The Effect of Antibiotics on the Infant Gut Fungal Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. DNA Extraction and MiSeq Library Preparation
2.3. Analysis of Sequencing Data
2.4. Statistical Analysis
2.5. Ethics Statement
3. Results
3.1. Patient Characteristics
3.2. Overview of the Gut Mycobiota Composition
3.3. Difference between the Control Group and the Group Treated with Antibiotics
3.4. Spearman Correlations between Fungi and Bacteria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Chehoud, C.; Albenberg, L.G.; Judge, C.; Hoffmann, C.; Grunberg, S.; Bittinger, K.; Baldassano, R.N.; Lewis, J.D.; Bushman, F.D.; Wu, G.D. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm. Bowel. Dis. 2015, 21, 1948–1956. [Google Scholar] [CrossRef] [PubMed]
- Standaert-Vitse, A.; Sendid, B.; Joossens, M.; Francois, N.; Vandewalle-El Khoury, P.; Branche, J.; Van Kruiningen, H.; Jouault, T.; Rutgeerts, P.; Gower-Rousseau, C.; et al. Candida albicans colonization and ASCA in familial Crohn’s disease. Am. J. Gastroenterol. 2009, 104, 1745–1753. [Google Scholar] [CrossRef]
- Kumamoto, C.A. Inflammation and gastrointestinal Candida colonization. Curr. Opin. Microbiol. 2011, 14, 386–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal microbiota dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef] [Green Version]
- El Mouzan, M.; Al-Hussaini, A.; Fanelli, B.; Assiri, A.; AlSaleem, B.; Al Mofarreh, M.; Al Sarkhy, A.; Alasmi, M. Fungal Dysbiosis in Children with Celiac Disease. Dig. Dis. Sci. 2021, 67, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Fan, C.; Yao, A.; Xu, X.; Zheng, G.; You, Y.; Jiang, C.; Zhao, X.; Hou, Y.; Hung, M.C.; et al. The adaptor protein CARD9 protects against colon cancer by restricting mycobiota-mediated expansion of myeloid-derived suppressor cells. Immunity 2018, 49, 504–514.e4. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, M.L.; Limon, J.J.; Bar, A.S.; Leal, C.A.; Gargus, M.; Tang, J.; Brown, J.; Funari, V.A.; Wang, H.L.; Crother, T.R.; et al. Immunological consequences of intestinal fungal dysbiosis. Cell Host. Microbe 2016, 19, 865–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boutin, R.C.; Sbihi, H.; McLaughlin, R.J.; Hahn, A.S.; Konwar, K.M.; Loo, R.S.; Dai, D.; Petersen, C.; Brinkman, F.S.L.; Winsor, G.L.; et al. Composition and Associations of the Infant Gut Fungal Microbiota with Environmental Factors and Childhood Allergic Outcomes. Mbio 2021, 12, e03396-20. [Google Scholar] [CrossRef] [PubMed]
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gesell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A.; et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017, 5, 153. [Google Scholar] [CrossRef] [PubMed]
- Santus, W.; Devlin, J.R.; Behnsen, J. Crossing Kingdoms: How the Mycobiota and Fungal-Bacterial Interactions Impact Host Health and Disease. Infect. Immun. 2021, 89, e00648-20. [Google Scholar] [CrossRef] [PubMed]
- Richard, M.L.; Sokol, H. The gut mycobiota: Insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 331–345. [Google Scholar] [CrossRef]
- Wu, X.; Xia, Y.; He, F.; Zhu, C.; Ren, W. Intestinal mycobiota in health and diseases: From a disrupted equilibrium to clinical opportunities. Microbiome 2021, 9, 60. [Google Scholar] [CrossRef] [PubMed]
- Huseyin, C.E.; O’Toole, P.W.; Cotter, P.D.; Scanlan, P.D. Forgotten fungi—The gut mycobiome in human health and disease. FEMS Microbiol. Rev. 2017, 41, 479–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schei, K.; Avershina, E.; Øien, T.; Rudi, K.; Follestad, T.; Salamati, S.; Ødegård, R.A. Early gut mycobiota and mother-offspring transfer. Microbiome 2017, 5, 107. [Google Scholar] [CrossRef] [Green Version]
- Henderickx, J.G.E.; de Weerd, H.; Groot Jebbink, L.J.; van Zoeren-Grobben, D.; Hemels, M.A.C.; van Lingen, R.A.; Knol, J.; Belzer, C. The first fungi: Mode of delivery determines early life fungal colonization in the intestine of preterm infants. Microbiome Res. Rep. 2022, 1, 7. [Google Scholar] [CrossRef]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Korpela, K.; Salonen, A.; Saxen, H.; Nikkonen, A.; Peltola, V.; Jaakkola, T.; de Vos, W.; Kolho, K.L. Antibiotics in early life associate with specific gut microbiota signatures in a prospective longitudinal infant cohort. Pediatr. Res. 2020, 88, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Bejaoui, S.; Poulsen, M. The impact of early life antibiotic use on atopic and metabolic disorders: Meta-analyses of recent insights. Evol. Med. Public Health 2020, 24, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Aires, J. First 1000 Days of Life: Consequences of Antibiotics on Gut Microbiota. Front. Microbiol. 2021, 12, 681427. [Google Scholar] [CrossRef]
