Abstract
Fungaria are an unmatched resource for providing genetic data from authoritative, taxonomically-correct fungal species, especially type specimens. These specimens serve to anchor species hypotheses by enabling the correct taxonomic placement of taxa in systematic studies. The DNA from ancient specimens older than 30 years is commonly fragmented, and sometimes highly contaminated by exogenous, non-target fungal DNA, making conventional PCR amplification and Sanger sequencing difficult or impossible. Here, we present the results of DNA extraction, PCR amplification of the ITS2 region, and Illumina MiSeq Nano sequencing of nine recent and 11 ancient specimens, including seven type specimens. The taxa sampled included a range of large and fleshy, to small and tough, or small, melanized specimens of Discina, Gyromitra, Propolis, Stictis, and Xerotrema, with a culture of Lasiosphaeria serving as a positive control. DNA was highly fragmented and in very low quantity for most samples, resulting in inconclusive or incorrect results for all but five samples. Taxonomically-correct sequences were generated from the holotype specimens of G. arctica, G. korshinskii, and G. leucoxantha, from the neotype of G. ussuriensis, and from the positive control. Taxonomic assignments were confirmed through morphology, top BLASTn hits, and maximum likelihood phylogenetic analyses. Though this study was not cost-effective due to the small number of samples submitted and few generating correct sequences, it did produce short DNA barcode fragments for four type specimens that are essential for their correct taxonomic placement in our ongoing systematic studies.
1. Introduction
Fungarium specimens offer an unprecedented diversity in the taxonomic breadth and depth of taxa that cannot be compared to or achieved through even the largest and broadest fieldwork studies aimed at obtaining fresh specimens, mainly due to the ephemeral nature of fungi. Fungaria are an untapped resource that contain the most precious and valuable specimens through their holdings of type specimens [1], which represent the only absolute link between a fungal species and its taxonomic name. However, mining genetic data from type specimens for DNA barcoding and genomic studies is often difficult due to the age, preservation, and storage of these often ancient (>30 years old) specimens [2]. Over time, DNA from century-old specimens becomes highly degraded into fragments that are generally less than 500 bp [3,4,5]. This fragmentation can be accelerated by the use of a variety of chemical pesticides that were often employed in fungaria to reduce the damage to the specimens by insects [6,7]. Another constraint is that the amount of physical material associated with type specimens is often limited, so some fungaria are restrictive on whether they allow destructive sampling for molecular studies. Lastly, specimens have historically been stored in compact conditions due to space limitations, and under environmental conditions to preserve their morphological features, not to optimize DNA quantity or quality [6,8]. This has led to intact exogenous DNA by other fungi in the form of conidia, spores, and hyphae contaminating ancient specimens. Universal primers used in DNA barcoding do not discriminate between target and contaminate DNA in the amplification process, and next-generation sequencing is very sensitive and highly impartial when generating its final products.
Attempts to DNA barcode large numbers of recent (<30 years old) fungarium specimens using Sanger sequencing have been relatively successful [9,10,11]. Next-generation sequencing is commonly employed for estimating fungal diversity from environmental samples, and for whole genome sequencing of recent and ancient fungarium specimens [12]. However, it has not been widely used for obtaining molecular barcode data from fungarium specimens, especially ancient type specimens that are potentially contaminated by other fungi, and in which the DNA is usually fragmented, making PCR amplification and traditional Sanger sequencing difficult or impossible [8]. Only a few other studies to date have attempted to use next-generation sequencing to obtain DNA barcode data from fungarium specimens [8,11,13,14]. Forin et al. [8] were moderately successful in using Illumina MiSeq technology to sequence the ITS2 region from ancient Peziza specimens, and Forin et al. [13] successfully sequenced the ITS1 and/or ITS2 regions from ancient Rosellinia specimens held in Saccardo’s mycological herbarium. Olds et al. [11] were highly successful in using the Illumina platform to sequence the ITS2 region from more recent specimens, mainly from the Denver Botanical Garden, but only moderately successful when sequencing ancient specimens collected between 1910–1990. Runnel et al. [14] were successful in using PacBio sequencing to generate long-read ITS-LSU sequences from recent specimens collected between 2015–2020.
The goal of this study was to generate ITS2 sequence data from recalcitrant ancient and recent dried voucher specimens, especially type material, employing Illumina next-generation sequencing technology. Providing DNA sequence data for authenticated voucher specimens connects the morphological descriptions with the molecular characters, which is vitally important for affixing taxonomic names in systematic studies. These short DNA barcode fragments are essential for the correct taxonomic placement of type specimens in our ongoing systematic studies, and provide a valuable confirmation of distributional patterns in fungi.
