005, 97, 534–548. [Google Scholar] [CrossRef]
- Gomes, R.R.; Glienke, C.; Viderira, S.I.R.; Lombard, L.; Groenewald, J.Z.; Crous, P.W. Diaporthe, a genus of endophytic, saprobic and plant pathogenic fungi. Persoonia 2013, 31, 1–41. [Google Scholar] [CrossRef]
- Kierpiec-Baran, B.; Zolna, M.; Kowalik, M. Diversity of fungi colonizing leaves of rhododendron (Rhododendron L.) cuttings. Acta Agrobot. 2014, 67, 21–26. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Huang, S.K.; Abdel-Wahab, M.A.; Daranagama, D.A.; Dayarathne, M.; D’souza, M.J.; Goonasekara, I.D.; et al. Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers. 2015, 72, 199–301. [Google Scholar] [CrossRef]
- Tanaka, K.; Endo, M.; Hirayama, K.; Okeanae, I.; Hosoya, T.; Sato, T. Phylogeny of Discosia and Seimatosporium, and introduction of Adisciso and Immersidiscosia genera nova. Persoonia 2011, 26, 85–98. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Bhat, J.D.; Gareth Jones, E.B.; McKenzie, E.H.C.; Dai, D.Q.; Daranagama, D.A.; Dayarathne, M.C.; Goonasekara, I.D.; et al. Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers. 2015, 73, 73–144. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Gardiennet, A.; Voglmayr, H. Resolution of morphology-based taxonomic delusions: Acrocordiella, Basiseptospora, Blogiascospora, Clypeosphaeria, Hymenopleella, Lepteutypa, Pseudapiospora, Requienella, Seiridium and Strickeria. Persoonia 2016, 37, 82–105. [Google Scholar] [CrossRef]
- Libert, M.A. Plantae Cryptogamae, quas in Arduenna collegit. Fasc 1837, 4, 301–400. [Google Scholar]
- Liu, F.; Bonthond, G.; Groenewald, J.Z.; Cai, L.; Crous, P.W. Sporocadaceae, a family of coelomycetous fungi with appendagebearing conidia. Stud. Mycol. 2019, 92, 287–415. [Google Scholar] [CrossRef]
- Index Fungorum. 2022. Available online: http://www.indexfungorum.org/names/Names.asp (accessed on 26 June 2022).
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Groenewald, J.Z.; Xu, J.; Crous, P.W. Pestalotiopsis revisited. Stud. Mycol. 2014, 79, 121–186. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of Fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Hyde, K.D.; Hongsanan, S.; Jeewon, R.; Bhat, D.J.; McKenzie, E.H.C.; Gareth Jones, E.B.; Phookamsak, R.; Ariyawansa, H.A.; Boonmee, S.; Zhao, Q.; et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 80, 1–270. [Google Scholar] [CrossRef]
- Gerardo-Lugo, S.S.; Tovar-Pedraza, J.M.; Maharachchikumbura, S.S.N.; Apodaca-Sánchez, M.A.; Correia, K.C.; Sauceda-Acosta, C.P.; Camacho-Tapia, M.; Hyde, K.D.; Marraiki, N.; Elgorban, A.M.; et al. Characterization of Neopestalotiopsis species associated with mango grey leaf spot disease in Sinaloa, Mexico. Pathogens 2020, 9, 788. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Bhat, D.J.; Dayarathne, M.C.; Huang, S.K.; Norphanphoun, C.; Senanayake, I.C.; Perera, R.H.; et al. Families of Sordariomycetes. Fungal Divers. 2016, 79, 1–317. [Google Scholar] [CrossRef]
- Hyfe, K.D.; Norphanphoun, C.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Jones, E.B.G.; Bundhun, D.; Chen, Y.J.; Bao, D.F.; Boonmee, S.; Calabon, M.S.; et al. Refined families of Sordariomycetes. Mycosphere 2020, 11, 305–1059. [Google Scholar]
