AGC/AKT Protein Kinase SCH9 Is Critical to Pathogenic Development and Overwintering Survival in Magnaporthe oryzae
Abstract
:1. Introduction
2. Materials and Methods
2.1. MoSch9 Identification and Sequence Alignment
2.2. Fungal Isolates, Growth, and Storage Conditions
2.3. Targeted Gene Deletion of MoSch9 Mutants and Generation of Complementation Strains
2.4. Assays for Conidial Production, Germination, and Appressorium Development
2.5. Pathogenicity, Rice Root Infection, and Penetration Assays
2.6. Stress and Freeze Tolerance Assay
2.7. RNA Extraction, Real-Time qPCR, and RNA Seq Analysis
2.8. Statistical Analysis
3. Results
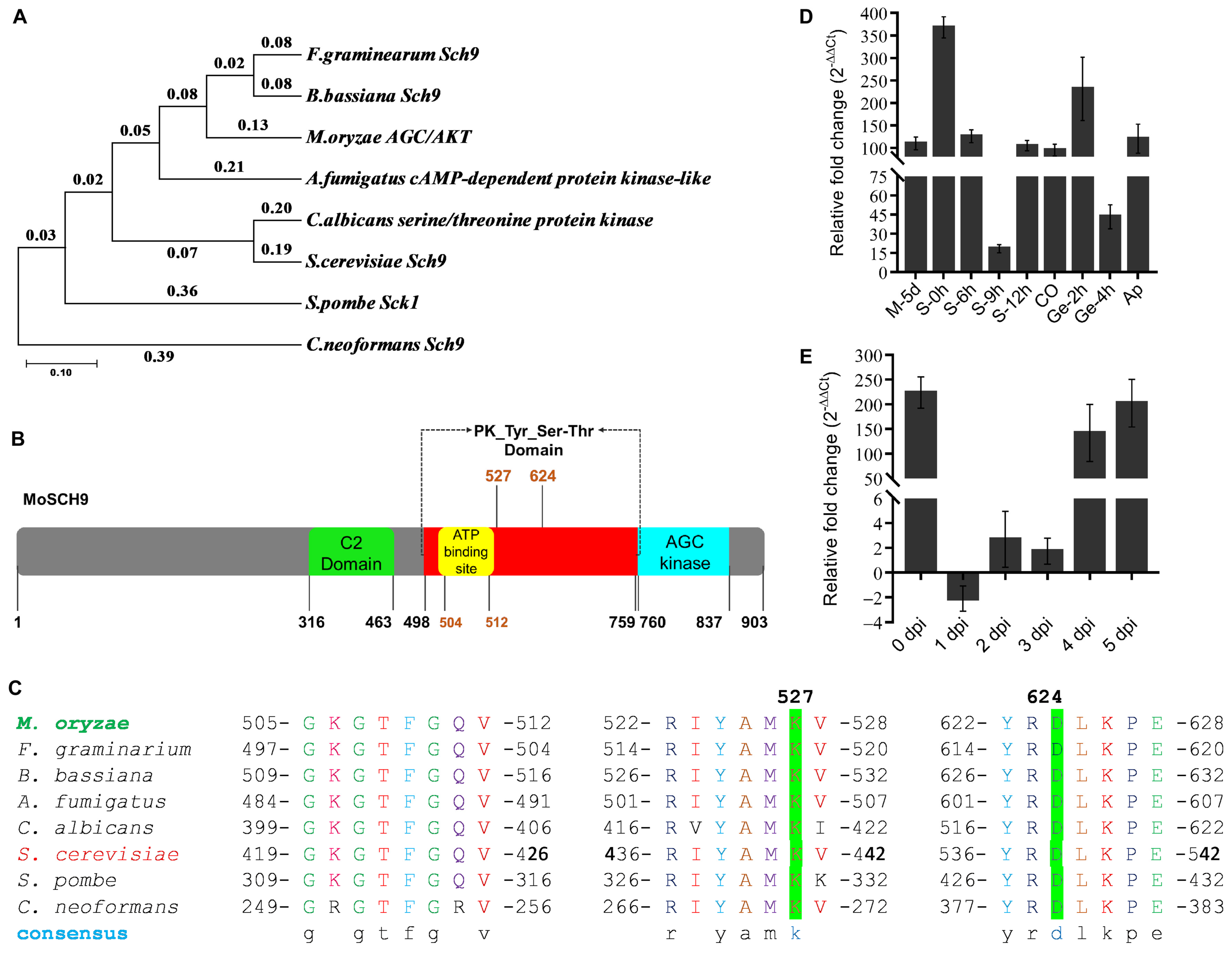
3.1. Phylogenetic Analysis and Expression Profiling of MoSCH9 in Magnaporthe Oryzae
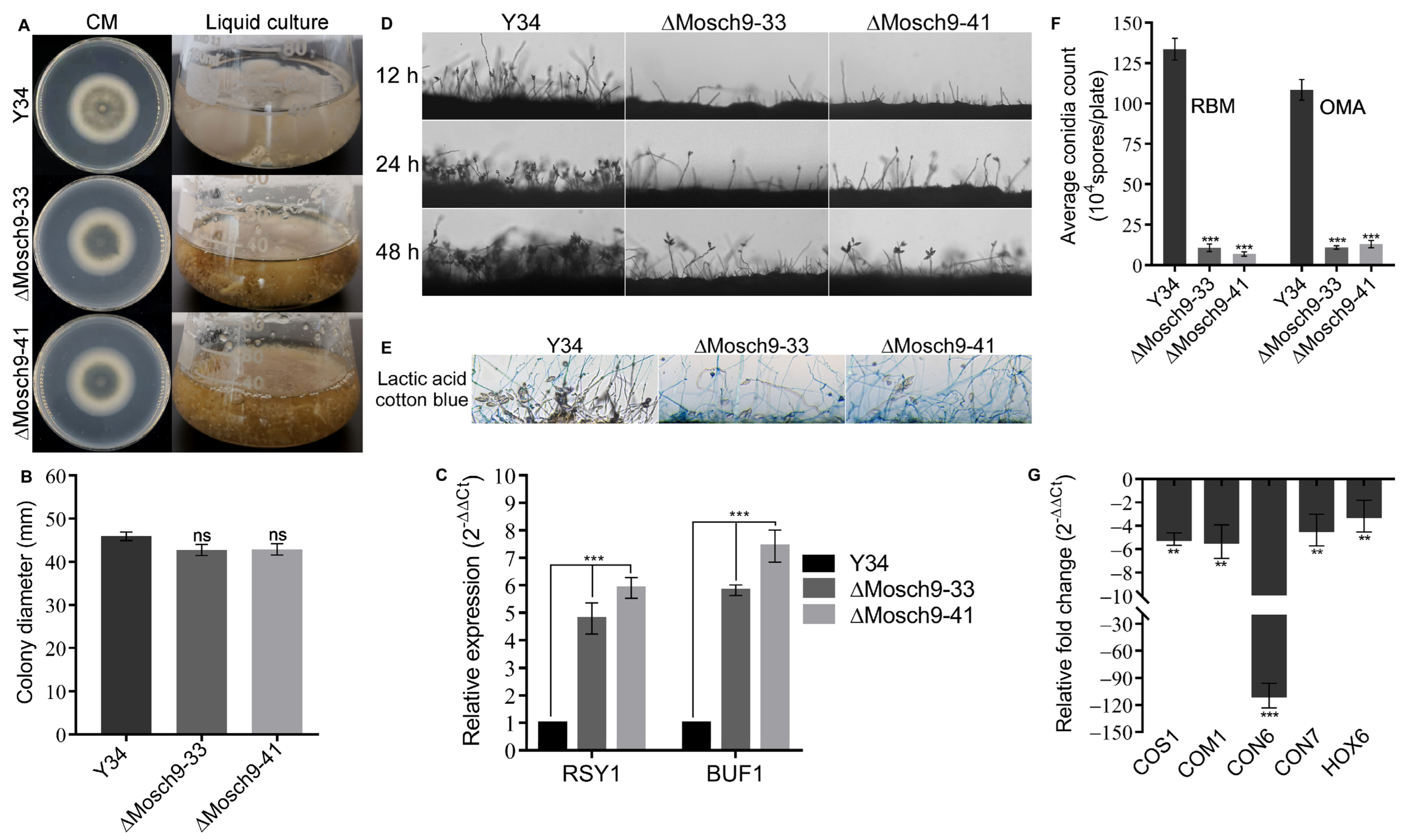