2. Materials and Methods
2.1. Taxa Selection
The taxa included in this study were selected based on one or more of the following conditions: (1) inclusion of both recent and ancient specimens; (2) sampling a range of large and fleshy, to small and tough, or small, highly melanized specimens; (3) the presumption that standard PCR amplification would fail based on ancient specimens; (4) documented previous PCR amplification failure regardless of age; and (5) the importance of obtaining even a small fragment of the DNA sequence for type specimens. As such, one specimen of Propolis, one specimen of Discina, three specimens of Stictis, six specimens of Gyromitra, and eight specimens of Xerotrema were selected, along with a culture of Lasiosphaeria lanuginosa, which served as the positive control (Table 1). Morphological analyses of the ascocarps, asci, and ascospores were conducted to confirm taxonomic identification for all specimens, except for five (G. arctica, G. korshinskii, G. leucoxantha, G. perlata, G. ussuriensis (LE 179636)) in which only enough material for DNA extraction was sent. Regardless, G. arctica, G. korshinskii, and G. leucoxantha represent type specimens, making confirmation of the taxonomic assignment based on morphology irrelevant. A morphological examination of G. perlata has already been thoroughly documented [15]. A morphological analysis of G. ussuriensis (LE 179636) would be pointless, since it is an immature specimen consisting of only a few pieces of a single, highly fragmented ascoma (Eugene Popov pers. comm.).

Table 1.
Samples used in this study with taxonomic name, fungarium number, and year the specimen was collected.
2.2. DNA Extraction, Quantification, and Fragmentation Analysis
Small samples (10–100 mg) of fungal tissue from dried voucher specimens (or a culture in the case of the positive control) were placed in 1.5 µL centrifuge tubes either by one of the authors or by the curator/collections manager at the lending institution. One of the following (or both for one sample) DNA extraction methods was used: an EZNA® Microelute Genomic DNA Kit (Omega Bio-tek), or modification of the NaOH extraction [16] (Table 2). Fungal tissue was repeatedly frozen at −20 °C and thawed at room temperature at least three times in either lysis buffer or 50–200 µL of 0.5 M NaOH to help break up fungal cell walls, and was then ground using a micropestle and a tissue grinder. For the EZNA kit, extraction followed the manufacturer’s instructions, except DNA was diluted in only 30 µL of DNA-free water in the final step. For the NaOH method, ground extract was incubated at 4 °C for 30 min, centrifuged at 16,800× g for 2 min, and 5 µL of the resulting supernatant was added to 45 µL of 100 mM Tris-HCl buffered with NaOH to pH 8.5–8.9. In one sample (CUP-A-030055), this 50 µL DNA solution was extracted a second time using the EZNA kit. The DNA for some samples (S-F-11771; UPS-F-144599; and all Propolis, Stictis, and Xerotrema) was further cleaned and concentrated using a Zymo DNA Clean & ConcentratorTM kit (Zymo Research) (Table 2). All procedures were conducted under standard conditions on a 10% bleach-disinfected laboratory benchtop using UV-sterilized materials. Final DNA extracts were subjected to quantification using a Qubit fluorometer (Thermo Fisher Scientific, Waltham, MA, USA), and DNA fragmentation was assessed on a 5300 high-sensitivity Fragment Analyzer (Advanced Analytical Technologies, Inc., Santa Clara, CA, USA).

Table 2.
Results of Illumina sequencing showing the taxonomic name assigned to the sample, the top BLASTn hit, type of DNA extraction, Qbit score, length of ITS2 sequences, number of sequences generated per samples, number and percent of sequences matching the target species, whether Sanger sequences of the entire ITS or ITS2 were generated, and GenBank accession number.
2.3. PCR Amplification and Sanger Sequencing
PCR amplification for eventual Sanger sequencing was completed in 25 µL total reaction volumes consisting of the following: 12.5 μL GoTaq® Green Master Mix (Promega); 2.5 µL bovine serum albumin (New England Biolabs); 2.5 µL of 50% dimethyl sulfoxide (Fisher Scientific); 1.5 μL of each 10 μM primer combination, ITS1F-ITS2 or ITS3-ITS4 [17,18]; and 5 μL of DNA. PCR was completed on a Bio-Rad C1000 thermal cycler under the following conditions: initial denaturation at 94 °C for 2 min, followed by 40 cycles of 94 °C for 30 s, 41 °C for 20 s, 72 °C for 1 min, with a final extension step of 72 °C for 10 min. Gel electrophoresis on a 1% TBE agarose gel stained with ethidium bromide was used to verify the presence of a PCR product. Purification was completed using a Wizard® SV Gel and PCR Clean-Up System (Promega) with the purified product diluted in 20–30 µL DNA-free water. A BigDye® Terminator 3.1 cycle sequencing kit (Applied Biosystems Inc., Waltham, MA, USA) was used to sequence the separate ITS1 and ITS2 regions in both directions using the above PCR primers on an Applied Biosystems 3730XL high-throughput capillary sequencer at the Roy J. Carver Biotechnology Center at the University of Illinois.
2.4. PCR Amplification and Next-Generation Sequencing
Comparisons between the ITS1 and ITS2 regions have shown the phylogenetic signal between these markers to be similar [19,20], but the ITS2 is often preferred, given the greater number of reference sequences available and the potential for introns in the ITS1 region [21,22]. Although the ITS1 region is more variable and better at resolving species in Gyromitra [23], it also contains large introns in some groups in this genus, making it very difficult to PCR-amplify ancient specimens with fragmented DNA (Miller unpub. data).
