Comparative Proteomics Study on the Postharvest Senescence of Volvariella volvacea
Abstract
:1. Introduction
2. Materials and Methods
2.1. V. volvacea Samples
2.2. Physiological, Phenotypic, and Texture Experiments
2.3. Protein Preparation, Protein Digestion, and iTRAQ Labeling
2.4. Reverse-Phase Liquid Chromatography (RPLC) and MS Analysis
2.5. Protein Identification and Quantification
2.6. Bioinformatics and Statistical Analyses
3. Results
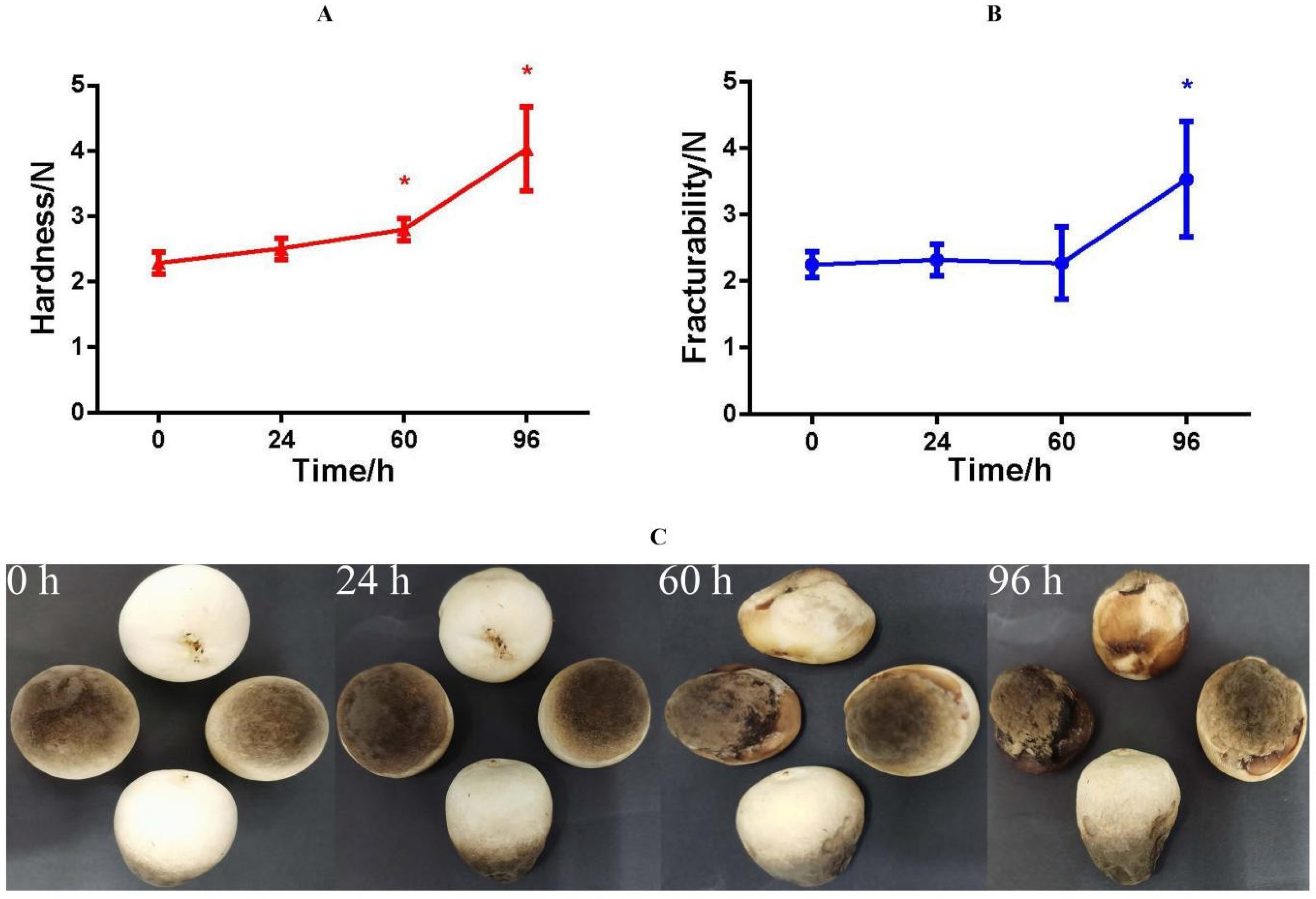
3.1. Physiological, Phenotypic, and Texture Analyses
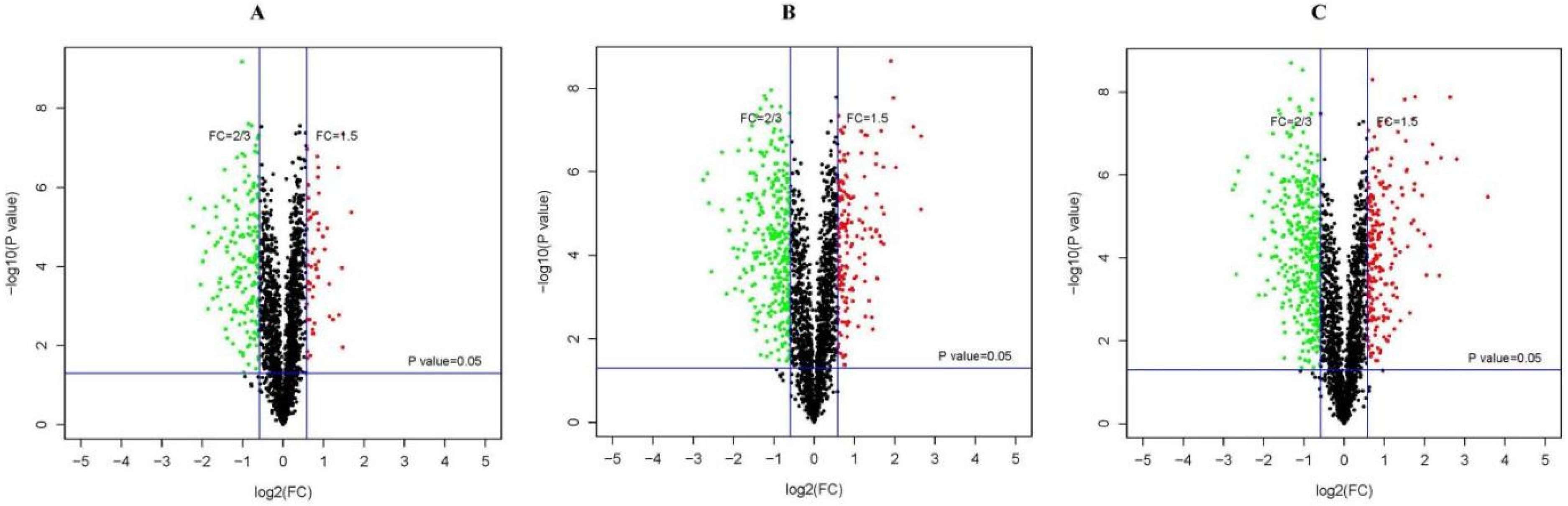