- Korpela, K.; Zijlmans, M.A.C.; Kuitunen, M.; Kukkonen, K.; Savilahti, E.; Salonen, A.; de Weerth, C.; de Vos, W.M. Childhood BMI in relation to microbiota in infancy and lifetime antibiotic use. Microbiome 2017, 5, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schei, K.; Simpson, M.R.; Avershina, E.; Rudi, K.; Øien, T.; Júlíusson, P.B.; Underhill, D.; Salamati, S.; Ødegård, R.A. Early Gut Fungal and Bacterial Microbiota and Childhood Growth. Front. Pediatr. 2020, 8, 572538. [Google Scholar] [CrossRef] [PubMed]
- Saari, A.; Virta, L.J.; Sankilampi, U.; Dunkel, L.; Saxen, H. Antibiotic exposure in infancy and risk of being overweight in the first 24 months of life. Pediatrics 2015, 135, 617–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korpela, K.; Salonen, A.; Virta, L.J.; Kekkonen, R.A.; Forslund, K.; Bork, P.; de Vos, W.M. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 2016, 7, 10410. [Google Scholar] [CrossRef] [PubMed]
- Korpela, K.; de Vos, W.M. Antibiotic use in childhood alters the gut microbiota and predisposes to overweight. Microb. Cell 2016, 3, 296–298. [Google Scholar] [CrossRef] [PubMed]
- Dollive, S.; Chen, Y.-Y.; Grunberg, S.; Bittinger, K.; Hoffmann, C.; Vandivier, L.; Cuff, C.; Lewis, J.D.; Wu, G.D.; Bushman, F.D. Fungi of the murine gut: Episodic variation and proliferation duringantibiotic treatment. PLoS ONE 2013, 8, e71806. [Google Scholar] [CrossRef]
- Noverr, M.C.; Noggle, R.M.; Toews, G.B.; Huffnagle, G.B. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect. Immun. 2004, 72, 4996–5003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noverr, M.C.; Falkowski, N.R.; McDonald, R.A.; McKenzie, A.N.; Huffnagle, G.B. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: Role of host genetics, antigen, and interleukin-13. Infect. Immun. 2005, 73, 30–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seelbinder, B.; Chen, J.; Brunke, S.; Vazquez-Uribe, R.; Santhaman, R.; Meyer, A.C.; de Oliveira Lino, F.S.; Chan, K.-F.; Loos, D.; Imamovic, L.; et al. Antibiotics create a shift from mutualism to competition in human gut communities with a longer-lasting impact on fungi than bacteria. Microbiome 2020, 8, 133. [Google Scholar] [CrossRef] [PubMed]
- Dadar, M.; Tiwari, R.; Karthik, K.; Chakraborty, S.; Shahali, Y.; Dhama, K. Candida albicans-Biology, molecular characterization, pathogenicity, and advances in diagnosis and control—An update. Microb. Pathog. 2018, 117, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Hani, U.; Shivakumar, H.G.; Vaghela, R.; Osmani, R.A.; Shrivastava, A. Candidiasis: A fungal infection—Current challenges and progress in prevention and treatment. Infect. Disord. Drug Targets 2015, 15, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Cabral, D.J.; Penumutchu, S.; Norris, C.; Morones-Ramirez, J.R.; Belenky, P. Microbial competition between Escherichia coli and Candida albicans reveals a soluble fungicidal factor. Microb. Cell 2018, 5, 249–255. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Hogan, D.A.; Mylonakis, E. Medically important bacterial–fungal interactions. Nat. Rev. Microbiol. 2010, 8, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Haak, B.W.; Argelaguet, R.; Kinsella, C.M.; Kullberg, R.F.; Lankelma, J.M.; Deijs, M.; Klein, M.; Jebbink, M.F.; Hugenholtz, F.; Kostidis, S.; et al. Integrative Transkingdom Analysis of the Gut Microbiome in Antibiotic Perturbation and Critical Illness. mSystems 2021, 6, e01148-20. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Elsevier Academic Press: Cambridge, MA, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Ventin-Holmberg, R.; Eberl, A.; Saqib, S.; Korpela, K.; Virtanen, S.; Sipponen, T.; Salonen, A.; Saavalainen, P.; Nissilä, E. Bacterial and Fungal Profiles as Markers of Infliximab Drug Response in Inflammatory Bowel Disease. J. Crohns. Colitis. 2020, 15, 1019–1031. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. BLAST PROGRAMS. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Korpela, K. Mare: Microbiota Analysis in R Easily; R Package Version 1.0.; GitHub: San Francisco, CA, USA, 2016; Available online: https://github.com/katrikorpela/mare (accessed on 18 January 2021).