Purified DNA samples were analyzed using a high-sensitivity gel on a 5300 Fragment Analyzer to visualize the degree of DNA fragmentation. The ITS2 region was amplified using Fluidigm reagents and barcodes in a two-step PCR procedure. In the first step, the ITS2 region was amplified for 35 cycles using the fITS7 plus CS1 Fluidigm primer pad and the ITS4 plus CS2 Fluidigm primer pad [18,24]. The second step consisted of 14 cycles attaching the 10 bp sample specific identification barcode and Illumina adaptor barcodes. The final amplicon construct consisted of the following: Illumina i5 adaptor—CS1—fITS7 primer—region of interest—ITS4 primer—CS2—sample specific 10 bp barcode—Illumina i7 adaptor. The 5300 Fragment Analyzer was used to visualize the amplicons after each PCR step to confirm the presence of amplicons. The final amplicons were sequenced on Illumina MiSeq v3 platform rapid 2 × 250 nt PE NANO v2 paired-end reads (San Diego, CA, USA). All PCR amplification and sequencing steps were performed at the Roy J. Carver Biotechnology Center at the University of Illinois.
2.5. Next-Generation Sequencing Data Processing, OTU Identification, and Taxonomic Assignment
The resulting Illumina sequences were processed with DADA2 [25] using both forward and reverse amplicons. The filterAndTrim function was set up as follows: maxN = 0, maxEE = c(2,2), truncQ = 1, and rm.phix = True. The LearnErrors function was completed on both forward and reverse reads. After running the core dada function, forward and reverse reads were merged with the mergePairs function. Chimeras were removed using the removeBimeraDenovo function with the method set to consensus. All steps were completed with R version 4.1.2 [26]. A FASTA file of the dada2 amplicon sequence variants (ASVs) was generated using SeaView [27]. The FASTA file was compared against the NCBI nucleotide database, excluding environmental sequences, using NCBI BLASTn [28] to determine the sequence identity. The NCBI XML file/s were downloaded and parsed using the NCBI BLAST parser tool [29], and the best NCBI BLASTn match was retained. In addition to ASVs (99% sequence similarity), we also examined the results via Organized Taxonomic Units (OTUs) at 97% sequence similarity using DECIPHER [30] in R statistical software. In short, an initial sequence alignment was completed with the function, AlignSeqs; a distance matrix was computed from the aligned sequences using the function, DistanceMatrix; and 97% OTU clustering was completed using the function, IdClusters, with method set to complete and the cutoff set to 0.03. Raw Illumina reads have been deposited into the NCBI Sequence Read Archive (SRA) database (BioProject ID: PRJNA870474, http://www.ncbi.nlm.nih.gov/bioproject/870474).
2.6. Phylogenetic Analyses
Verification of the taxonomic name assigned to each specimen can only be completed if well-annotated, taxonomically-accurate sequences for that taxon occur in GenBank. Since only four of our 12 sampled taxa have available sequences in GenBank (Table 2), it was necessary to conduct phylogenetic analyses to verify the taxonomic assignment suggested by morphology and BLASTn sequence similarity. Alignments of the ITS2 region (or the entire ITS for G. korshinskii) were assembled using MUSCLE®, as implemented in Sequencher 5.4.6 (Gene Codes Corporation, Ann Arbor, MI, USA) for each target species and its most closely-related taxa based on BLASTn results and previous phylogenetic analyses ([31,32] Miller unpub. data). The best-fit model of evolution was determined to be the general time reversible (GTR) model [33] by jModeltest [34,35] based on the Akaike information criterion (AIC) [36]. A maximum likelihood (ML) analysis with 1000 bootstrap replicates was performed using PhyML, as implemented in Seaview 5.0.5 [37], with all parameters optimized and the GTR model. Clades with bootstrap values (BV) ≥70% were considered significant and strongly supported [38].
3. Results and Discussion
3.1. Taxa Sampled
A range of recent-to-ancient specimens that varied from large and fleshy (i.e., Discina, Gyromitra), to small and tough (i.e., Propolis, Stictis), or minute and melanized (i.e., Xerotrema), were selected for this study. Specimens were collected between 1863–2009, except for the positive control culture isolated in 2021, and, therefore, ranged in age from 13–159 years (Table 1). It was presumed that all specimens have been stored under typical fungarium conditions of moderate temperature and low humidity at the 11 institutions from which the specimens were loaned. No obvious signs of additional fungal growth contaminating the target specimens was observed on the outside of the sampled tissue. The taxonomic names listed in Table 1 were confirmed through morphological examination when possible. Gyromitra ussuriensis (LE 179636) consists of a highly fragmented specimen that is immature (Eugene Popov, pers. comm.), so it could only be identified through molecular data.
3.2. DNA Extraction, Quantification, and Fragmentation
Extraction using either the EZNA kit, which results in lower amounts of higher-quality DNA, or the NaOH method, which results in higher amounts of lower-quality DNA, did not seem to affect the overall quantity of the final DNA, although the sample size was very limited (Table 2). Cleaning and concentrating the DNA after extraction with a Zymo kit did not increase the success of PCR amplification and sequencing. Quantification using Qubit was too low to detect for most samples, but ranged from 1.65–6.56 ng/µL for the four samples with detectable DNA. Fragment analyses were run for 11 samples (Figure 1a–l), and all were fragmented except for the control, which showed no fragmentation between 20–800 bp (Figure 1e). DNA was undetectable and highly fragmented with no discernible peaks between 20–6000 bp, and the resulting sequences were inconclusive for specimens of S. fulva, X. megalospora, and X. quericola (Figure 1j–l). Although G. perlata had the highest amount of DNA, it also was the most fragmented, explaining why PCR amplification failed for this sample (Figure 1d). Fries’ material showed no signs of ever being treated with chemicals (Åsa Kruys pers. comm.), so age is the most likely explanation for its high rate of DNA fragmentation. The remaining Discina and five Gyromitra specimens had fragmented DNA, but obvious peaks between 20–300 bp, allowing the generation of successful PCR products and taxonomically correct ITS2 sequences for all five Gyromitra specimens (Figure 1b,c,f–i).