3.2. Deletion of MoSch9 Gene Has a Significant Effect on Hyphal Melanization and Conidiogenesis in M. Oryzae
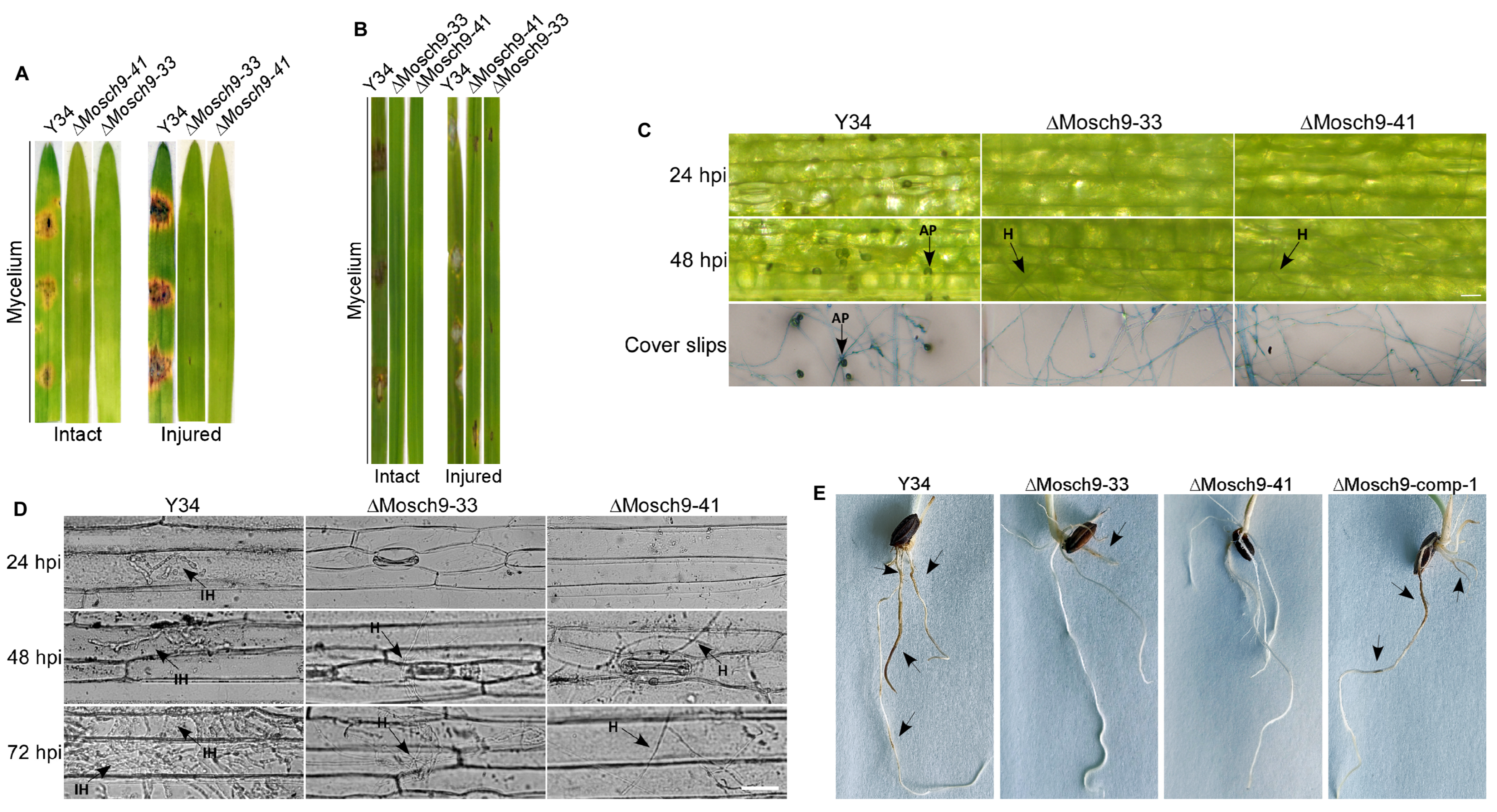
3.3. Deletion of MoSch9 Completely Abolished Hyphal Penetration of M. Oryzae into Host Tissues
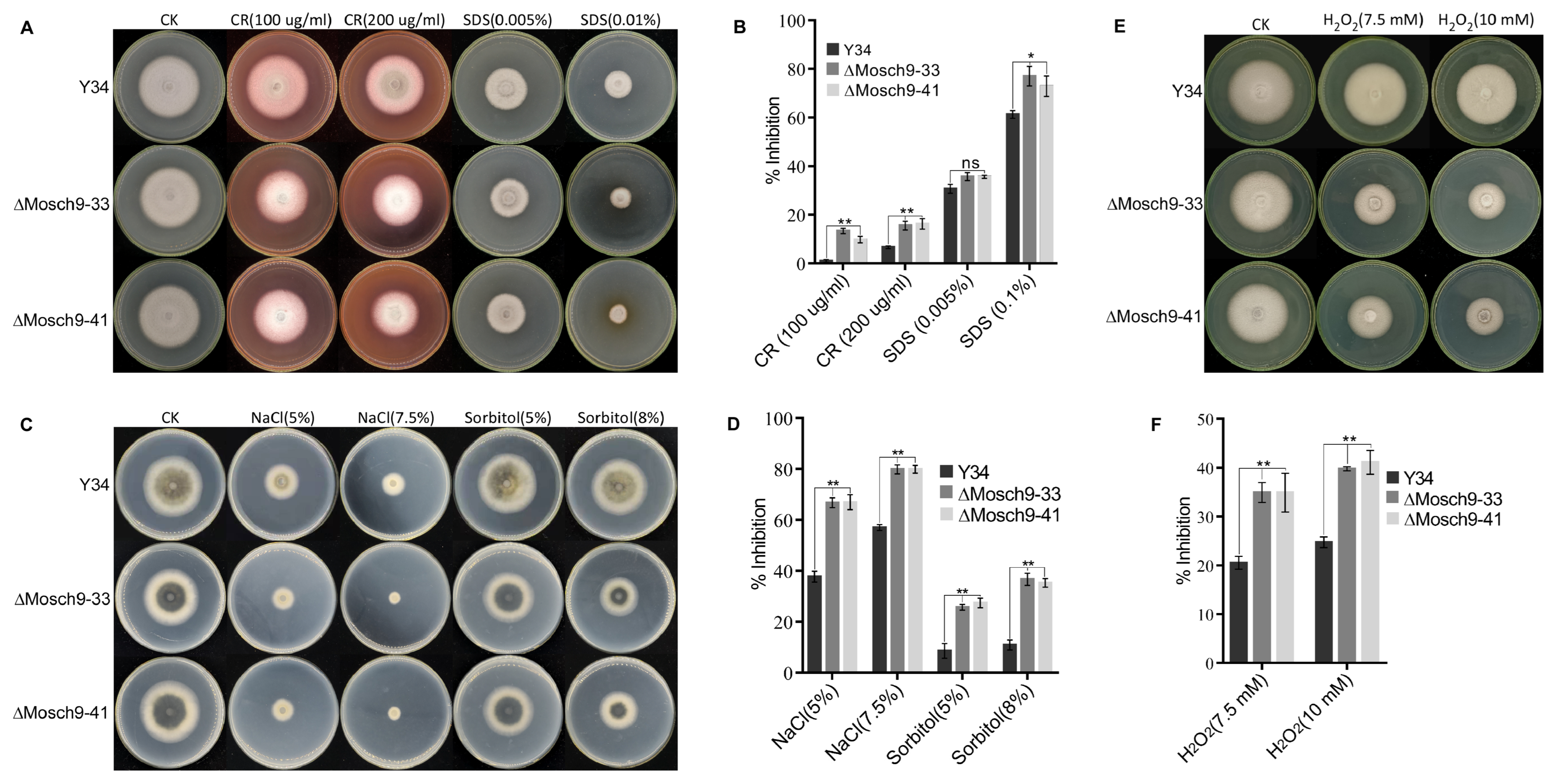
3.4. MoSch9 Deletion Affects Multiple Environmental Stress Tolerance of Magnaporthe Oryzae
3.5. MoSch9 Is Associated with Cold Tolerance and Critical for M. Oryzae Overwintering Survival
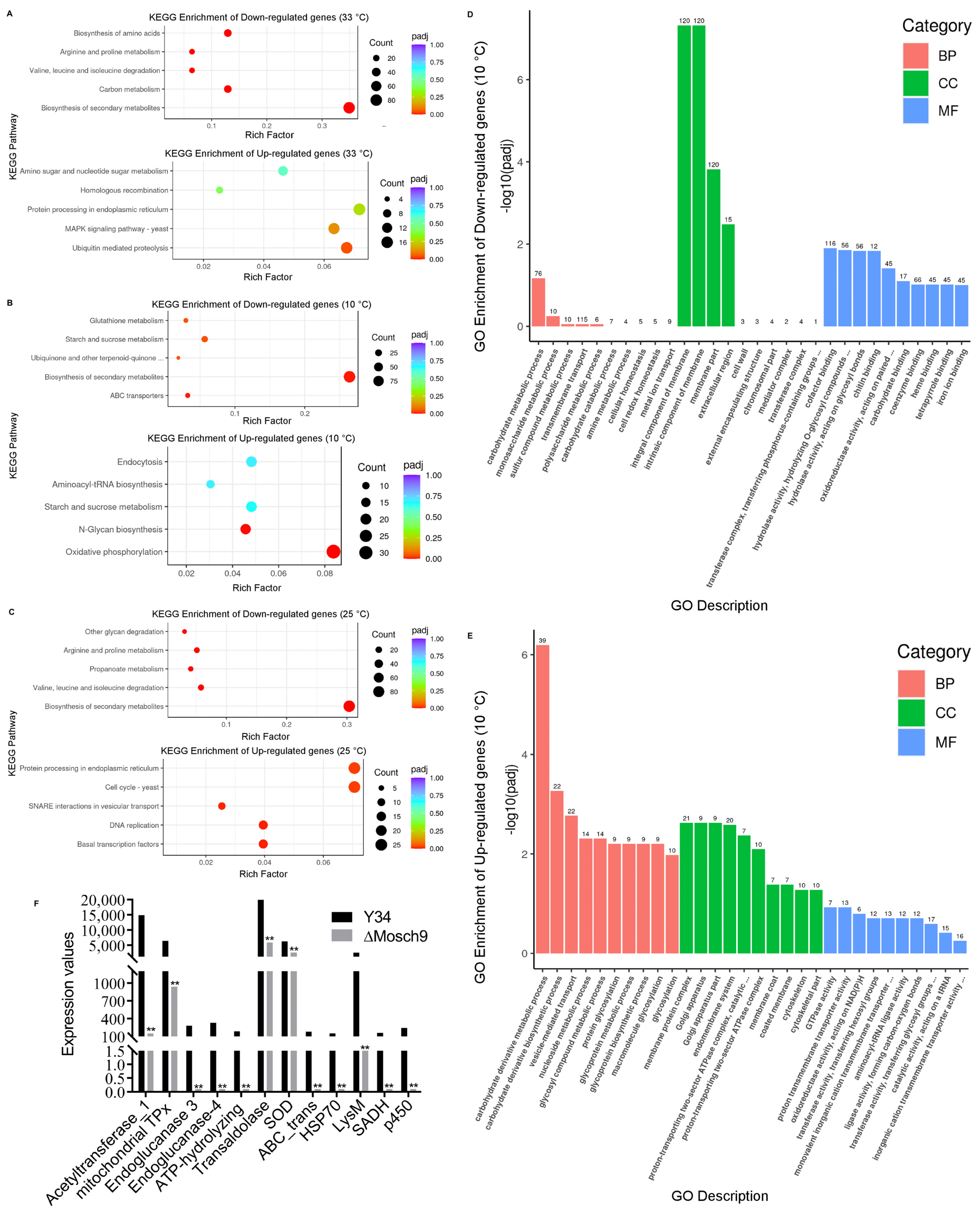
3.6. Transcriptomic Profile of M. Oryzae Wild Type and ΔMoSch9 Mutant Strains during Cold Temperature Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giraud, T.; Gladieux, P.; Gavrilets, S. Linking the emergence of fungal plant diseases with ecological speciation. Trends Ecol. Evol. 2010, 25, 387–395. [Google Scholar] [CrossRef] [Green Version]