- Norphanphoun, C.; Jayawardena, R.S.; Chen, Y.; Wen, T.C.; Meepol, W.; Hyde, K.D. Morphological and phylogenetic characterization of novel pestalotioid species associated with mangroves in Thailand. Mycosphere 2019, 10, 531–578. [Google Scholar] [CrossRef]
- Liu, X.; Tibpromma, S.; Zhang, F.; Xu, J.; Kandawatte, T.C.; Karunarathna, S.C.; Mortimer, P.E. Neopestalotiopsis cavernicola sp. nov. from Gem Cave in Yunnan Province, China. Phytotaxa 2021, 512, 1–27. [Google Scholar] [CrossRef]
- Nitschke, T. Pyrenomycetes Germanici; Eduard Trewendt Breslau: Germany, 1870; Volume 2, pp. 161–320. [Google Scholar]
- Kishi, K. (Ed.) Plant Diseases in Japan; Zenkoku Noson Kyoiku Kyokai: Tokyo, Japan, 1998. (In Japanese) [Google Scholar]
- Kajitani, Y.; Kanematsu, S. Diaporthe kyushuensis sp. nov., the teleomorph of the causal fungus of grapevine swelling arm in Japan, and its anamorph Phomopsis vitimegaspora. Mycoscience 2000, 41, 111–114. [Google Scholar] [CrossRef]
- Katsumoto, K. List of Fungi Recorded in Japan; Kanto Branch of the Mycological Society of Japan: Chiba, Japan, 2010. (In Japanese) [Google Scholar]
- Ando, Y.; Masuya, H.; Aikawa, T.; Ichihara, M. Diaporthe toxicodendri sp. nov., a causal fungus of the canker disease on Toxicodendron vernicifluum in Japan. Mycosphere 2017, 8, 1157–1167. [Google Scholar] [CrossRef]
- Li, Y.; Tan, P.; Zhao, D.G. Diaporthe nobilis, a new record on Camellia sinensis in Guizhou Province, China. Mycosphere 2017, 8, 1–8. [Google Scholar] [CrossRef]
- Guarnaccia, V.; Groenewald, J.Z.; Woodhall, J.; Armengol, J.; Cinelli, T.; Eichmerier, A.; Ezra, D.; Fontaine, F.; Gramaje, D.; Gutierrez-Aguirregabiria, A. Diaporthe diversity and pathogenicity revealed from a broad survey of grapevine diseases in Europe. Persoonia 2018, 40, 135–153. [Google Scholar] [CrossRef]
- Sessa, L.; Abreo, E.; Bettucci, L.; Lupo, S. Diversity and virulence of Diaporthe species associated with wood disease symptoms in deciduous fruit trees in Uruguay. Phytopathol. Mediterr. 2017, 56, 431–444. [Google Scholar]
- Udayanga, D.; Liu, X.Z.; Crous, P.W.; McKenzie, E.H.C.; Chukeatirote, E.; Hyde, K.D. A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal Divers. 2012, 56, 157–171. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Camporesi, E.; Hyde, K.D.; Zhang, W.; Yan, J.Y.; Li, X.H. Molecular phylogenetic analysis reveals seven new Diaporthe species from Italy. Mycosphere 2017, 8, 853–877. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Hernandez-Restrepo, M.; Wingfield, M.J.; Akulov, A.; Carnegie, A.J.; Cheewangkoon, R.; Gramaje, D.; Groenewald, J.Z.; Guarnaccia, V.; Halleen, F. Genera of phytopathogenic fungi: GOPHY 2. Stud. Mycol. 2019, 92, 47–133. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Pem, D.; Dissanayake, L.; Wijesinghe, S.N.; Bundhun, D.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.A.; Bhat, D.J.; Buyck, B.; Cai, L.; Dai, Y.C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The faces of fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- Jeewon, R.; Hyde, K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosp 2016, 11, 1669–1677. [Google Scholar] [CrossRef]