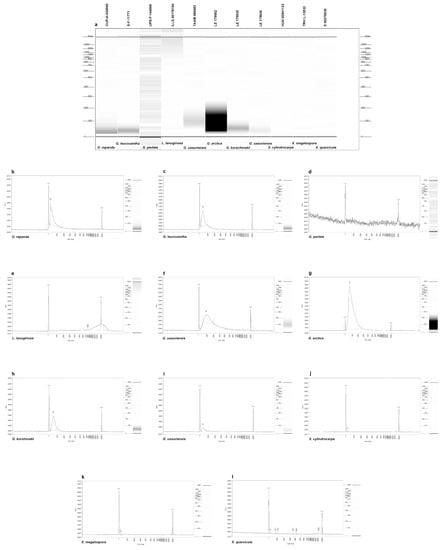
Figure 1.
Gel image and graphs showing DNA fragmentation of 11 samples from fragment analyzer. Scales on left and rights sides of gel image indicate base pairs; scale on left side of charts indicates amount of fragmentation in relative fluorescence units; size of fragments is shown in base pairs along bottom; gel image for each sample is repeated to the right of chart with scale in base pairs. (a) Gel image showing DNA fragmentation of 11 samples. (b) Discina repanda. (c) Gyromitra leucoxantha. (d) Gyromitra perlata. (e) Lasiosphaeria lanuginosa. (f) Gyromitra ussuriensis. (g) Gyromitra arctica. (h) Gyromitra korshinskii. (i) Gyromitra ussuriensis. (j) Sticta cylindrocarpa. (k) Xerotrema megalospora. (l) Xerotrema quericola.
3.3. Sanger Sequencing
Although PCR amplification was attempted for all samples, only G. arctica, G. korshinskii, and the control successfully PCR-amplified and produced subsequent Sanger sequences in both directions. Whereas the entire ITS region was PCR-amplified in two pieces and sequenced for G. korshinskii and the control, only the ITS2 region was successfully PCR-amplified and was sequenced for G. arctica (Table 2). These three consensus Sanger sequences, which contain accompanying chromatograms, had zero or one nucleotide difference between them and their Illumina-generated sequences, which lack chromatograms. Sanger sequences provide additional data to support our taxonomic assignments, and serve as a proof-of-concept for the Illumina sequencing.
3.4. Next-Generation Sequencing
PCR amplification was successful for all samples except G. perlata, which happens to be the oldest specimen, from around 1863 (Table 1). The Illumina MiSeq Nano sequencing generated a total of 891,996 sequences: 445,998 in the forward direction and 445,998 in the reverse, of which, 77% of the sequences passed quality filtering. In total, there were 312,683 contig sequences, with an average of 16,457 contig sequences per sample (Table 2). Prior to removing chimeras, the unique contig ASVs per sample ranged from 6 to 93 per sample. In total, 88 bimeras were detected, resulting in 609 unique ASVs across all samples. These ASVs were clustered into 283 OTUs at 97% similarity. After OTU clustering, it was noted that every OTU composed of multiple ASVs contained consistent final taxonomic determinations. The final sequence length for the ITS2 region ranged from 253 bp to 446 bp (Figure 2).
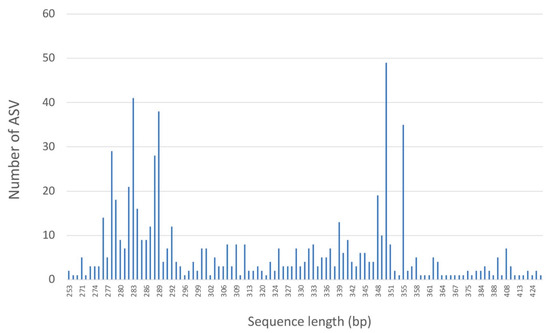
Figure 2.
Distribution of ITS2 sequence lengths across all samples.
3.5. Contamination
Ancient, and even sometimes recent, specimens can be heavily contaminated with exogenous fungal material, including conidia, spores, and hyphae. These fungal contaminates are PCR-amplified and sequenced, typically in low abundance, during Illumina sequencing. All of our samples contained sequences from contaminate fungi (Figure 3), including Wallemia spp., which are highly xerotolerant or xerophilic basidiomycetes that commonly occur in osmotically-challenged environments, as would be found in modern fungaria [39]. The fact that 67% of the sequences from Gyromitra ussuriensis were Wallemia mellicola (Figure 3e) suggests better PCR amplification, possibly due to less DNA fragmentation of the contaminate or a high amount of contaminate occurring on the specimen compared to the target. However, since 23% of the sequences were the target fungus (Table 2), with the third highest level of sequences at 2.5% (Figure 3e), we feel confident making this taxonomic assignment. All samples, except G. leucoxantha, contained Monilia fructicola, which is not surprising, since our lab was working with cultures of this peach pathogen at the time we conducted this study (Figure 3). We were also sequencing isolates of Collectotrichum siamense, a pathogen of apples, which also shows up in the control, although at a very low abundance (Figure 3g). Sequences of target species, albeit it in a very low abundance, occurred in two other unrelated samples (Figure 3c,e), indicating either the possibility of minor cross-over occurred during our molecular procedures, or cross-contamination of spores in the fungarium.