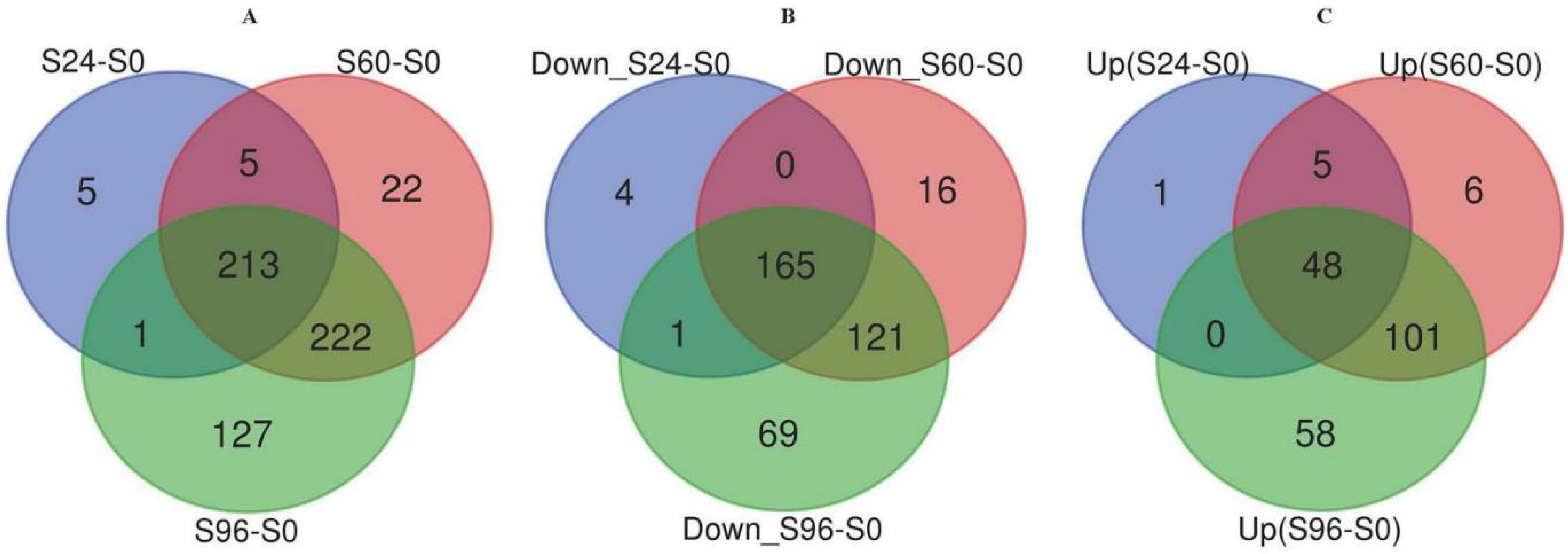
3.2. Identification of DEPs
3.3. Hierarchical Clustering Analysis
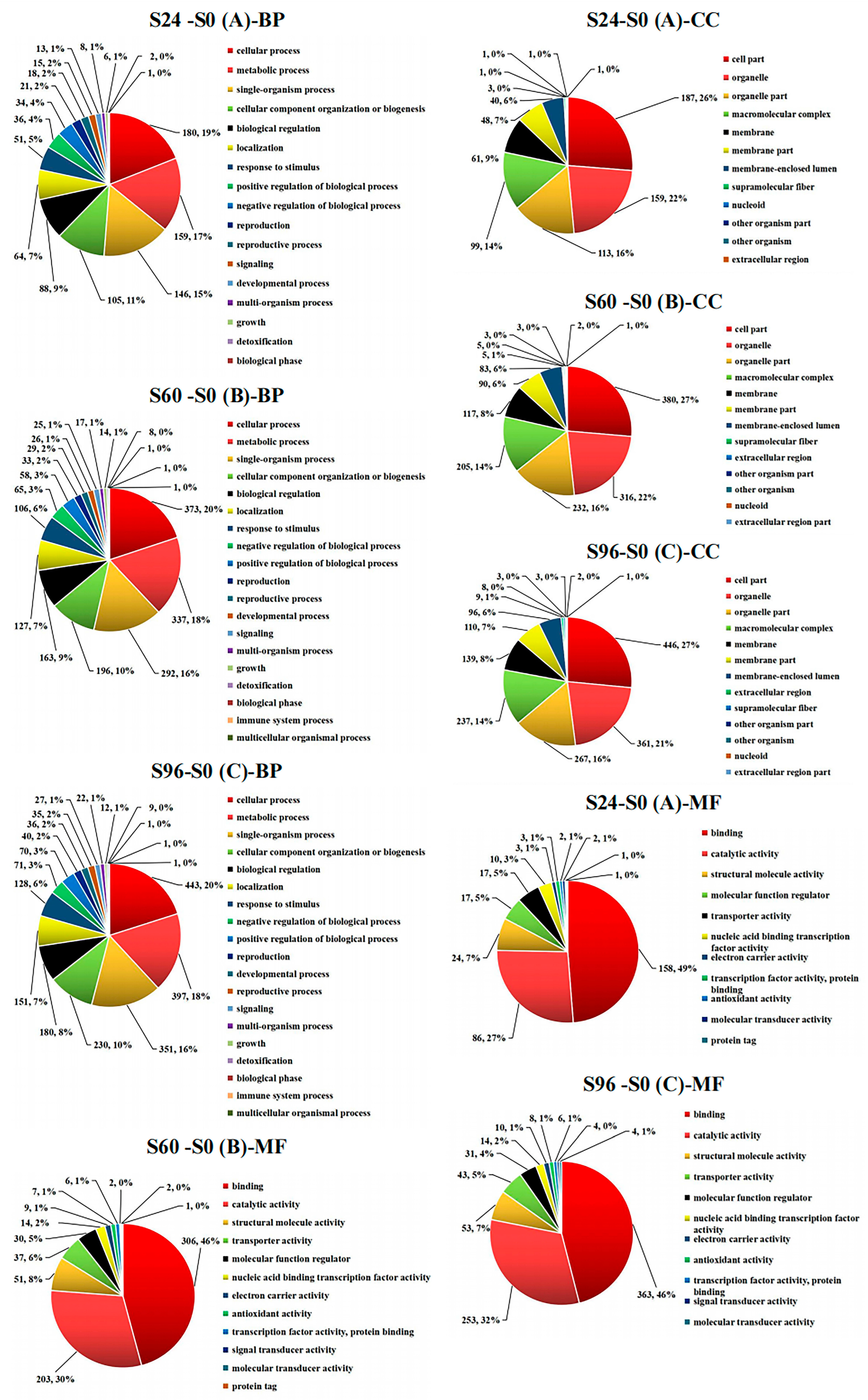
3.4. GO Analysis of DEPs
3.5. KEGG Pathway Analysis
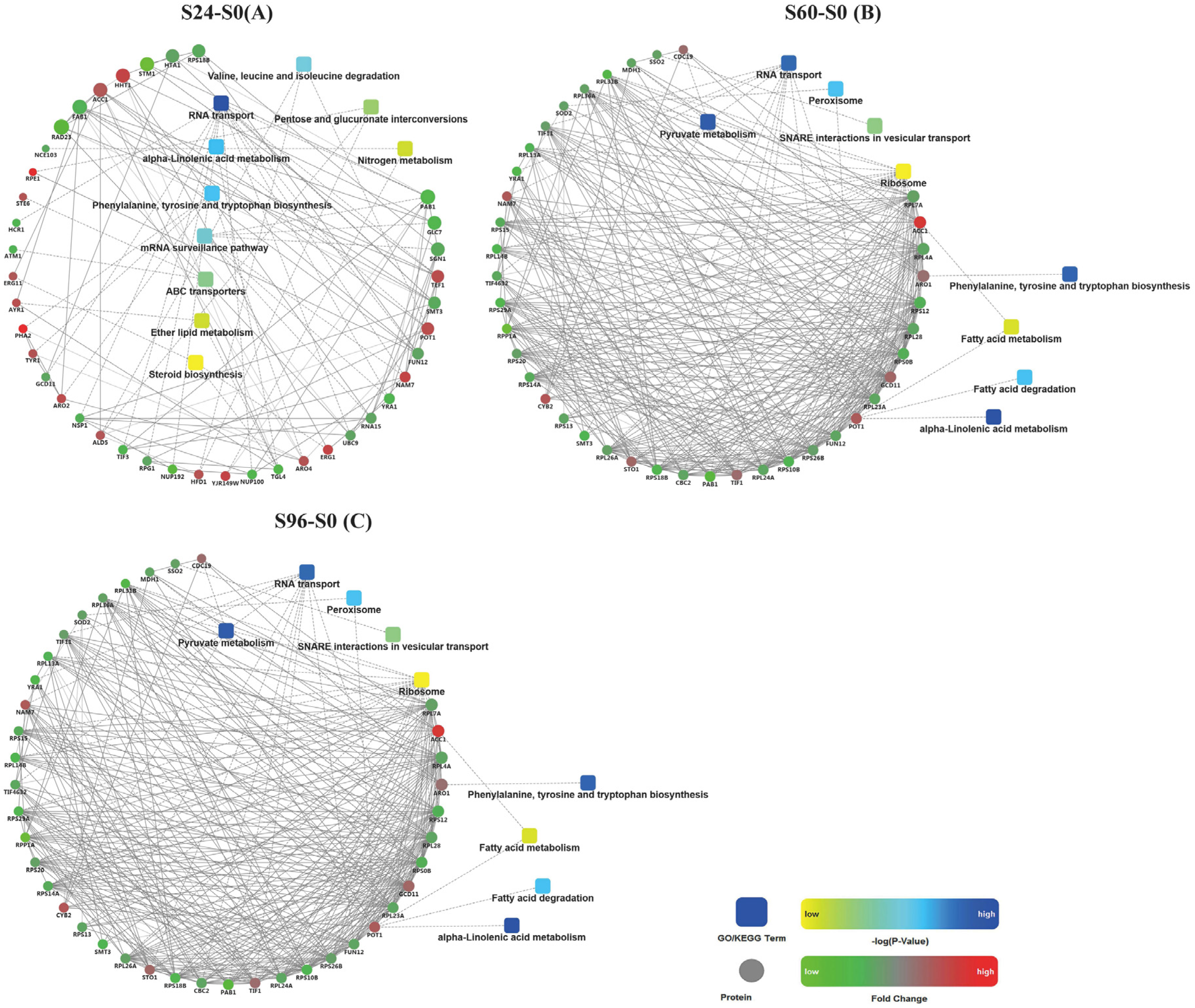
3.6. Analysis of Protein–Protein Interactions (PPIs) among DEPs
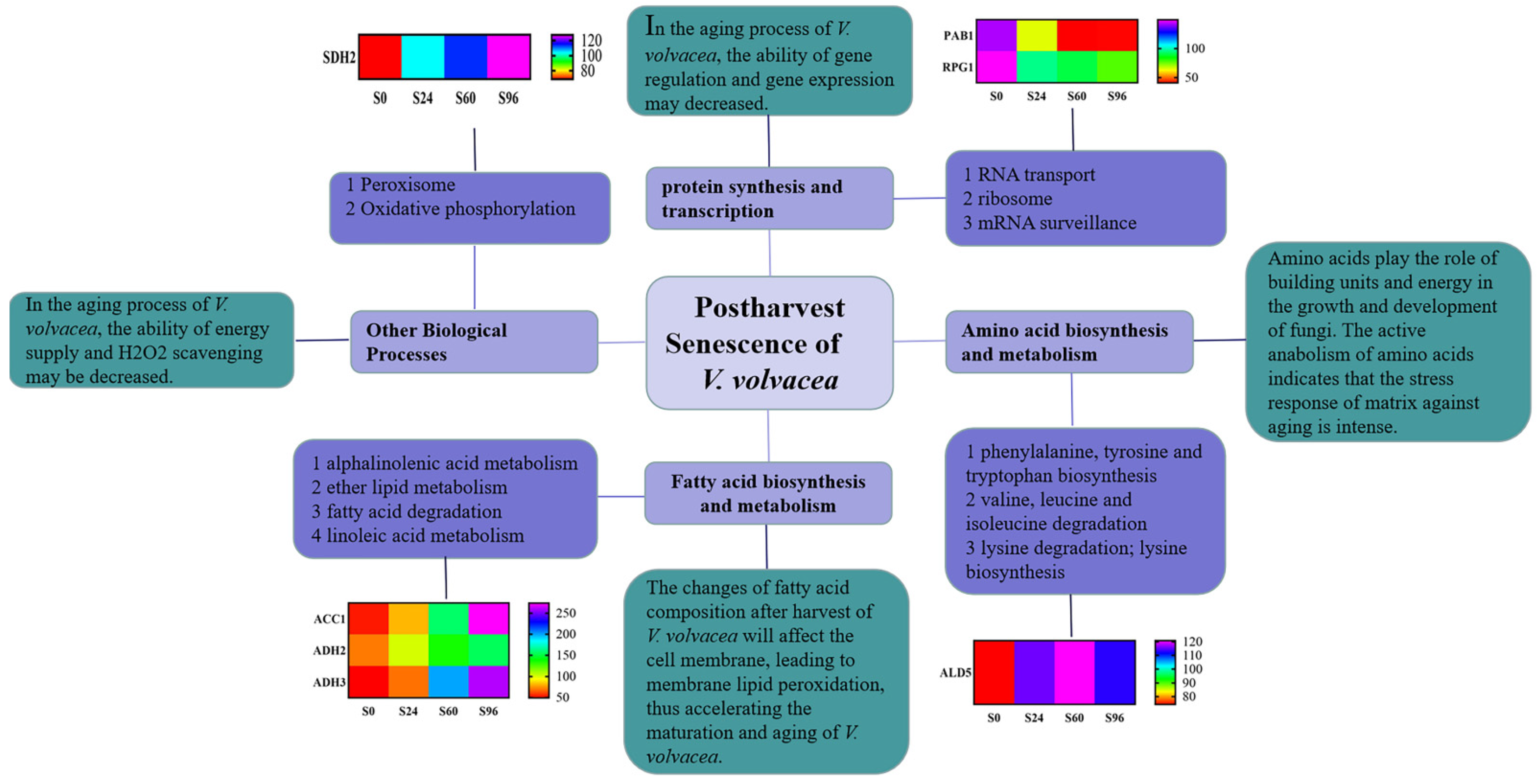
4. Discussion
4.1. Physiological Responses of V. volvacea to Suitable-Temperature Storage Conditions
4.2. DEPs in V. volvacea Stored at 15 °C after Harvest
4.2.1. DEPs Associated with RNA Transport
4.2.2. DEPs Associated with Fatty Acid Biosynthesis and Metabolism