- White, T.J.; Burns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- O’Donnell, K.; Cigelnik, E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef]
- Chaiwan, N.; Maharachchikumbura, S.S.N.; Wanasinghe, D.N.; Doilom, M.; Jayaeardena, R.S.; Hyde, K.D. First sexual morph record of Sarcopodium vanilla. Mycotaxon 2020, 134, 707–717. [Google Scholar] [CrossRef]
- Dissanayake, A.J.; Bhunjun, C.S.; Maharachchikumbura, S.S.N.; Liu, J.K. Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 2020, 11, 2653–2677. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2017, 30, 772–780. [Google Scholar] [CrossRef]
- Bundhun, D.; Jeewon, R.; Senanayake, I.C.; Camporesi, E.; Aluthmuhandiram, J.V.S.; Tang, A.M.C.; Kang, J.C.; Bhoyroo, V.; Hyde, K.D. Morpho-molecular characterization of Discosia ravennica sp. nov. and a new host record for Sporocadus rosigena. MycoKeys 2021, 79, 173–192. [Google Scholar] [CrossRef]
- Rannala, B.; Yang, Z. Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J. Mol. Evol. 1996, 43, 304–311. [Google Scholar] [CrossRef]
- Kishino, H.; Hasegawa, M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 1989, 29, 170–179. [Google Scholar] [CrossRef]
- Zhaxybayeva, O.; Gogarten, J.P. Bootstrap, Bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. BMC Genom. 2002, 3, 4. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J. Tracer v1.4. 2007. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 30 June 2022).
- Rambaut, A. FigTree v1.4: Tree Figure Drawing Tool. 2014. Available online: http://treebio.ed.ac.uk/software/figtree/ (accessed on 30 June 2022).
- Bruen, T.C.; Philippe, H.; Bryant, D. A simple and robust statistical test for detecting the presence of recombination. Genetics 2006, 172, 2665–2681. [Google Scholar] [CrossRef]
- Huson, D.H. Splits Tree: Analyzing and visualizing evolutionary data. Bioinformatics 1998, 14, 68–73. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Quaedvlieg, W.; Binder, M.; Groenewald, J.Z.; Summerell, B.A.; Carnegie, A.J.; Burgess, T.I.; Crous, P.W. Introducing the consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia 2014, 33, 1–40. [Google Scholar] [CrossRef]
- Farr, D.F.; Rossman, A.Y. Fungal Databases, Systematic Mycology and Microbiology Laboratory, ARS, USDA. 2022. Available online: http://nt.arsgrin.gov/fungaldatabases (accessed on 10 June 2022).
- Tangthirasunun, N.; Silar, P.; Bhat, D.J.; Chukeatirote, E.; Wijayawardene, D.N.N.; Maharachchikumbura, S.S.N.; Hyde, K.D. Morphology and phylogeny of Pseudorobillarda eucalypti sp. nov., from Thailand. Phytotaxa 2014, 176, 251–259. [Google Scholar] [CrossRef]
- Li, W.J.; Liu, J.K.; Bhat, D.J.; Camporesi, E.; Dai, D.Q.; Mortimer, P.E.; Xu, J.; Hyde, K.D.; Chomnunti, P. Molecular phylogenetic analysis reveals two new species of Discosia from Italy. Phytotaxa 2015, 203, 37–46. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Hardy, G.S.J.; Gene, J.; Guarro, J.; Baseia, I.G.; Garcia, D.; Gusmao, L.F.P.; Souza-Motta, C.M.; et al. Fungal Planet description sheets: 716–784. Persoonia 2018, 40, 240–393. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Wingfield, M.J.; Guarro, J.; Cheewangkoon, R.; van der Bank, M.; Swart, W.J.; Stchigel, A.M.; Cano-Lira, J.F.; Roux, J.; Madrid, H.; et al. Fungal Planet description sheets: 154–213. Persoonia 2013, 31, 188–296. [Google Scholar] [CrossRef]