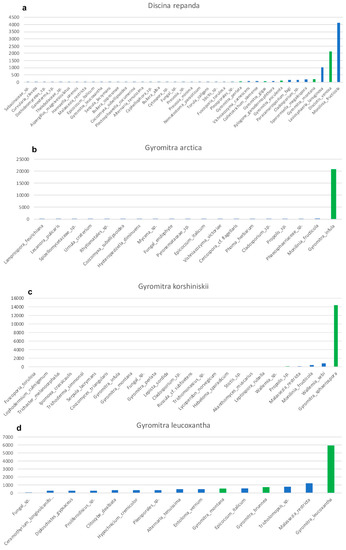
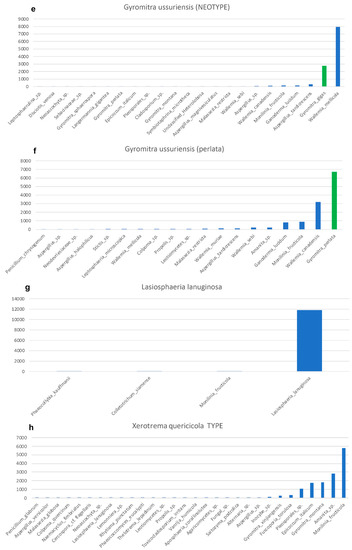
Figure 3.
Taxonomic assignments shown as increasing number of Illumina-generated ITS2 sequences for each sample. Bars are shown in green for all species in the genus, Gyromitra. (a) Discina repanda. (b) Gyromitra arctica. (c) Gyromitra korshinskii. (d) Gyromitra leucoxantha. (e) Gyromitra ussuriensis. (f) Gyromitra ussuriensis. (g) Lasiosphaeria lanuginosa. (h) Xerotrema quericola.
3.6. Phylogenetic Analyses
- Gyromitra arctica
Gyromitra arctica produced sufficient DNA after an EZNA extraction, resulting in a DNA concentration of 3.08 ng/µL based on Qubit analysis. Sanger sequences of the ITS2 region in both directions were obtained, and these had zero bp differences compared to the Illumina sequences. The target species represented 97% of the Illumina sequences (Table 2). Since no sequences of G. arctica are available in GenBank, this species BLASTed to G. infula (Table 2), which occurs as a closely-related species in our ML analysis (Figure 4a). Although some authors have placed G. arctica as a synonym under G. ambigua [40,41,42], Species Fungorum and MycoBank recognize it as a separate species, which agrees with our analyses.
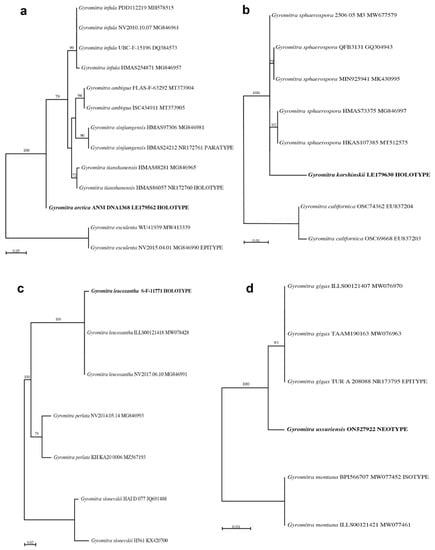
Figure 4.
Phylogenetic trees showing the taxonomic placement of the target species based on maximum likelihood analyses of Illumina-generated ITS2 (or ITS in (b)) sequences. The taxonomic name followed by the fungarium number and GenBank accession number are given for each taxon. The target species are shown in bold font. Bootstrap values > 50% are shown above branches. (a) Gyromitra arctica. (b) Gyromitra korshinskii. (c) Gyromitra leucoxantha. (d) Gyromitra ussuriensis.
- Gyromitra korshinskii
Although the EZNA DNA extraction of G. korshinskii resulted in a low amount of DNA, PCR amplification and subsequent Sanger and Illumina sequencing produced sequences that did not differ from one another. The target species represented 87% of the Illumina sequences (Table 2). Since no sequences of G. korshinskii are available in GenBank, this species BLASTed to G. sphaerospora (Table 2), which occurs in a strongly-supported clade with G. korshinskii in our ML analysis (Figure 4b). Although G. korshinskii has been placed as a synonym under G. sphaerospora [43], Species Fungorum and MycoBank recognize it as a separate species, and additional studies are needed to determine if G. korshinskii should be recognized as a distinct species.