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; O’Hara, B.; Stevens, M.H.H.; Oksanen, M.J. Vegan: Community Ecology Package; Version 2.5-7; R Core Team: Vienna, Austria, 2020; Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 2 February 2022).
- Venables, W.; Ripley, B. Modern Applied Statistics with S; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar] [CrossRef] [Green Version]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.R. Nlme: Linear and Nonlinear Mixed Effects Models; R Package Version 3.1-14.; R Core Team: Vienna, Austria, 2022; Available online: https://cran.r-project.org/web/packages/nlme/index.html (accessed on 2 February 2022).
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar]
- Harrell, F.E. Hmisc: Harrell Miscellaneous; R Package Version 4.6-0; R Core Team: Vienna, Austria, 2021; Available online: https://CRAN.R-project.org/package=Hmisc (accessed on 2 February 2022).
- Borali, E.; De Giacomo, C. Clostridium difficile infection in children: A review. J. Pediatr. Gastroenterol. Nutr. 2016, 63, e130–e140. [Google Scholar] [CrossRef] [PubMed]
- Korpela, K.; de Vos, W.M. Early life colonization of the human gut: Microbes matter everywhere. Curr. Opin. Microbiol. 2018, 44, 70–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strati, F.; Cavalieri, D.; Albanese, D.; De Felice, C.; Donati, C.; Hayek, J.; Jousson, O.; Leoncini, S.; Pindo, M.; Renzi, D.; et al. Altered gut microbiota in Rett syndrome. Microbiome 2016, 4, 41. [Google Scholar] [CrossRef] [Green Version]
- James, S.A.; Phillips, S.; Telatin, A.; Baker, D.; Ansorge, R.; Clarke, P.; Hall, L.J.; Carding, S.R. Preterm Infants Harbour a Rapidly Changing Mycobiota That Includes Candida Pathobionts. J. Fungi 2020, 6, 273. [Google Scholar] [CrossRef] [PubMed]