- Hyde, K.D.; Chaiwan, N.; Norphanphoun, C.; Boonmee, S.; Camporesi, E.; Chethana, K.W.T.; Dayarathne, M.C.; de Silva, N.I.; Dissanayake, A.J.; Ekanayaka, A.H.; et al. Mycosphere notes 169–224. Mycosphere 2018, 9, 271–430. [Google Scholar] [CrossRef]
- Greengarten, P.J.; Tuininga, A.R.; Morath, S.U.; Falco, R.C.; Norelus, H.; Daniels, T.J. Occurrence of Soil- and Tick-Borne Fungi and Related Virulence Tests for Pathogenicity to Ixodes scapularis (Acari: Ixodidae). J. Med. Entomol. 2011, 48, 337–344. [Google Scholar] [CrossRef]
- Rahi, P.; Vyas, P.; Sharma, S.; Gulati, A.; Gulati, A. Plant growth promoting potential of the fungus Discosia sp. FIHB 571 from tea rhizosphere tested on chickpea, maize and pea. Indian J. Microbiol. 2009, 49, 128–133. [Google Scholar] [CrossRef]
- Jeewon, R.; Liew, E.C.Y.; Hyde, K.D. Phylogenetic relationships of Pestalotiopsis and allied genera inferred from ribosomal DNA sequences and morphological characters. Mol. Phylogenet. Evol. 2002, 25, 378–392. [Google Scholar] [CrossRef]
- Pereira, J.S.; Costa, R.R.; Nagamoto, N.S.; Forti, L.C.; Pagnocca, F.C.; Rodrigues, A. Comparative analysis of fungal communities in colonies of two leafcutting ant species with different substratum preferences. Fungal Ecol. 2016, 21, 68–75. [Google Scholar] [CrossRef]
- Marčiulynas, A.; Marčiulynienė, D.; Lynikiene, J.; Gedminas, A.; Vaiciukyne, M.; Menkis, A. Fungi and Oomycetes in the Irrigation Water of Forest Nurseries. Forests 2020, 11, 459. [Google Scholar] [CrossRef]
- Wijayawardene, D.N.N.; Goonasekara, I.D.; Camporesi, E.; Wang, Y.; An, Y.L. Two new Seimatosporium species from Italy. Mycosphere 2016, 7, 204–213. [Google Scholar] [CrossRef]
- Kumar, V.; Cheewangkoon, R.; Gentekaki, E.; Maharachchikumbura, S.S.N.; Brahmanage, R.S.; Hyde, K.D. Neopestalotiopsis alpapicalis sp. nov. a new endophyte from tropical mangrove trees in Krabi Province (Thailand). Phytotaxa 2019, 393, 251–262. [Google Scholar] [CrossRef]
- Bezerra, J.D.P.; Machado, A.R.; Firmino, A.L.; Rosado, A.W.C.; de Souza, C.A.F.; de Souza-Motta, C.M.; de Sousa Freire, K.T.L.; Paiva, L.M.; Magalhaes, O.M.C.; Pereira, O.L. Mycological diversity description I. Acta Bot. Bras. 2018, 32, 656–666. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; McKenzie, E.H.C.; Bhat, D.J.; Phillips, A.J.L.; Wanasinghe, D.N.; Samarakoon, M.C.; Jayawardena, R.S.; Dissanayake, A.J.; Tennakoon, D.S.; et al. Fungal diversity notes 840-928: Micro-fungi associated with Pandanaceae. Fungal Divers. 2018, 93, 1–160. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Guo, L.D.; Lei, C.; Chukeatirote, E.; Wu, W.; Sun, X.; Crous, P.W.; Bhat, D.J.; McKenzie, E.; Bahkali, A. A multi-locus backbone tree for Pestalotiopsis, with a polyphasic characterization of 14 new species. Fungal Divers. 2012, 56, 95–129. [Google Scholar] [CrossRef]