- Gyromitra leucoxantha
An NaOH DNA extraction followed by a Zymo clean and concentrate of G. leucoxantha resulted in a low amount of DNA, but PCR amplification and Illumina sequencing were still successful, with 46% of the sequences representing the target organism (Table 2). Other sequences of G. leucoxantha are available in GenBank, and our holotype sequence occurred in a highly-supported clade with these taxonomically-correct sequences (Figure 4c).
- Gyromitra ussuriensis
Gyromitra ussuriensis produced sufficient DNA after an EZNA extraction, resulting in a DNA concentration of 2.02 ng/µL based on Qubit analysis. Although this specimen was heavily contaminated with Wallemia mellicola, 23% of the sequences were the target fungus (Table 2, Figure 3e). No sequences of G. ussuriensis are currently available in GenBank. It was thought to be a synonym of G. gigas [44,45,46], but occurs as a distinct species in these analyses (Figure 4d). Since the holotype specimen at VLA of G. ussuriensis is missing and presumably lost, and no original material or illustration exists (Eugenis Bulakh, pers. comm.), a neotype has been designated [47].
3.7. Costs
The average cost in our lab for the generation of an ITS2 sequence that is Sanger sequenced in both directions is US$6, which includes the in-house costs associated with DNA extraction, PCR amplification, PCR clean-up, generation of fluorescently-labeled sequences and ETOH clean-up of Big Dyes, and the cost for Sanger sequencing on an ABI 3730 at the University of Illinois. This is compared to the per-sample costs at our sequencing center associated with a 2-step PCR amplification ($20), quality check ($4), and the Illumina MiSeq Nano sequencing ($37), or $61 per sample; fragment analysis was an additional $32 per sample. Obviously, more than 20 samples could have been Illumina sequenced, and the 2-step PCR amplification could have been performed in our lab to reduce costs. However, even at ~10X the costs of Sanger sequencing, the ability to obtain an ITS2 sequence from a type specimen is worth the cost for our studies. In the case of G. leucoxantha and G. ussuriensis, sequences from type specimens were only obtained through Illumina sequencing, justifying the higher cost.
3.8. Comparisons with Other Studies
This is one of the first studies to definitively show that Illumina sequencing can provide taxonomically-correct sequences for ancient, highly-fragmented, contaminated type specimens. Though Forin et al. [8] successfully sequenced 23 of 36 ancient Peziza specimens from the Saccardo Mycological Herbarium, no type specimens were included in their study. However, Forin et al. [13] successfully sequenced five of nine type specimens of Rosellinia from the same herbarium. Both studies used the CTAB method for DNA extraction, which may explain their greater success in sequencing smaller, melanized specimens, versus the NaOH or EZNA kit methods utilized in this study. Though Old et al. [11] did not attempt to sequence any type specimens, their success in obtaining ITS2 sequences from 766 specimens was much greater with recent specimens from 1991–2020 vs. ancient specimens from 1910–1990. Runnel et al. [14] did not include any type specimens in their study of 423 specimens, but showed that PacBio high-throughput sequencing was more successful than Sanger sequencing at a comparable cost.
4. Conclusions
Our study attempted to sequence the ITS2 region from recent and ancient DNA from both large and fleshy, and small and tough specimens. Unfortunately, DNA was highly-fragmented in the ancient, smaller specimen of S. fulva, and the recent, smaller specimens of X. megalospora and X. quericola, resulting in inconclusive sequences. However, DNA from the ancient, larger specimens of Gyromitra was less fragmented, and we were successful in obtaining taxonomically-correct sequences from four type specimens. The cost of generating ITS2 sequences for only four type specimens in this study was justified by the value we believe these bring for the correct taxonomic placement of names in our systematic studies.
Author Contributions
Conceptualization, A.N.M. and D.B.R.; methodology, A.N.M. and D.B.R.; formal analysis, A.N.M. and D.B.R.; investigation, A.N.M., J.K. and D.B.R.; resources, A.N.M. and J.K.; data curation, A.N.M. and J.K.; writing—original draft preparation, A.N.M.; writing—review and editing, A.N.M., J.K. and D.B.R.; visualization, A.N.M.; project administration, A.N.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The Sanger sequence data generated in this study are openly available in NCBI at: https://www.ncbi.nlm.nih.gov/. Raw Illumina reads have been deposited into the NCBI Sequence Read Archive (SRA) database (BioProject ID: PRJNA870474), and can be accessed via the following: http://www.ncbi.nlm.nih.gov/bioproject/870474.