4.2.3. DEPs Associated with Amino Acid Biosynthesis and Metabolism
4.2.4. DEPs Associated with Other Biological Processes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giovannoni, J. Molecular Biology of fruits Maturation and Ripening. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 725–749. [Google Scholar] [CrossRef] [PubMed]
- Paliyath, G.; Murr, D.P.; Handa, A.K.; Lurie, S. Postharvest Biology and Technology of Fruits, Vegetables and Flowers; Wiley-Blackwell Publishing: New Delhi, India, 2008; pp. 154–196. [Google Scholar]
- Tian, S.; Qin, G.; Li, B. Reactive oxygen species involved in regulating fruits senescence and fungal pathogenicity. Plant Mol. Biol. 2013, 82, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.L.; Huang, Y.; Wang, Z.J. Proteomic Analysis of V. volvacea in Response to Cold Stress. Food Sci. 2016, 41, 212–220. [Google Scholar] [CrossRef]
- Zha, L.; Chen, M.J.; Yu, C.X.; Guo, Q.; Zhao, X.; Li, Z.P.; Zhao, Y.; Li, C.H.; Yang, H.L. Differential proteomics study of postharvest V. volvacea during storage at 4 °C. Sci. Rep. 2020, 10, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Hnseler, E.; Nyhlén, L.E.; Rast, D.M. Isolation and properties of chitin synthetase from Agaricus bisporus mycelium. Exp. Mycol. 1983, 7, 17–30. [Google Scholar] [CrossRef]
- Schwalb, M.N. Developmentally regulated proteases from the basidiomycete Schizophyllum commune. J. Biol. Chem. 1977, 252, 8435–8439. [Google Scholar] [CrossRef]
- Chen, B.; Zhong, Y.; Chen, X.; Wu, G.; Lin, S.; Xie, B.; Jiang, Y. Study on the effect of ethephon and 1-MCP on postharvest preservation process of V. volvacea. Edible Med. Mushrooms 2018, 26, 40–43. Available online: http://en.cnki.com.cn/Article_en/CJFDTOTAL-ZSYC201801022.htm (accessed on 19 January 2018).