- Song, Y.; Geng, N.; Hyde, K.D.; Zhao, W.S.; Wei, J.G.; Kang, J.C.; Wang, Y. Two new species of Pestalotiopsis from Southern China. Phytotaxa 2013, 126, 22–30. [Google Scholar] [CrossRef]
- Ma, X.Y.; Maharachchikumbura, S.S.N.; Chen, B.W.; Hyde, K.D.; McKenzie, E.H.C.; Chomnunti, P.; Kang, J.C. Endophytic pestaloid taxa in Dendrobium orchids. Phytotaxa 2019, 419, 268–286. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Le Roux, J.J.; Richardson, D.M.; Strasberg, D.; Shivas, R.G.; Alvardo, P.; Edwards, J.; Moreno, G.; Sharma, R.; et al. Fungal Planet description sheets: 371–399. Persoonia 2015, 35, 264–327. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Guo, L.D.; Chukeatirote, E.; Hyde, K.D. Improving the backbone tree for the genus Pestalotiopsis; addition of P. steyaertii and P. magna sp. nov. Mycol. Prog. 2014, 13, 617–624. [Google Scholar] [CrossRef]
- Huanluek, N.; Jjayawardena, R.S.; Maharachchikumbura, S.S.N.; Harishchandra, D.L. Additions to pestalotioid fungi in Thailand: Neopestalotiopsis hydeana sp. nov. and Pestalotiopsis hydei sp. nov. Phytotaxa 2021, 479, 23–43. [Google Scholar] [CrossRef]
- Ayoubi, N.; Soleimani, M.J. Strawberry fruit rot caused by Neopestalotiopsis iranensis sp. nov. and N. mesopotamica. Curr. Microbiol. 2016, 72, 329–336. [Google Scholar] [CrossRef]
- Akinsanmi, O.A.; Nisa, S.; Jeff-Ego, O.S.; Shivas, R.G.; Drenth, A. Dry flower disease of Macadamia in Australia caused by Neopestalotiopsis macadamiae sp. nov. and Pestalotiopsis macadamiae sp. nov. Plant Dis. 2016, 101, 45–53. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Chooi, Y.H.; Gilchrist, C.L.M.; Lacey, E.; Pitt, J.I.; Roets, F.; Swart, W.J.; Cano-Lira, J.F.; Valenzuela-Lopez, N.; et al. Fungal Planet description sheets: 1042–1111. Persoonia 2020, 44, 301–459. [Google Scholar] [CrossRef]
- Silvério, M.L.; Cavalcanti, M.A.Q.; Silva, G.A.; Oliveira, R.J.V.; Bezerra, J.L. A new epifoliar species of Neopestalotiopsis from Brazil. Agrotropica 2016, 28, 151–158. [Google Scholar] [CrossRef]
- Jiang, N.; Bonthond, G.; Fan, X.L.; Tian, C.M. Neopestalotiopsis rosicola sp. nov. causing stem canker of Rosa chinensis in China. Mycotaxon 2018, 133, 271–283. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Guo, L.D.; Chukeatirote, E.; Mckenzie, E.H.C.; Hyde, K.D. A destructive new disease of Syzygium samarangense in Thailand caused by the new species Pestalotiopsis amarangensis. Trop. Plant Pathol. 2013, 38, 227–235. [Google Scholar] [CrossRef]
- Jiang, N.; Fan, X.L.; Tian, C.M. Identification and Characterization of Leaf-Inhabiting Fungi from Castanea Plantations in China. J. Fungi 2021, 7, 64. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Liu, M.; Maharachchikumbura, S.S.N.; Zhang, W.; Xing, Q.; Hyde, K.D.; Nilthong, S.; Li, X.; Yan, J. Neopestalotiopsis vitis sp. nov. causing grapevine leaf spot in China. Phytotaxa 2016, 258, 63–74. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Zhang, Y.M.; Wang, Y.; Hyde, K.D. Pestalotiopsis anacardiacearum sp. nov. (Amphisphaeriaceae) has an intricate relationship with Penicillaria jocosatrix, the mango tip borer. Phytotaxa 2013, 99, 49–57. [Google Scholar] [CrossRef]