Acknowledgments
We thank the following curators and collections managers for the loan of specimens for destructive sampling: Arne Anderberg and Johannes Lundberg (S), Åsa Kruys (UPS), Betsy Arnold and Joseph Meyer (ASU), David Harris (E), Eugene Popov (LE), Jamie Minnaert-Grote (ILLS), Kadri Pärtel (TAAM), Kathie Hodge and Teresa Iturriaga (CUP), Kristian Hassel (TRH), Mary Berbee and Karen Golinski (UBC), and Michaela Schmull (HUH). Mark Band is thanked for the PCR amplification and fragment analyses of next-generation sequencing samples, and Chris Wright and Alvero Hernandez are thanked for the Illumina MiSeq Nano next-generation sequencing at the Roy J. Carver Biotechnology Center at the University of Illinois.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Andrew, C.; Diez, J.; James, T.Y.; Kauserud, H. Fungarium specimens: A largely untapped source in global change biology and beyond. Phil. Trans. R. Soc. B 2018, 374, 20170392. [Google Scholar] [CrossRef] [PubMed]
- Dentinger, B.T.; Margaritescu, S.; Moncalvo, J.M. Rapid and reliable high-throughput methods of DNA extraction for use in barcoding and molecular systematics of mushrooms. Mol. Ecol. Resour. 2010, 10, 628–633. [Google Scholar] [CrossRef]
- Taylor, J.W.; Swann, E.C. DNA from herbarium specimens. In Ancient DNA; Herrmann, B., Hummel, S., Eds.; Springer: New York, NY, USA, 1994; pp. 166–181. [Google Scholar] [CrossRef]
- Pääbo, S.; Poinar, H.; Serre, D.; Jaenicke-Després, V.; Hebler, J.; Rohland, N.; Kuch, M.; Krause, J.; Vigilant, L.; Hofreiter, M. Genetic analyses from ancient DNA. Annu. Rev. Genet. 2004, 38, 645–679. [Google Scholar] [CrossRef] [PubMed]
- Dabney, J.; Meyer, M.; Paabo, S. Ancient DNA damage. CSH Perspect. Biol. 2013, 5, a012567. [Google Scholar] [CrossRef] [PubMed]
- Mueller, G. A New Challenge for Mycological Herbaria: Destructive Sampling of Specimens for Molecular Data. In Managing the Modern Herbarium; Metsger, D.A., Byers, S.C., Eds.; Elton-Wolfe Publishing: Vancouver, BC, Canada, 1999; pp. 287–300. [Google Scholar]
- Kigawa, R.; Nochide, H.; Kimura, H.; Miura, S. Effects of various fumigants, thermal methods and carbon dioxide treatment on DNA extraction and amplification: A case study on freeze-dried mushroom and freeze-dried muscle specimens. Collect. Forum 2003, 18, 74–85. [Google Scholar]
- Forin, N.; Nigris, S.; Voyron, S.; Girlanda, M.; Vizzini, A.; Casadoro, G.; Baldan, B. Next Generation Sequencing of Ancient Fungal Specimens: The Case of the Saccardo Mycological Herbarium. Front. Ecol. Evol. 2018, 6, 1–19. [Google Scholar] [CrossRef]
- Bradshaw, M.; Tobin, P. Sequencing herbarium specimens of a common detrimental plant pathogen (powdery mildew). Phytopathology 2020, 110, 1248–1254. [Google Scholar] [CrossRef]
- Osmundson, T.W.; Robert, V.A.; Schoch, C.L.; Baker, L.J.; Smith, A.; Robich, G.; Mizzan, L.; Garbelotto, M.M. Filling Gaps in Biodiversity Knowledge for Macrofungi: Contributions and Assessment of an Herbarium Collection DNA Barcode Sequencing Project. PLoS ONE 2013, 8, e62419; [Google Scholar] [CrossRef]
- Olds, G.C.; Berta-Thompson, J.W.; Loucks, J.J.; Levy, R.A.; Wilson, A.W. Applying a Modified Metabarcoding Approach for the Sequencing of Macrofungal Specimens from Fungarium Collections. Master’s Thesis, University of Colorado, Denver, CO, USA, 2021. [Google Scholar] [CrossRef]
- Dentinger, B.T.M.; Gaya, E.; O’Brien, H.; Suz, L.M.; Lachlan, R.; Diaz-Valderrama, J.R.; Koch, R.A.; Aime, C.M. Tales from the crypt: Genome mining from fungarium specimens improves resolution of the mushroom tree of life. Biol. J. Linn. Soc. 2016, 117, 11–32. [Google Scholar] [CrossRef]
- Forin, N.; Vizzini, A.; Fainelli, F.; Ercole, E.; Baldan, B. Taxonomic re-examination of nine Rosellinia types (Ascomycota, Xylariales) stored in the Saccardo mycological collection. Microorganisms 2021, 9, 666. [Google Scholar] [CrossRef]
- Runnel, K.; Abarenkov, K.; Copot, O.; Mikryukov, V.; Kõljalg, U.; Saar, I.; Tedersoo, L. DNA barcoding of fungal specimens using long-read high-throughput sequencing. bioRxiv 2022. [Google Scholar] [CrossRef]
- Van Vorren, N. Typification of Gyromitra perlata, type-species of the subgenus Discina (Discinaceae). Ascomycete.org 2017, 9, 19–22. [Google Scholar] [CrossRef]
- Osmundson, T.W.; Eyre, C.A.; Hayden, K.; Dhillon, K.; Garbelotto, M. Back to basics: An evaluation of NaOH and alternative rapid DNA extraction protocols for DNA barcoding, geno-typing, and disease diagnostics from fungal and oomycete samples. Mol. Ecol. Resour. 2012, 13, 66–74; [Google Scholar] [CrossRef] [PubMed]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118; [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols, A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; San Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Bazzicalupo, A.L.; Bálint, M.; Schmitt, I. Comparison of ITS1 and ITS2 rDNA in 454 sequencing of hyperdiverse fungal communities. Fungal Ecol. 2013, 6, 102–109. [Google Scholar] [CrossRef]
- Blaalid, R.; Kumar, S.; Nilsson, R.H.; Abarenkov, K.; Kirk, P.; Kauserud, H. ITS 1 versus ITS2 as DNA metabarcodes for fungi. Mol. Ecol. Resour. 2013, 13, 218–224. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Kristiansson, E.; Ryberg, M.; Hallenberg, N.; Larsson, K.-H. Intraspecific ITS variability in the Kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol. Bioinfor. 2008, 4, 193–201. [Google Scholar] [CrossRef]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Ariyawansa, H.A.; Aoki, T.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M.; et al. Unambiguous Identification of Fungi: Where Do We Stand and How Accurate and Precise is Fungal DNA Barcoding? IMA Fungus 2020, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.N.; Yoon, A.; Gulden, G.; Stensholt, Ø.; Van Vooren, N.; Ohenoja, E.; Methven, A.S. Studies in Gyromitra I: The Gyromitra gigas species complex. Mycol. Prog. 2020, 19, 1459–1473. [Google Scholar] [CrossRef]
- Ihrmark, K.; Bödeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region—evaluation by 454-sequencing of artificial and natural communities. FEMS Micro. Ecol. 2012, 82, 666–677; [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, 2021. Available online: https://www.r-project.org/ (accessed on 2 December 2021).