- Fischer, N.M.; Dool, S.E.; Puechmaille, S.J. Seasonal patterns of Pseudogymnoascus destructans germination indicate host–pathogen coevolution. Biol. Lett. 2020, 16, 20200177. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, G.; Lin, C.; He, C. Conidiophore Stalk-less1 Encodes a Putative Zinc-Finger Protein Involved in the Early Stage of Conidiation and Mycelial Infection in Magnaporthe oryzae. Mol. Plant-Microbe Interact. 2009, 22, 402–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soubeyrand, S.; Laine, A.-L.; Hanski, I.; Penttinen, A. Spatiotemporal Structure of Host-Pathogen Interactions in a Metapop-ulation. Am. Nat. 2009, 174, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Stack, R.W. A Comparison of the Inoculum Potential of Ascospores and Conidia of Gibberella Zeae. Can. J. Plant Pathol. 1989, 11, 137–142. [Google Scholar] [CrossRef]
- Xu, X.; Ma, L.; Hu, X. Overwintering of Wheat Stripe Rust Under Field Conditions in the Northwestern Regions of China. Plant Dis. 2019, 103, 638–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerling, M.; Pätzig, M.; Hempel, L.; Büttner, C.; Müller, M.E.H. Arable Weeds at the Edges of Kettle Holes as Overwintering Habitat for Phytopathogenic Fungi. Agronomy 2022, 12, 823. [Google Scholar] [CrossRef]
- Buruchara, R.; Mukaruziga, C.; Ampofo, K.O. Bean Disease and Pest Identification and Management; Centro Internacional de Agricultura Tropical International Center for Tropical Agriculture: Kamplala, Uganda, 2010.