- Amenyogbe, N.; Adu-Gyasi, D.; Enuameh, Y.; Asante, K.P.; Konadu, D.G.; Kaali, S.; Dosoo, D.; Panigrahi, P.; Kollmann, T.R.; Mohn, W.W.; et al. Bacterial and fungal gut community dynamics over the first five years of life in predominantly rural communities in Ghana. Front. Microbiol. 2021, 12, 664407. [Google Scholar] [CrossRef] [PubMed]
- Mok, K.; Suratanon, N.; Roytrakul, S.; Charoenlappanit, S.; Patumcharoenpol, P.; Chatchatee, P.; Vongsangnak, W.; Nakphaichit, M. ITS2 Sequencing and Targeted Meta-Proteomics of Infant Gut Mycobiome Reveal the Functional Role of Rhodotorula sp. during Atopic Dermatitis Manifestation. J. Fungi 2021, 7, 748. [Google Scholar] [CrossRef] [PubMed]
- Harding, J.N.; Siefker, D.; Vu, L.; You, D.; DeVincenzo, J.; Pierre, J.F.; Cormier, S.A. Altered gut microbiota in infants is associated with respiratory syncytial virus disease severity. BMC Microbiol. 2020, 20, 140. [Google Scholar] [CrossRef] [PubMed]
- d’Enfert, C.; Kaune, A.K.; Alaban, L.R.; Chakraborty, S.; Cole, N.; Delavy, M.; Kosmala, D.; Marsaux, B.; Fróis-Martins, R.; Morelli, M.; et al. The impact of the Fungus-Host-Microbiota interplay upon Candida albicans infections: Current knowledge and new perspectives. FEMS Microbiol. Rev. 2020, 45, fuaa060. [Google Scholar] [CrossRef] [PubMed]
- Allonsius, C.N.; Vandenheuvel, D.; Oerlemans, E.F.M.; Petrova, M.I.; Donders, G.G.G.; Cos, P.; Delputte, P.; Lebeer, S. Inhibition of Candida albicans morphogenesis by chitinase from Lactobacillus rhamnosus GG. Sci. Rep. 2019, 9, 2900. [Google Scholar] [CrossRef] [PubMed]