- Galtier, N.; Guouy, M.; Goutier, C. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny CABIOS. Comput. Appl. Biosci. 1996, 12, 543–548. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Ream, D.; Kiss, A.J. NCBI/GenBank BLAST Output XML Parser Tool. 2013. Available online: https://s3-us-west-2.amazonaws.com/oww-files-public/4/43/NCBI_XML_Parser.pdf (accessed on 2 January 2018).
- Wright, E.S. Using DECIPHER v2.0 to Analyze Big Biological Sequence Data in R. R J. 2016, 8, 352–359. [Google Scholar] [CrossRef]
- Methven, A.S.; Zelski, S.E.; Miller, A.N. A molecular phylogenetic assessment of the genus Gyromitra in North America. Mycologia 2013, 105, 1306–1314. [Google Scholar] [CrossRef][Green Version]
- Wang, X.-C.; Zhuang, W.-Y. A three-locus phylogeny of Gyromitra (Discinaceae, Pezizales) and discovery of two cryptic species. Mycologia 2018, 111, 69–77. [Google Scholar] [CrossRef]
- Rodríguez, F.; Oliver, J.L.; Marin, A.; Medina, J.R. The general stochastic model of nucleotide substitutions. J. Theor. Biol. 1990, 142, 485–501. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772; [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704; [Google Scholar] [CrossRef]
- Posada, D.; Buckley, T.R. Model selection and model averaging in phylogenetics: Advantages of Akaike Information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 2004, 53, 793–808. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224; [Google Scholar] [CrossRef] [PubMed]
- Hillis, D.M.; Bull, J.J. An Empirical Test of Bootstrapping as a Method for Assessing Confidence in Phylogenetic Analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Zajc, J.; Gunde-Cimerman, N. The Genus Wallemia-From Contamination of Food to Health Threat. Microorganisms 2018, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Benedix, E.H. Art- und Gattungsgrenzen bei höheren Discomyceten. III. Die Kult. 1969, 17, 253–284. [Google Scholar] [CrossRef]
- Kotlaba, F.; Pouzar, Z. Additionnal localities of Gyromitra fastigiata with notes on the generic classification of Gyromitra. Česká Mykol. 1974, 28, 84–95. [Google Scholar]
- Van Vooren, N.; Moreau, P.-A. Essai taxinomique sur le genre Gyromitra Fr. sensu lato (Pezizales). 2. Le genre Gyromitra Fr., sous-genre Gyromitra. Ascomycete.org 2009, 1, 7–14. [Google Scholar] [CrossRef]
- Popov, E.; Svetasheva, T. Gyromitra korshinskii. The IUCN Red List of Threatened Species 2019: E.T75118940A75118943. Available online: http://dx.doLorg/l0.2305/IUCN.UK.2019-2.RlTS,VSll8940A75118943.en (accessed on 25 October 2021).
- Raitviir, A. Once more on Neogyromitra caroliniana. Botaanika-Alased Tõõd 1970, 9, 364–373. [Google Scholar]
- Van Vooren, N.; Moreau, P.-A. Essai taxinomique sur le genre Gyromitra Fr. sensu lato (Pezizales). 3. Le genre Gyromitra Fr., sous-genre Discina. Ascomycete.org 2009, 1, 3–13. [Google Scholar] [CrossRef]
- Carbone, M.; Van Vooren, N.; Klener, V.; Alvarado, P. Preliminary phylogenetic and morphological studies in the Gyromitra gigas lineage (Pezizales): Epitypification of Gyromitra gigas and G. ticiniana. Ascomycete.org 2018, 10, 187–199. [Google Scholar] [CrossRef]
- Miller, A.N.; Dirks, A.C.; Filippova, N.; Popov, E.; Methven, A.S. Studies in Gyromitra II: Cryptic speciation in the Gyromitra gigas species complex; rediscovery of G. ussuriensis and G. americanigigas sp. nov. Mycol. Prog. 2022, 21, 81. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).