- Ye, H.; Chen, J.X.; Yu, R.C.; Chen, Q.L.; Liu, W. The effect of irradiation on stored V. volvacea and the physiological mechanism. Acta Agric. Nucleatae Sinic. 2000, 14, 24–28. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?FileName=HNXB200001005&DbName=CJFQ2000 (accessed on 20 February 2000).
- Dhalsamant, K.; Dash, S.K.; Bal, L.M.; Panda, M.K. Effect of perforation mediated MAP on shelf life of mushroom (V. volvacea). Sci. Hortic. 2015, 189, 41–50. [Google Scholar] [CrossRef]
- Wu, G. Effect of microwave treatment, coating with chitosan and xanthan gum, and storage temperature on the strage quality of V. volvacea fruits bodies. Acta Edulis Fungi 2009, 16, 45–50. [Google Scholar] [CrossRef]
- Gong, M.; Wang, H.; Chen, M.; Bao, D.; Zhu, Q.; Tan, Q. A newly discovered ubiquitin-conjugating enzyme E2 correlated with the cryogenic autolysis of V. volvacea. Gene 2016, 583, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.; Wang, H.; Chen, M.; Xue, C.; Li, Z.; Bian, Y.; Bao, D. Application of mating type genes in molecular marker-assisted breeding of the edible V. volvacea. Sci. Hortic. 2014, 180, 59–62. [Google Scholar] [CrossRef]
- Palma, J.M.; Corpas, F.J.; Río, L.A.D. Proteomics as an approach to the understanding of the molecular physiology of fruits development and ripening. China Fruits Veg. 2011, 74, 1230–1243. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.H.; Tan, Q.; Bao, D.P.; Zhang, X.H.; Jian, H.H.; Li, Y.; Yang, R.H.; Wang, Y. Comparative Proteomic analysis of light-Induced mycelial brown film formation in Lentinula edodes. BioMed Res. Int. 2016, 2016, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y.; Chen, H.; Tang, M.; Shen, S. Proteome analysis of an ectomycorrhizal fungus Boletus edulis under salt shock. Mycol. Res. 2007, 111, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Men, J.L.; Chang, M.C.; Feng, C.P.; Yuan, L.G. iTRAQ-based quantitative proteome revealed metabolic changes of Flammulina velutipes mycelia in response to cold stress. J. Proteom. 2017, 156, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, Y. Protein expression during Flammulina velutipes fruiting bodies formation. Mycoence 2010, 51, 163–169. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, B.B.; Cheung, P.C.K. Comparative proteomic analysis of mushroom cell wall proteins among the different developmental stages of Pleurotus tuber-regium. J. Agric. Food Chem. 2012, 60, 6173–6182. [Google Scholar] [CrossRef] [PubMed]
- Rahmad, N.; Al-Obaidi, J.R.; Nor Rashid, N.M.; Zean, N.B.; Mohd Yusoff, M.H.Y.; Shaharuddin, N.S.; Mohd Jamil, N.A.; Mohd Saleh, N. Comparative proteomic analysis of different developmental stages of the edible mushroom Termitomyces heimii. Biol. Res. 2014, 47, 2–8. [Google Scholar] [CrossRef] [Green Version]
- Horie, K.; Rakwal, R.; Hirano, M.; Shibato, J.; Nam, H.W.; Kim, Y.S.; Kouzuma, Y.; Agrawal, G.K.; Masuo, Y.; Yonekura, M. Proteomics of two cultivated mushrooms Sparassis crispa and Hericium erinaceum provides insight into their numerous functional protein components and diversity. J. Proteome Res. 2008, 7, 1819–1835. [Google Scholar] [CrossRef] [PubMed]
- Zha, L.; Chen, M.J.; Yan, S.H.; Li, C.H.; Wang, H.; Xi, L.P.; Wang, C.G.; Zhao, Y. Morphological and physiological indexes of V. volvacea during storage at 15 °C. Food Mach. 2018, 34, 110–114. [Google Scholar] [CrossRef]
- Wu, J.; Fu, Q.L.; Li, J.J.; Zhu, X.F.; Li, M. The physiology effect of selenium on postharvest tomato fruits ripening andsenescence. J. Anhui Agric. Univ. 2020, 47, 155–160. [Google Scholar] [CrossRef]
- Isaacson, T.; Damasceno, C.M.B.; Saravanan, R.S.; He, Y.; Catalá, C.; Saladié, M.; Rose, J.K.C. Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat. Protoc. 2006, 1, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Cui, W.; Pan, J.; Xie, Y.; Wang, J.; Shen, W. Proteomic analysis provides insights into the molecular bases of hydrogen gas-induced cadmium resistance in Medicago sativa. J. Proteom. 2017, 152, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.P.; Li, X.H.; Yang, L.J.; Pan, Y.F.; Tang, X.P.; Tang, Y. Research on optimum storage temperatures and characteristics of fresh Siraitia grosvenorii. Food Sci. Technol. 2017, 42, 44–48. [Google Scholar] [CrossRef]
- Zhou, Y.J.; Gao, H.Y.; Chen, H.J.; Tao, F.; Fang, X.J. Effects of hypobaric storage on reactive oxygen species metabolism of Pleurotus eryngii during cold storage. J. Nucl. Agric. Sci. 2015, 29, 1108–1113. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?FileName=HNXB201506013&DbName=CJFQ2015 (accessed on 27 June 2015).