- Fourie, D. Distribution and Severity of Bacterial Diseases on Dry Beans (Phaseolus vulgaris L.) in South Africa. J. Phytopathol. 2002, 150, 220–226. [Google Scholar] [CrossRef]
- Endert, E.; Ritchie, D.F. Overwintering and Survival of Pseudomonas syringae Pv. Syringae and Symptom Development in Peach Trees. Plant Dis. 1984, 68, 468–470. [Google Scholar] [CrossRef] [Green Version]
- Sundin, G.W.; Jones, A.L.; Olson, B.D. Overwintering and Population Dynamics of Pseudomonas syringae Pv. Syringae and Ps Pv. morsprunorum on Sweet and Sour Cherry Trees. Can. J. Plant Pathol. 1988, 10, 281–288. [Google Scholar] [CrossRef]
- Raj, K.; Pal, V. Overwintering of Xanthomonas campestris Pv. Oryzae. Int. Rice Res. Newsl. 1988, 13, 22–23. [Google Scholar]
- Schaechter, M. Encyclopedia of Microbiology; Academic Press: Cambridge, MA, USA, 2009; ISBN 0123739446. [Google Scholar]
- Staib, P.; Morschhäuser, J. Chlamydospore Formation in Candida albicans and Candida dubliniensis–an Enigmatic Developmental Programme. Mycoses 2007, 50, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Elad, Y.; Baker, R. The Role of Competition for Iron and Carbon in Suppression of Chlamydospore Germination of Fusarium Spp. by Pseudomonas spp. Phytopathology 1985, 75, 1053–1059. [Google Scholar] [CrossRef]
- Wagner, L.; Stielow, B.; Hoffmann, K.; Petkovits, T.; Papp, T.; Vágvölgyi, C.; De Hoog, G.S.; Verkley, G.; Voigt, K. A Com-prehensive Molecular Phylogeny of the Mortierellales (Mortierellomycotina) Based on Nuclear Ribosomal DNA. Per-Soonia-Mol. Phylogeny Evol. Fungi 2013, 30, 77–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dennis, R.L. Fossil Mycelium with Clamp Connections from the Middle Pennsylvanian. Science 1969, 163, 670–671. [Google Scholar] [CrossRef]
- Griffith, M.; Yaish, M.W. Antifreeze proteins in overwintering plants: A tale of two activities. Trends Plant Sci. 2004, 9, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Crocker, E.V.; Lanzafane, J.J.; Karp, M.A.; Nelson, E.B. Overwintering seeds as reservoirs for seedling pathogens of wetland plant species. Ecosphere 2016, 7, e01281. [Google Scholar] [CrossRef] [Green Version]
- Summerell, B.; Burgess, L. Factors influencing survival of Pyrenophora tritici-repentis: Stubble management. Mycol. Res. 1989, 93, 38–40. [Google Scholar] [CrossRef]
- Pereyra, S.A.; Dill-Macky, R.; Sims, A.L. Survival and Inoculum Production of Gibberella zeae in Wheat Residue. Plant Dis. 2004, 88, 724–730. [Google Scholar] [CrossRef] [Green Version]
- Raveloson, H.; Ramonta, I.R.; Tharreau, D.; Sester, M. Long-term survival of blast pathogen in infected rice residues as major source of primary inoculum in high altitude upland ecology. Plant Pathol. 2017, 67, 610–618. [Google Scholar] [CrossRef] [Green Version]
- Faivre-Rampant, O.; Geniès, L.; Piffanelli, P.; Tharreau, D. Transmission of rice blast from seeds to adult plants in a non-systemic way. Plant Pathol. 2012, 62, 879–887. [Google Scholar] [CrossRef]
- Akase, K.; Kusaba, M. Overwintering of Pyricularia oryzae in wild infected foxtails. J. Gen. Plant Pathol. 2017, 83, 197–204. [Google Scholar] [CrossRef]
- Borgia, P.T. Roles of the orlA, tsE, and bimG genes of Aspergillus nidulans in chitin synthesis. J. Bacteriol. 1992, 174, 384–389. [Google Scholar] [CrossRef] [Green Version]
- Suresh, K.; Subramanyam, C. A Putative Role for Calmodulin in the Activation of Neurospora crassa Chitin Synthase. FEMS Microbiol. Lett. 1997, 150, 95–100. [Google Scholar] [CrossRef]
- Manning, G.; Plowman, G.D.; Hunter, T.; Sudarsanam, S. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 2002, 27, 514–520. [Google Scholar] [CrossRef]
- Bao, J.; Chen, M.; Zhong, Z.; Tang, W.; Lin, L.; Zhang, X.; Jiang, H.; Zhang, D.; Miao, C.; Tang, H.; et al. PacBio Sequencing Reveals Transposable Elements as a Key Contributor to Genomic Plasticity and Virulence Variation in Magnaporthe oryzae. Mol. Plant 2017, 10, 1465–1468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinha, A.K.; Jaggi, M.; Raghuram, B.; Tuteja, N. Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal. Behav. 2011, 6, 196–203. [Google Scholar] [CrossRef] [Green Version]
- Hamel, L.-P.; Nicole, M.-C.; Duplessis, S.; Ellis, B.E. Mitogen-Activated Protein Kinase Signaling in Plant-Interacting Fungi: Distinct Messages from Conserved Messengers. Plant Cell 2012, 24, 1327–1351. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Wang, Y.; Zhou, X.; Wang, Y.; Xu, J.-R. The Sch9 Kinase Regulates Conidium Size, Stress Responses, and Pathogenesis in Fusarium graminearum. PLoS ONE 2014, 9, e105811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toda, T.; Cameron, S.; Sass, P.; Wigler, M. SCH9, a gene of Saccharomyces cerevisiae that encodes a protein distinct from, but functionally and structurally related to, cAMP-dependent protein kinase catalytic subunits. Genes Dev. 1988, 2, 517–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterson, P.P.; Liu, Z. Identification and Characterization of Rapidly Accumulating Sch9Δ Suppressor Mutations in Saccha-romyces cerevisiae. G3 2021, 11, jkab134. [Google Scholar] [CrossRef] [PubMed]