- Mason, K.L.; Erb Downward, J.R.; Falkowski, N.R.; Young, V.B.; Kao, J.Y.; Huffnagle, G.B. Interplay between the gastric bacterial microbiota and Candida albicans during post antibiotic recolonization and gastritis. Infect. Immun. 2012, 80, 150–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acosta-Rodriguez, E.V.; Rivino, L.; Geginat, J.; Jarrossay, D.; Gattorno, M.; Lanzavecchia, A.; Sallusto, F.; Napolitani, G. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 2007, 8, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Bacher, P.; Hohnstein, T.; Beerbaum, E.; Röcker, M.; Blango, M.G.; Kaufmann, S.; Röhmel, J.; Eschenhagen, P.; Grehn, C.; Seidel, K.; et al. Human Anti-fungal Th17 Immunity and Pathology Rely on Cross-Reactivity against Candida albicans. Cell 2019, 176, 1340–1355.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schirmer, M.; Garner, A.; Vlamakis, H.; Xavier, R.J. Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 2019, 17, 497–511. [Google Scholar] [CrossRef] [PubMed]
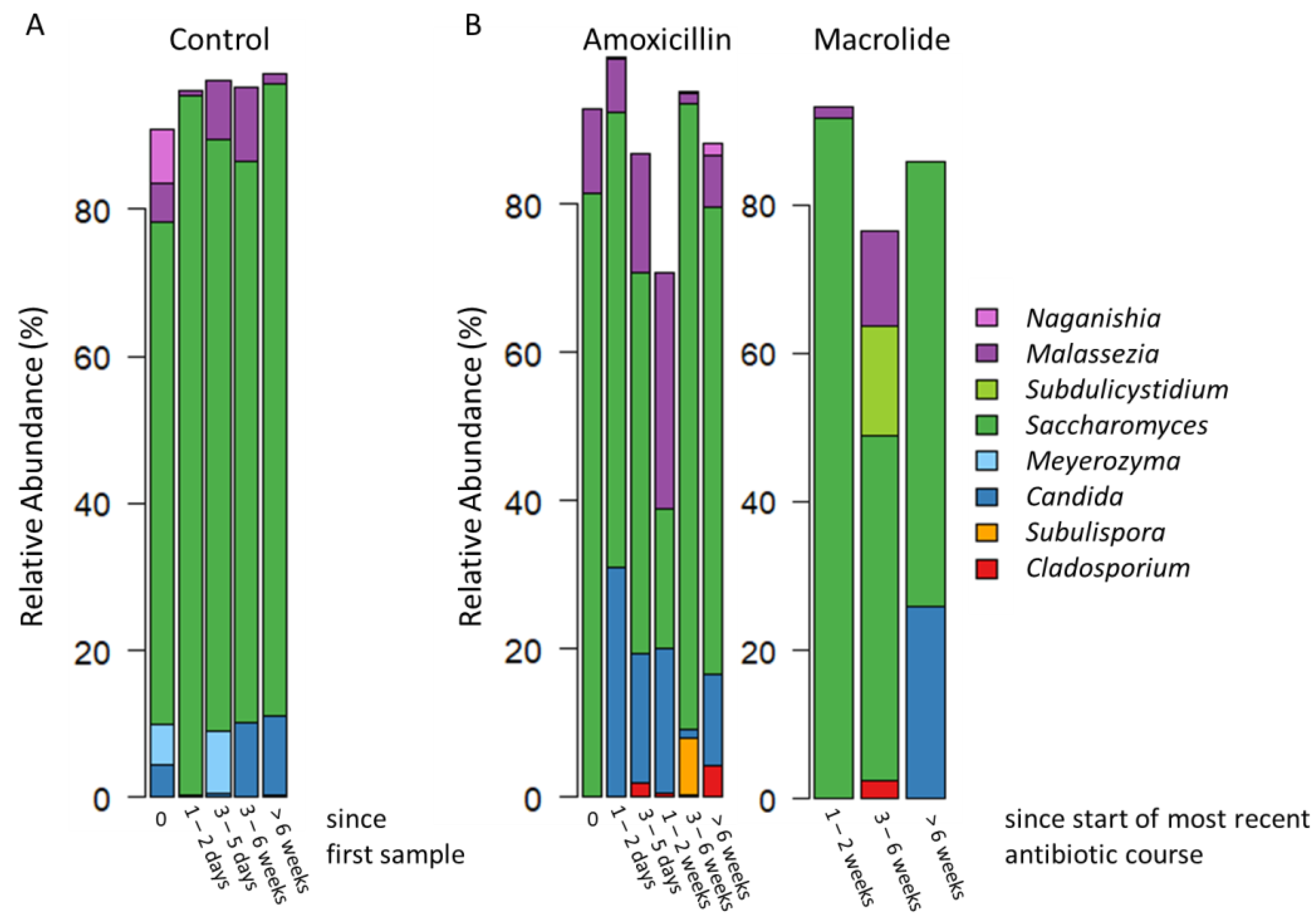
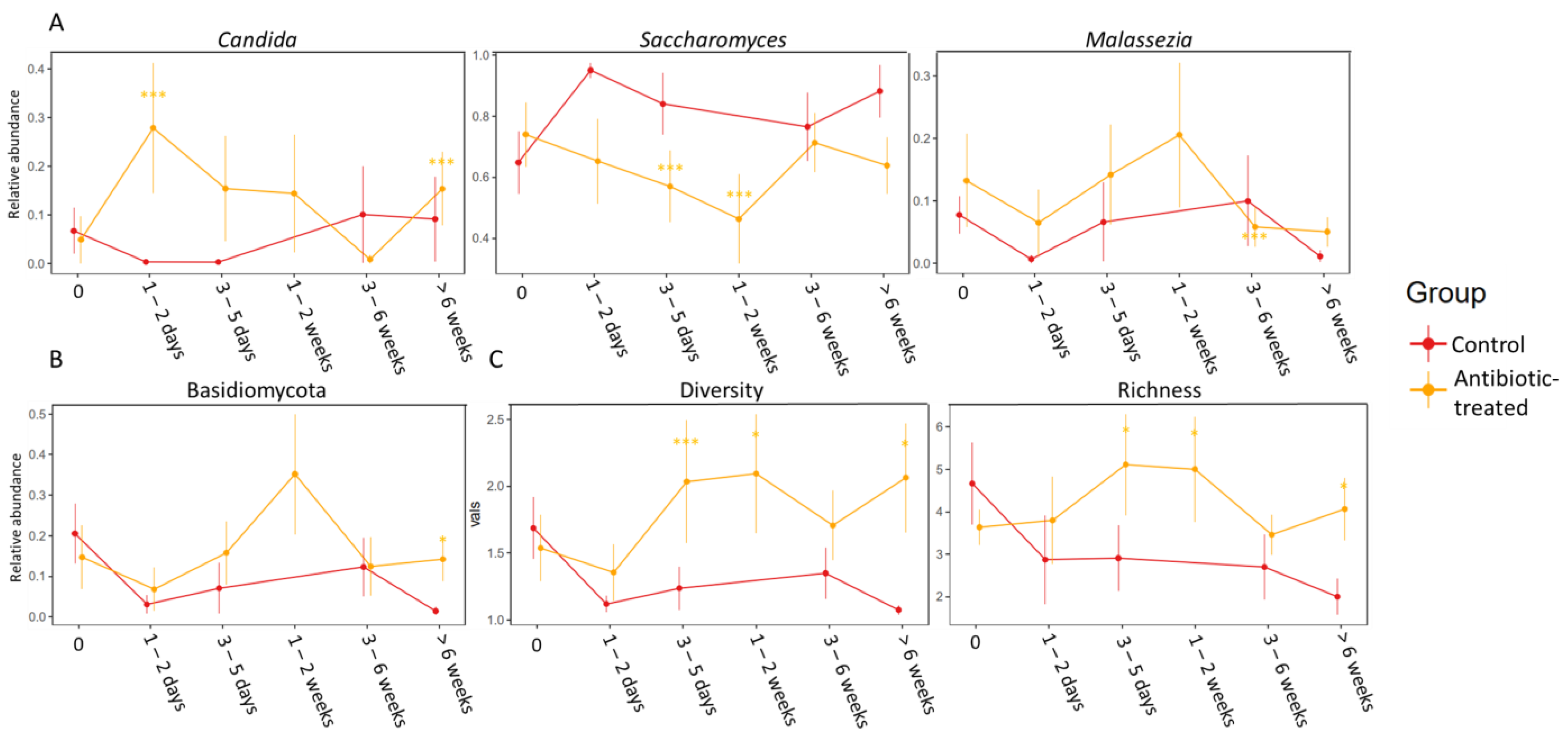
| Time Since Most Recent Course of Antibiotics or Since First Sample | 0 Days | 1–2 Days | 3–5 Days | 1–2 Weeks | 3–6 Weeks | >6 Weeks |
|---|---|---|---|---|---|---|
| Control (no. of infants—16; no. of samples—50) | 14 | 8 | 9 | 0 | 10 | 9 |
| Amoxicillin (no. of infants—21; no. of samples—51) | 10 | 9 | 8 | 5 | 9 | 10 |
| Macrolide (no. of infants—4; no. of samples—12) | 0 | 0 | 0 | 3 * | 5 * | 4 |
| Patients | All (N = 37) | Antibiotics during Admission (N = 21) | No Antibiotics during Admission (N = 16) |
|---|---|---|---|
| Antibiotic-naïve at recruitment, N(%) | 37 (100) | 21 (100) | 16 (100) |
| Female, N(%) | 15 (40.5) | 7 (33) | 8 (50) |