- Tian, H.Y.; Rao, J.P. Fresh-keeping effect of chlorine dioxide on Hayward Kiwi fruits with mechanical damage. Food Sci. 2012, 33, 298–302. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?FileName=SPKX201218065&DbName=CJFQ2012 (accessed on 25 September 2012).
- Yang, X.X. Related Genes and Regularity Effect Stress Granules Formation in Saccharomyces cerevisiae Hansen. Ph.D. Thesis, Harbin Institute of Technology, Harbin, China, 2015. Available online: https://kns.cnki.net/KCMS/detail/detail.aspx?dbname=CDFDLAST2016&filename=1015957481.nh (accessed on 1 June 2015).
- Guan, Y.F.; Huang, X.Y.; Zhu, J.; Gao, J.F.; Zhang, H.X.; Yang, Z.N. Ruptured pollen grain1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol. 2008, 147, 852–863. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grigull, J.; Mnaimneh, S.; Pootoolal, J.; Robinson, M.D.; Hughes, T.R. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays revea. J. Mol. Cell Biol. 2004, 24, 5534–5547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warringer, J.; Hult, M.; Regot, S.; Posas, F.; Sunnerhagen, P. The HOG pathway dictates the short-term translational response after hyperosmotic shock. Mol. Biol. Cell. 2010, 21, 3080–3092. [Google Scholar] [CrossRef] [PubMed]
- Salas, J.J.; Ohlrogge, J.B. Characterization of substrate specificity of plant FatA and FatB acyl-ACP thioesterases. Arch. Biochem. Biophys. 2002, 403, 25–34. [Google Scholar] [CrossRef]
- Li, Z.G.; Yin, W.B.; Song, L.Y.; Chen, Y.H.; Guan, R.Z.; Wang, J.Q.; Wang, R.R.C.; Hu, Z.M. Genes encoding the biotin carboxylase subunit of acetyl-CoA carboxylase from Brassica napus and parental species: Cloning, expression patterns, and evolution. Genome 2011, 53, 360–370. [Google Scholar] [CrossRef]
- Andre, C.; Haslam, R.P.; Shanklin, J. Feedback regulation of plastidic acetyl-CoA carboxylase by 18:1-acyl carrier protein in Brassica napus. Proc. Natl. Acad. Sci. USA 2012, 109, 10107–10112. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.J.; Zhang, L.L.; Zhang, Y. Study on major fatty acids during rapeseed changes of main maturing process. J. Chin. Cereals Oils Assoc. 2016, 31, 82–88. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?FileName=ZLYX201607017&DbName=CJFQ2016 (accessed on 25 July 2016).
- Huang, W.; Ma, X.; Wang, Q.; Gao, Y.; Xue, Y.; Niu, X.; Yu, G.; Liu, Y. Significant improvement of stress tolerance in tobacco plants by overexpressing a stress-responsive aldehyde dehydrogenase gene from maize (Zea mays). Plant Mol. Biol. 2008, 68, 451–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y. Expression of alcohol dehydrogenase 3 in young roots of different salt sensitive rice varieties under Salt Stress. Jiangsu Agric. Sci. 2013, 41, 33–35. [Google Scholar] [CrossRef]
- Pezeshki, S.R.; Delaune, R.D.; Patrick, W.H. Differential response of selected mangroves to soil flooding and salinity: Gas exchange and biomass partitioning. Can. J. For. Res. 1990, 20, 869–874. [Google Scholar] [CrossRef]
- Zhao, S.; Chen, Y.H.; Chen, H.; Xiao, G.Y. Dynamic analysis of Adh2 gene of rice (Oryza sativa L.) under submergence stress using real-time quantitative PCR. Chin. J. Eco-Agric. 2008, 16, 455–458. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?FileName=ZGTN200802038&DbName=CJFQ2008 (accessed on 18 March 2008). [CrossRef]
- Yan, J.P.; Liang, Y.; Tan, X.L. Expression of ADHa and BADH in young root of cotton (Gossypium hirsutum) under waterlogged stress. China Cotton. 2011, 38, 22–24. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?FileName=ZMZZ201110008&DbName=CJFQ2011 (accessed on 15 October 2011).