- Udayanga, D.; Castlebury, L.A.; Rossman, A.Y.; Hyde, K.D. Species limits in Diaporthe: Molecular re-assessment of D. citri, D. cytosporella, D. foeniculina and D. rudis. Persoonia 2014, 32, 83–101. [Google Scholar] [CrossRef]
- Tan, Y.P.; Edwards, J.; Grice, K.R.E.; Shivas, R.G. Molecular phylogenetic analysis reveals six new species of Diaporthe from Australia. Fungal Divers. 2013, 61, 251–260. [Google Scholar] [CrossRef]
- Santos, J.M.; Vrandecic, K.; Cosic’, J.; Duvnjak, T.; Phillips, A.J.L. Resolving the Diaporthe species occurring on soybean in Croatia. Persoonia 2011, 27, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Lombard, L.; van Leeuwen, G.C.M.; Guarnaccia, V.; Polizzi, G.; van Rijswick, P.C.J.; Rosendahl, K.; Gabler, J.; Crous, P.W. Diaporthe species associated with Vaccinium, with specific reference to Europe. Phytopathol. Mediterr. 2014, 53, 85–97. [Google Scholar]
- Huang, F.; Hou, X.; Dewdney, M.M.; Fu, Y.; Chen, G.; Hyde, K.D.; Li, H. Diaporthe species occurring on citrus in China. Fungal Divers. 2013, 61, 237–250. [Google Scholar] [CrossRef]
- Toulouse, N. Morelet in Ann. Soc. Sci. nat. 1968, Archeol, Toulon and Var, 20, p. 104. Available online: http://sftp.kew.org/pub/data-repositories/LibriFungorum/IXF3/IXF3-550.jpg (accessed on 26 June 2022).
- Shaw, C.G. Host fungus index for the Pacific Northwest. I. Hosts. Wash. State Univ. Agric. Exp. Sta. Bull. 1973, 765, 1–121. [Google Scholar]
- Eglitis, M.; Gould, C.J.; Johnson, F. Fungi found on Ericaceae in the Pacific coastal area. Wash. State Univ. Agric. Exp. Sta. Bull. 1966, 675, 21. [Google Scholar]
- United States Agricultural Research Service; Crops Research Division. Index of Plant Diseases in the United States; Agriculture handbook; US Department of Agriculture: Annapolis, MD, USA, 1960; pp. 1–531.
- Tassi, F. Origine e sviluppo delle Leptostromacee e loro rapporti con le famiglie affini. Bull. Lab. Orto Bot. Reale Univ. Siena 1904, 6, 3–124. [Google Scholar]
- Huseyinov, E.; Selcuk, F. Contribution to study of mycoflora of Turkey I. Coelomycetes of orders Melanconiales and Sphaeropsidales on forest trees and shrubs in the Black Sea coast (Rize and Trabzon Provinces). Mikol. Fitopatol. 2001, 35, 28–33. [Google Scholar]
- Dennis, R.W.G. Fungi of the Hebrides; Royal Botanic Gardens, Kew: Richmond, UK, 1986; pp. 1–383. [Google Scholar]
- Nag Raj, T.R. Coelomycetous Anamorphs with Appendage-Bearing Conidia; Mycologue Publications: Waterloo, ON, Canada, 1993; pp. 1–1101. [Google Scholar]
- Sydow, P.; Sydow, H.; Butler, E.J. Fungi Indiae Orientalis. Ann. Mycol. 1916, 14, 177–220. [Google Scholar]
- Mathur, R.S. The Coelomycetes of India; Bishen Singh Mahendra Pal Singh: Delhi, India, 1979; pp. 1–460. [Google Scholar]
- Kobayashi, T. Index of Fungi Inhabiting Woody Plants in Japan; Host, Distribution and Literature; Zenkoku-Noson-Kyoiku Kyokai Publishing Co., Ltd.: Tokyo, Japan, 2007; pp. 1–1227. [Google Scholar]
- Kirk, P.M.; Spooner, B.M. An account of the fungi of Arran, Gigha and Kintyre. Kew Bull. 1984, 38, 503–597. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).