- Deprez, M.-A.; Eskes, E.; Wilms, T.; Ludovico, P.; Winderickx, J. PH Homeostasis Links the Nutrient Sensing PKA/TORC1/Sch9 Menage-a-Trois to Stress Tolerance and Longevity. Microb. Cell 2018, 5, 119. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Moir, R.D.; Willis, I.M. Regulation of RNA Polymerase III Transcription Involves SCH9-dependent and SCH9-independent Branches of the Target of Rapamycin (TOR) Pathway. J. Biol. Chem. 2009, 284, 12604–12608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batool, W.; Shabbir, A.; Lin, L.; Chen, X.; An, Q.; He, X.; Pan, S.; Chen, S.; Chen, Q.; Wang, Z.; et al. Translation Initiation Factor eIF4E Positively Modulates Conidiogenesis, Appressorium Formation, Host Invasion and Stress Homeostasis in the Filamentous Fungi Magnaporthe oryzae. Front. Plant Sci. 2021, 12, 646343. [Google Scholar] [CrossRef]
- Aron, O.; Wang, M.; Mabeche, A.W.; Wajjiha, B.; Li, M.; Yang, S.; You, H.; Cai, Y.; Zhang, T.; Li, Y.; et al. MoCpa1-mediated arginine biosynthesis is crucial for fungal growth, conidiation, and plant infection of Magnaporthe oryzae. Appl. Microbiol. Biotechnol. 2021, 105, 5915–5929. [Google Scholar] [CrossRef]
- Norvienyeku, J.; Zhong, Z.; Lin, L.; Dang, X.; Chen, M.; Lin, X.; Zhang, H.; Anjago, W.M.; Lin, L.; Abdul, W.; et al. Methylmalonate-semialdehyde dehydrogenase mediated metabolite homeostasis essentially regulate conidiation, polarized germination and pathogenesis inMagnaporthe oryzae. Environ. Microbiol. 2017, 19, 4256–4277. [Google Scholar] [CrossRef]
- Aliyu, S.R.; Lin, L.; Chen, X.; Abdul, W.; Lin, Y.; Otieno, F.J.; Shabbir, A.; Batool, W.; Zhang, Y.; Tang, W. Disruption of Putative Short-Chain Acyl-CoA Dehydrogenases Compromised Free Radical Scavenging, Conidiogenesis, and Pathogenesis of Mag-naporthe oryzae. Fungal Genet. Biol. 2019, 127, 23–34. [Google Scholar] [CrossRef]
- Abdul, W.; Aliyu, S.R.; Lin, L.; Sekete, M.; Chen, X.; Otieno, F.J.; Yang, T.; Lin, Y.; Norvienyeku, J.; Wang, Z. Family-Four Aldehyde Dehydrogenases Play an Indispensable Role in the Pathogenesis of Magnaporthe oryzae. Front. Plant Sci. 2018, 9, 980. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Cao, J.; Du, A.; An, Q.; Chen, X.; Yuan, S.; Batool, W.; Shabbir, A.; Zhang, D.; Wang, Z. EIF3k Domain-Containing Protein Regulates Conidiogenesis, Appressorium Turgor, Virulence, Stress Tolerance, and Physiological and Pathogenic De-velopment of Magnaporthe oryzae. Front. Plant Sci. 2021, 2316, 748120. [Google Scholar] [CrossRef]
- Aron, O.; Wang, M.; Lin, L.; Batool, W.; Lin, B.; Shabbir, A.; Wang, Z.; Tang, W. MoGLN2 Is Important for Vegetative Growth, Conidiogenesis, Maintenance of Cell Wall Integrity and Pathogenesis of Magnaporthe oryzae. J. Fungi 2021, 7, 463. [Google Scholar] [CrossRef]
- Mir, A.A.; Park, S.-Y.; Sadat, A.; Kim, S.; Choi, J.; Jeon, J.; Lee, Y.-H. Systematic characterization of the peroxidase gene family provides new insights into fungal pathogenicity in Magnaporthe oryzae. Sci. Rep. 2015, 5, 11831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Jia, B.; Liang, X.; Liu, J.; Wang, Y.; Liang, X.; Yan, H.; Wang, Y.; Zhang, S. An Adenosine Kinase in Apoplastic Location Is Involved in Magnaporthe oryzae Cold Acclimation. J. Basic Microbiol. 2014, 54, 269–277. [Google Scholar] [CrossRef]
- Hoi, J.W.S.; Beau, R.; Latgé, J.-P. A novel dehydrin-like protein from Aspergillus fumigatus regulates freezing tolerance. Fungal Genet. Biol. 2012, 49, 210–216. [Google Scholar] [CrossRef]
- Dang, Y.; Wei, Y.; Batool, W.; Sun, X.; Li, X.; Zhang, S.-H. Contribution of the Mitochondrial Carbonic Anhydrase (MoCA1) to Conidiogenesis and Pathogenesis in Magnaporthe oryzae. Front. Microbiol. 2022, 13, 845570. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhao, J.; Li, X.; Li, Y.; Jiang, L. The protein kinase CaSch9p is required for the cell growth, filamentation and virulence in the human fungal pathogen Candida albicans. FEMS Yeast Res. 2010, 10, 462–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, J.; Orth, K. Rise of a Cereal Killer: The Biology of Magnaporthe oryzae Biotrophic Growth. Trends Microbiol. 2018, 26, 582–597. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, T.T.; Wösten, H.A.B.; Dijksterhuis, J. Fungal Spores for Dispersion in Space and Time. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherlands, 2013; Volume 85, pp. 43–91. ISBN 0065-2164. [Google Scholar]
- Li, X.; Gao, C.; Li, L.; Liu, M.; Yin, Z.; Zhang, H.; Zheng, X.; Wang, P.; Zhang, Z. MoEnd3 regulates appressorium formation and virulence through mediating endocytosis in rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2017, 13, e1006449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, L.-A.; Li, G.-T.; Liu, Y.; Liu, M.-G.; Zhang, S.-J.; Yang, J.; Zhou, X.-Y.; Peng, Y.-L.; Xu, J.-R. Differences between Ap-pressoria Formed by Germ Tubes and Appressorium-like Structures Developed by Hyphal Tips in Magnaporthe oryzae. Fungal Genet. Biol. 2013, 56, 33–41. [Google Scholar] [CrossRef]
- Talbot, N.J. On the Trail of a Cereal Killer: Exploring the Biology of Magnaporthe grisea. Annu. Rev. Microbiol. 2003, 57, 177–202. [Google Scholar] [CrossRef] [Green Version]
- Que, Y.; Xu, Z.; Wang, C.; Lv, W.; Yue, X.; Xu, L.; Tang, S.; Dai, H.; Wang, Z. The putative deubiquitinating enzyme MoUbp4 is required for infection-related morphogenesis and pathogenicity in the rice blast fungus Magnaporthe oryzae. Curr. Genet. 2019, 66, 561–576. [Google Scholar] [CrossRef]