| Age at hospitalization, median (range, months) | 2.3 (0.8–9.3) | 2.5 (0.8–9.3) | 2.1 (0.8–6.8) |
| Vaginal delivery, N(%) | 31 (83.8) | 17 (81.0) | 14 (87.5) |
| Exclusive or partial breast-feeding, N(%) | 32 (86.5) | 17 (81.0) | 15 (93.8) |
| Birth weight, median (range, kg) | 3.7 (2.5–4.9) | 3.5 (2.5–4.9) | 3.8 (2.7–4.5) |
| Length of hospital stay, median (range, days) | 4 (1–9) | 4 (2–9) | 4 (1–7) |
| Annotated samples with >100 reads, N | 113 | 63 | 50 |
| Fecal samples obtained and successfully annotated, median (range, N/patient) | 3 (1–6) | 3 (1–5) | 3.5 (1–6) |
| Length of follow-up, median (range, months) | 3 (0–9.5) | 3 (0–9.5) | 2 (0–6) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ventin-Holmberg, R.; Saqib, S.; Korpela, K.; Nikkonen, A.; Peltola, V.; Salonen, A.; de Vos, W.M.; Kolho, K.-L. The Effect of Antibiotics on the Infant Gut Fungal Microbiota. J. Fungi 2022, 8, 328. https://doi.org/10.3390/jof8040328
Ventin-Holmberg R, Saqib S, Korpela K, Nikkonen A, Peltola V, Salonen A, de Vos WM, Kolho K-L. The Effect of Antibiotics on the Infant Gut Fungal Microbiota. Journal of Fungi. 2022; 8(4):328. https://doi.org/10.3390/jof8040328
Chicago/Turabian StyleVentin-Holmberg, Rebecka, Schahzad Saqib, Katri Korpela, Anne Nikkonen, Ville Peltola, Anne Salonen, Willem M. de Vos, and Kaija-Leena Kolho. 2022. "The Effect of Antibiotics on the Infant Gut Fungal Microbiota" Journal of Fungi 8, no. 4: 328. https://doi.org/10.3390/jof8040328
APA StyleVentin-Holmberg, R., Saqib, S., Korpela, K., Nikkonen, A., Peltola, V., Salonen, A., de Vos, W. M., & Kolho, K.-L. (2022). The Effect of Antibiotics on the Infant Gut Fungal Microbiota. Journal of Fungi, 8(4), 328. https://doi.org/10.3390/jof8040328