- Wang, N.B.; Zhao, J.; He, X.Y.; Sun, H.Y.; Zhang, G.P.; Wu, F.B. Comparative proteomic analysis of drought tolerance in the two contrasting Tibetan wild genotypes and cultivated genotype. BMC Genom. 2015, 16, 432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, V. Recent progress in deciphering the biosynthesis of aspartate-derived amino acids in plants. Mol. Plant 2010, 3, 54–65. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef]
- Yu, H.; Wang, T. Proteomic dissection of endosperm starch granule associated proteins reveals a network coordinating starch biosynthesis and amino acid metabolism and glycolysis in rice endosperms. Front. Plant Sci. 2016, 1, 707. [Google Scholar] [CrossRef]
- Reinbothe, C.; Pollmann, S.; Reinbothe, S. Singlet oxygen signaling links photosynthesis to translation and plant growth. Trends Plant Sci. 2010, 15, 499–506. [Google Scholar] [CrossRef]
- Rothman, S. How is the balance between protein synthesis and degradation achieved. Theor. Biol. Med. Model. 2010, 7, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Fang, D.; Yang, W.; Deng, Z.; An, X.; Zhao, L.; Hu, Q. Proteomic investigation of metabolic changes of mushroom (Flammulina velutipes) packaged with nanocomposite material during cold storage. J. Agric. Food Chem. 2017, 65, 10368–10381. [Google Scholar] [CrossRef]
- Saint-Prix, F.; Boenquist, L.; Dequin, S. Functional analysis of the ALD gene family of Saccharomyces cerevisiae during anaerobic growth on glucose: The NADP+-dependent Ald6p and Ald5p isoforms play a major role in acetate formation. Microbiology 2004, 150, 2209–2220. [Google Scholar] [CrossRef]
- Blomberg, A.; Adler, L. Roles of glycerol and glycerol-3-phosphate dehydrogenase (NAD+) in acquired osmotolerance of Saccharomyces cerevisiae. J. Bacteriol. 1989, 171, 1087–1092. [Google Scholar] [CrossRef] [Green Version]
- Akhtar, N.; Blomberg, A.; Adler, L. Osmoregulation and protein expression in a pbsv2 mutant of Saccharomyces cerevisiae during adaptation to hypersaline stress. FEBS Lett. 1997, 403, 173–180. [Google Scholar] [CrossRef] [Green Version]
- Osamu, K.; Yoshio, N. Involvement of mitochondrial aldehyde dehydrogenase ALD5 in maintenance of the mitochondrial electron transport chain in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 1999, 181, 281–287. [Google Scholar] [CrossRef]
- Zhan, Y.; Liu, D.W.; Li, L.; Jiang, W.; Liu, Z.H. Response of respiratory enzyme activity to waterlogging in peanut seedlings. J. Peanut 2019, 48, 63–66, 74. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?FileName=PEAN201904011&DbName=CJFQ2019 (accessed on 15 December 2019).
- Zhao, Y.Y.; Chen, J.J.; Jin, P.; Yuan, R.X.; Li, H.H.; Zheng, Y.H. Effects of low temperature conditioning on chilling injury and energy status in cold-stored peach fruit. J. Food Sci. 2012, 33, 276–281. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?FileName=SPKX201204060&DbName=CJFQ2012 (accessed on 25 February 2012).
- Lombardo, A.; Carine, K.; Scheffler, I.E. Cloning and characterization of the iron-sulfur subunit gene of succinate dehydrogenase from Saccharomyces cerevisiae. J. Biol. Chem. 1990, 265, 10419–10423. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zha, L.; Chen, M.; Guo, Q.; Tong, Z.; Li, Z.; Yu, C.; Yang, H.; Zhao, Y. Comparative Proteomics Study on the Postharvest Senescence of Volvariella volvacea. J. Fungi 2022, 8, 819. https://doi.org/10.3390/jof8080819
Zha L, Chen M, Guo Q, Tong Z, Li Z, Yu C, Yang H, Zhao Y. Comparative Proteomics Study on the Postharvest Senescence of Volvariella volvacea. Journal of Fungi. 2022; 8(8):819. https://doi.org/10.3390/jof8080819
Chicago/Turabian StyleZha, Lei, Mingjie Chen, Qian Guo, Zongjun Tong, Zhengpeng Li, Changxia Yu, Huanling Yang, and Yan Zhao. 2022. "Comparative Proteomics Study on the Postharvest Senescence of Volvariella volvacea" Journal of Fungi 8, no. 8: 819. https://doi.org/10.3390/jof8080819
APA StyleZha, L., Chen, M., Guo, Q., Tong, Z., Li, Z., Yu, C., Yang, H., & Zhao, Y. (2022). Comparative Proteomics Study on the Postharvest Senescence of Volvariella volvacea. Journal of Fungi, 8(8), 819. https://doi.org/10.3390/jof8080819