- Sinha, R.P.; Singh, S.P.; Häder, D.-P. Database on mycosporines and mycosporine-like amino acids (MAAs) in fungi, cyanobacteria, macroalgae, phytoplankton and animals. J. Photochem. Photobiol. B: Biol. 2007, 89, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Pusztahelyi, T.; Holb, I.J.; Pã³Csi, I. Secondary metabolites in fungus-plant interactions. Front. Plant Sci. 2015, 6, 573. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.-R. MAP Kinases in Fungal Pathogens. Fungal Genet. Biol. 2000, 31, 137–152. [Google Scholar] [CrossRef]
- Johnson, G.L.; Lapadat, R. Mitogen-Activated Protein Kinase Pathways Mediated by ERK, JNK, and p38 Protein Kinases. Science 2002, 298, 1911–1912. [Google Scholar] [CrossRef] [Green Version]
- Zhou, T.; Dagdas, Y.F.; Zhu, X.; Zheng, S.; Chen, L.; Cartwright, Z.; Talbot, N.J.; Wang, Z. The glycogen synthase kinase MoGsk1, regulated by Mps1 MAP kinase, is required for fungal development and pathogenicity in Magnaporthe oryzae. Sci. Rep. 2017, 7, 945. [Google Scholar] [CrossRef] [PubMed]
- Bahn, Y.-S.; Xue, C.; Idnurm, A.; Rutherford, J.C.; Heitman, J.; E Cardenas, M. Sensing the environment: Lessons from fungi. Nat. Rev. Genet. 2007, 5, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Guan, K.-L. The mitogen activated protein kinase signal transduction pathway: From the cell surface to the nucleus. Cell. Signal. 1994, 6, 581–589. [Google Scholar] [CrossRef] [Green Version]
- Faure, M.; Voyno-Yasenetskaya, T.; Bourne, H. cAMP and beta gamma subunits of heterotrimeric G proteins stimulate the mitogen-activated protein kinase pathway in COS-7 cells. J. Biol. Chem. 1994, 269, 7851–7854. [Google Scholar] [CrossRef]
- Lv, X.; Zhang, W.; Chen, G.; Liu, W. Trichoderma reesei Sch9 and Yak1 regulate vegetative growth, conidiation, and stress response and induced cellulase production. J. Microbiol. 2015, 53, 236–242. [Google Scholar] [CrossRef]
- Wang, P.; Cox, G.M.; Heitman, J. A Sch9 protein kinase homologue controlling virulence independently of the cAMP pathway in Cryptococcus neoformans. Curr. Genet. 2004, 46, 247–255. [Google Scholar] [CrossRef]
- Alves de Castro, P.; Dos Reis, T.F.; Dolan, S.K.; Oliveira Manfiolli, A.; Brown, N.A.; Jones, G.W.; Doyle, S.; Riaño-Pachón, D.M.; Squina, F.M.; Caldana, C. The Aspergillus fumigatus SchASCH9 Kinase Modulates SakAHOG1 MAP Kinase Activity, and It Is Essential for Virulence. Mol. Microbiol. 2016, 102, 642–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fillinger, S.; Chaveroche, M.-K.; Shimizu, K.; Keller, N.; D’Enfert, C. cAMP and ras signalling independently control spore germination in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 2002, 44, 1001–1016. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-L.; Shi, T.; Yang, J.; Shi, W.; Gao, X.; Chen, D.; Xu, X.; Xu, J.-R.; Talbot, N.J.; Peng, Y.-L. N-Glycosylation of Effector Proteins by an α-1,3-Mannosyltransferase Is Required for the Rice Blast Fungus to Evade Host Innate Immunity. Plant Cell 2014, 26, 1360–1376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roux, A.E.; Quissac, A.; Chartrand, P.; Ferbeyre, G.; Rokeach, L.A. Regulation of chronological aging in Schizosaccharomyces pombe by the protein kinases Pka1 and Sck2. Aging Cell 2006, 5, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Sohail, M.A.; Yuan, Q.; Yan, Y.; Huang, J.; Hsiang, T.; Zheng, L. ChSch9 Is Required for Infection-Related Morphogenesis and Pathogenicity in Colletotrichum higginsianum. Can. J. Plant Pathol. 2021, 43, 871–885. [Google Scholar] [CrossRef]
- Goswami, R.S.; Kistler, H.C. Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 2004, 5, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Lee, M.R.; Kim, S.; Kim, J.C.; Park, S.E.; Li, D.; Shin, T.Y.; Nai, Y.-S.; Kim, J.S. Genomic Analysis of the Insect-Killing Fungus Beauveria bassiana JEF-007 as a Biopesticide. Sci. Rep. 2018, 8, 12388. [Google Scholar] [CrossRef]
- Mitchell, T.K.; Dean, R.A. The CAMP-Dependent Protein Kinase Catalytic Subunit Is Required for Appressorium Formation and Pathogenesis by the Rice Blast Pathogen Magnaporthe grisea. Plant Cell 1995, 7, 1869–1878. [Google Scholar]
- Dickman, M.B.; Ha, Y.-S.; Yang, Z.; Adams, B.; Huang, C. A Protein Kinase from Colletotrichum trifolii Is Induced by Plant Cutin and Is Required for Appressorium Formation. Mol. Plant-Microbe Interact. 2003, 16, 411–421. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.-H.; Lin, F.-C. Investigation of the biological roles of autophagy in appressorium morphogenesis in Magnaporthe oryzae. J. Zhejiang Univ. Sci. B 2008, 9, 793–796. [Google Scholar] [CrossRef] [Green Version]
- Cousin, A.; Mehrabi, R.; Guilleroux, M.; Dufresne, M.; Van der Lee, T.; Waalwijk, C.; Langin, T.; Kema, G.H.J. The MAP Kinase-encoding Gene MgFus3 of the Non-appressorium Phytopathogen Mycosphaerella graminicola Is Required for Penetration and in Vitro Pycnidia Formation. Mol. Plant Pathol. 2006, 7, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-H.; Lu, J.-P.; Zhang, L.; Dong, B.; Min, H.; Lin, F.-C. Involvement of a Magnaporthe grisea Serine/Threonine Kinase Gene, Mg ATG1, in Appressorium Turgor and Pathogenesis. Eukaryot. Cell 2007, 6, 997–1005. [Google Scholar] [CrossRef] [Green Version]
- Panikov, N.S.; Sizova, M. V Growth Kinetics of Microorganisms Isolated from Alaskan Soil and Permafrost in Solid Media Frozen Down To− 35 C. FEMS Microbiol. Ecol. 2007, 59, 500–512. [Google Scholar] [CrossRef]
- Santoro, M. Heat shock factors and the control of the stress response. Biochem. Pharmacol. 1999, 59, 55–63. [Google Scholar] [CrossRef]
- Kroll, K.; Pähtz, V.; Kniemeyer, O. Elucidating the fungal stress response by proteomics. J. Proteom. 2014, 97, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Tesei, D.; Marzban, G.; Zakharova, K.; Isola, D.; Selbmann, L.; Sterflinger, K. Alteration of protein patterns in black rock inhabiting fungi as a response to different temperatures. Fungal Biol. 2012, 116, 932–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oda, Y.; Totsuka, Y.; Wakabayashi, K.; Guengerich, F.P.; Shimada, T. Activation of Aminophenylnorharman, Ami-nomethylphenylnorharman and Aminophenylharman to Genotoxic Metabolites by Human N-Acetyltransferases and Cyto-chrome P450 Enzymes Expressed in Salmonella typhimurium Umu Tester Strains. Mutagenesis 2006, 21, 411–416. [Google Scholar] [CrossRef] [Green Version]
- Abu Bakar, N.; Karsani, S.A.; Alias, S.A. Fungal survival under temperature stress: A proteomic perspective. PeerJ 2020, 8, e10423. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Yamamoto, K.; Hamamoto, H.; Nakaune, R.; Hibi, T. A novel ABC transporter gene ABC2 involved in multidrug susceptibility but not pathogenicity in rice blast fungus, Magnaporthe grisea. Pestic. Biochem. Physiol. 2005, 81, 13–23. [Google Scholar] [CrossRef]
- Do, T.H.T.; Martinoia, E.; Lee, Y. Functions of ABC transporters in plant growth and development. Curr. Opin. Plant Biol. 2018, 41, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Gow, N.A.R.; Latge, J.-P.; Munro, C.A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5, 3–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latgé, J.-P. The cell wall: A carbohydrate armour for the fungal cell. Mol. Microbiol. 2007, 66, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Haslbeck, M.; Buchner, J. The Heat Shock Response: Life on the Verge of Death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef]
- Verghese, J.; Abrams, J.; Wang, Y.; Morano, K.A. Biology of the Heat Shock Response and Protein Chaperones: Budding Yeast (Saccharomyces cerevisiae) as a Model System. Microbiol. Mol. Biol. Rev. 2012, 76, 115–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaner, L.; Gibney, P.A.; Morano, K.A. The Hsp110 protein chaperone Sse1 is required for yeast cell wall integrity and morphogenesis. Curr. Genet. 2008, 54, 1. [Google Scholar] [CrossRef] [Green Version]
- Pandian, B.A.; Sathishraj, R.; Djanaguiraman, M.; Prasad, P.V.; Jugulam, M. Role of Cytochrome P450 Enzymes in Plant Stress Response. Antioxidants 2020, 9, 454. [Google Scholar] [CrossRef] [PubMed]
- Yu, E.; Owttrim, G.W. Characterization of the Cold Stress-Induced Cyanobacterial DEAD-Box Protein CrhC as an RNA Hel-icase. Nucleic Acids Res. 2000, 28, 3926–3934. [Google Scholar] [CrossRef] [PubMed]
- Chelysheva, V.V.; Smolenskaya, I.N.; Trofimova, M.C.; Babakov, A.V.; Muromtsev, G.S. Role of the 14-3-3 proteins in the regulation of H+-ATPase activity in the plasma membrane of suspension-cultured sugar beet cells under cold stress. FEBS Lett. 1999, 456, 22–26. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, T.; Bremer, E. Protection of Bacillus subtilis against Cold Stress via Compatible-Solute Acquisition. J. Bacteriol. 2011, 193, 1552–1562. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.; Song, B.; Tang, S.; He, J.; Zhou, Y.; Feng, J.; Shi, S.; Xu, X. Overexpression of the ABC transporter gene TsABCG11 increases cuticle lipids and abiotic stress tolerance in Arabidopsis. Plant Biotechnol. Rep. 2018, 12, 303–313. [Google Scholar] [CrossRef]
- Tanaka, K.; Gilroy, S.; Jones, A.M.; Stacey, G. Extracellular ATP signaling in plants. Trends Cell Biol. 2010, 20, 601–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batool, W.; Liu, C.; Fan, X.; Zhang, P.; Hu, Y.; Wei, Y.; Zhang, S.-H. AGC/AKT Protein Kinase SCH9 Is Critical to Pathogenic Development and Overwintering Survival in Magnaporthe oryzae. J. Fungi 2022, 8, 810. https://doi.org/10.3390/jof8080810
Batool W, Liu C, Fan X, Zhang P, Hu Y, Wei Y, Zhang S-H. AGC/AKT Protein Kinase SCH9 Is Critical to Pathogenic Development and Overwintering Survival in Magnaporthe oryzae. Journal of Fungi. 2022; 8(8):810. https://doi.org/10.3390/jof8080810
Chicago/Turabian StyleBatool, Wajjiha, Chang Liu, Xiaoning Fan, Penghui Zhang, Yan Hu, Yi Wei, and Shi-Hong Zhang. 2022. "AGC/AKT Protein Kinase SCH9 Is Critical to Pathogenic Development and Overwintering Survival in Magnaporthe oryzae" Journal of Fungi 8, no. 8: 810. https://doi.org/10.3390/jof8080810
APA StyleBatool, W., Liu, C., Fan, X., Zhang, P., Hu, Y., Wei, Y., & Zhang, S.-H. (2022). AGC/AKT Protein Kinase SCH9 Is Critical to Pathogenic Development and Overwintering Survival in Magnaporthe oryzae. Journal of Fungi, 8(8), 810. https://doi.org/10.3390/jof8080810





