Regulation of the Leucine Metabolism in Mortierella alpina
Abstract
1. Introduction
| Protein Function | Homolog in S. cerevisiae [1] | Homolog in P. oryzae [3] | Homolog in A. fumigatus [2,15] | Homolog in P. blakesleeanus [4] | Predicted Protein in M. alpina | e Value to S. cerevisiae Homologs | Encoding Contig | Signal Peptide Target-P [16] | Signal Pept. Mitofate [17] | |
|---|---|---|---|---|---|---|---|---|---|---|
| early steps in Leu biosynthesis | threonine deaminase | ILV1 | IlvA | 1.05 × 10−158 | ADAG01001032 | yes | no | |||
| acetohydroxy acid synthase catalytic subunit | ILV2 | ILV2 * | IlvB | 0 | ADAG01000936 | IRF-RS | yes | |||
| acetohydroxy acid synthase regulatory subunit | ILV6 | ILV6 * | IlvF1 IlvF2 | 8.91 × 10−73 8.91 × 10−69 | ADAG01000936 ADAG01001016 | no yes | no yes | |||
| bifunctional acetohydroxy acid reductoisomerase | ILV5 | ILV5 * | IlvE1 IlvE2 | 2.32 × 10−159 5.25 × 10−157 | ADAG01000936 ADAG01001016 | TRG-VK TRG-VK | yes yes | |||
| dihydroxy acid dehydratase | ILV3 | ILV3 * | IlvC | 1.56 × 10−153 | ADAG01001099 | HRY-ST | no | |||
| late steps in Leu biosynthesis | α-isopropylmalate synthase I | LEU4S/L | LEU4 | LeuC | LeuA | LeuA1 | 8.14 × 10−23 | ADAG01001098 | no | no |
| α-isopropylmalate synthase II | LEU9 | LEU9 | ||||||||
| α-isopropylmalate isomerase/dehydratase | LEU1 | LEU1 | LeuA | LeuA2 | 0 | ADAG01001081 | no | no | ||
| β-isopropylmalate dehydrogenase | LEU2 | Leu2A | Leu2A | LeuB1 LeuB2 | 1.38 × 10−112 1.83 × 10−36 | ADAG01001016 ADAG01000512 | no no | no no | ||
| Zn2Cys6 transcription factor | LEU3 | LEU3 | LeuB | LeuC1 LeuC2 | 5.57 × 10−2 5.78 × 10−2 | ADAG01000955 ADAG01000881 | no no | no no | ||
| CoA importer | LEU5 | LeuE1 LeuE2 | 2.33 × 10−60 1.45 × 10−47 | ADAG01001074 ADAG01001044 | no no | no no | ||||
| Iron-sulfur cluster transporter | ATM1 | AtmA1 AtmA2 | 2.19 × 10−154 1.50 × 10−92 | ADAG01000995 ADAG01001098 | no no | no no | ||||
| BCAA aminotransferase | BAT1 BAT2 - | BAT1 BAT2 BAT3 * | BatA - BatC | 1.69 × 10−52 - 1.61 × 10−32 | ADAG01001070 - ADAG01000958 | RFY-YR - no | yes - no |
2. Materials and Methods
2.1. Microbiological Methods
2.2. gDNA and RNA Isolation, cDNA Synthesis and Expression Analysis
2.3. Heterologous Protein Production and Size Exclusion Chromatography
2.4. Chromatographical Quantification of α-amino Acids, α-keto Acids and Malpinins
2.5. Determination of Enzyme Activities
3. Results
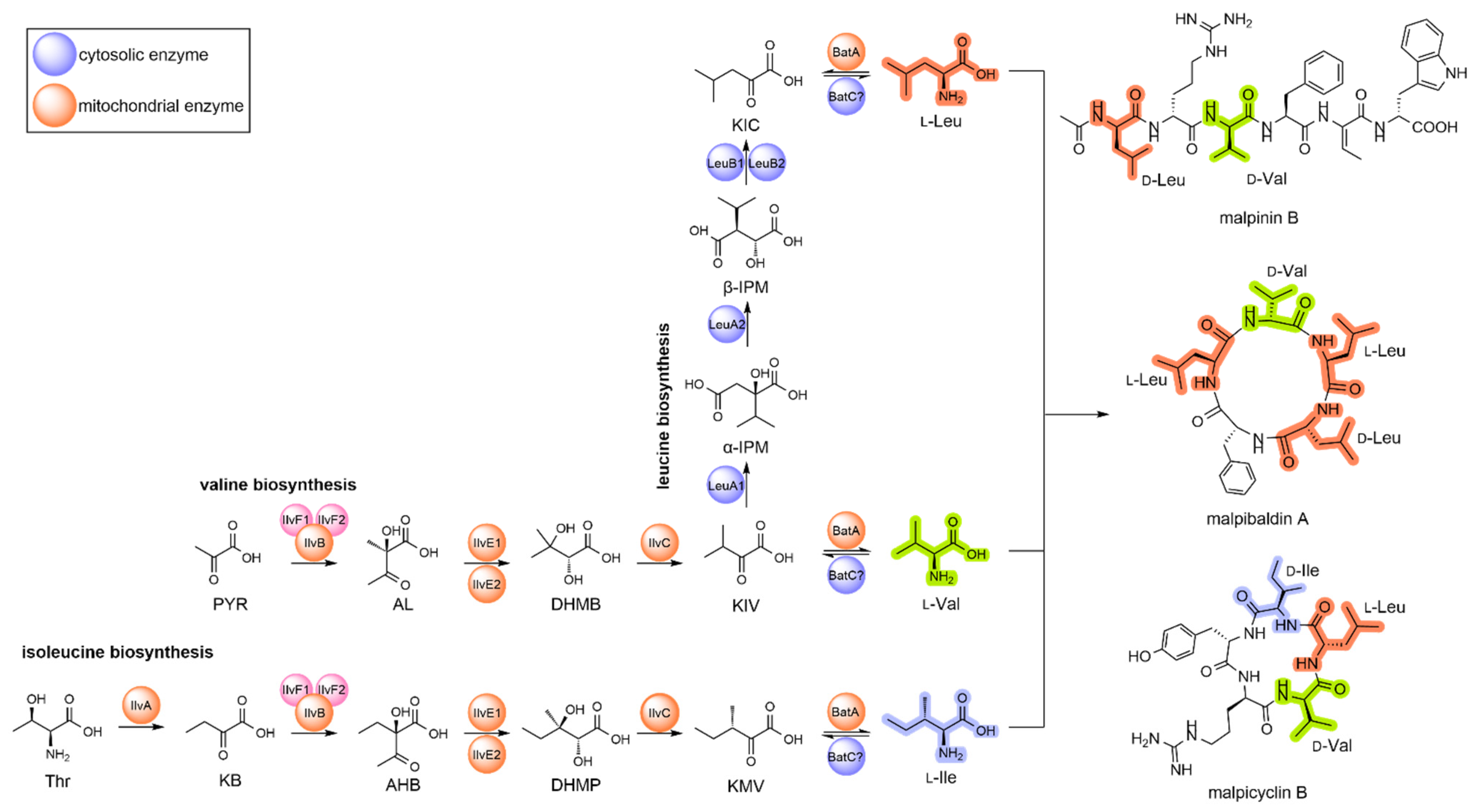
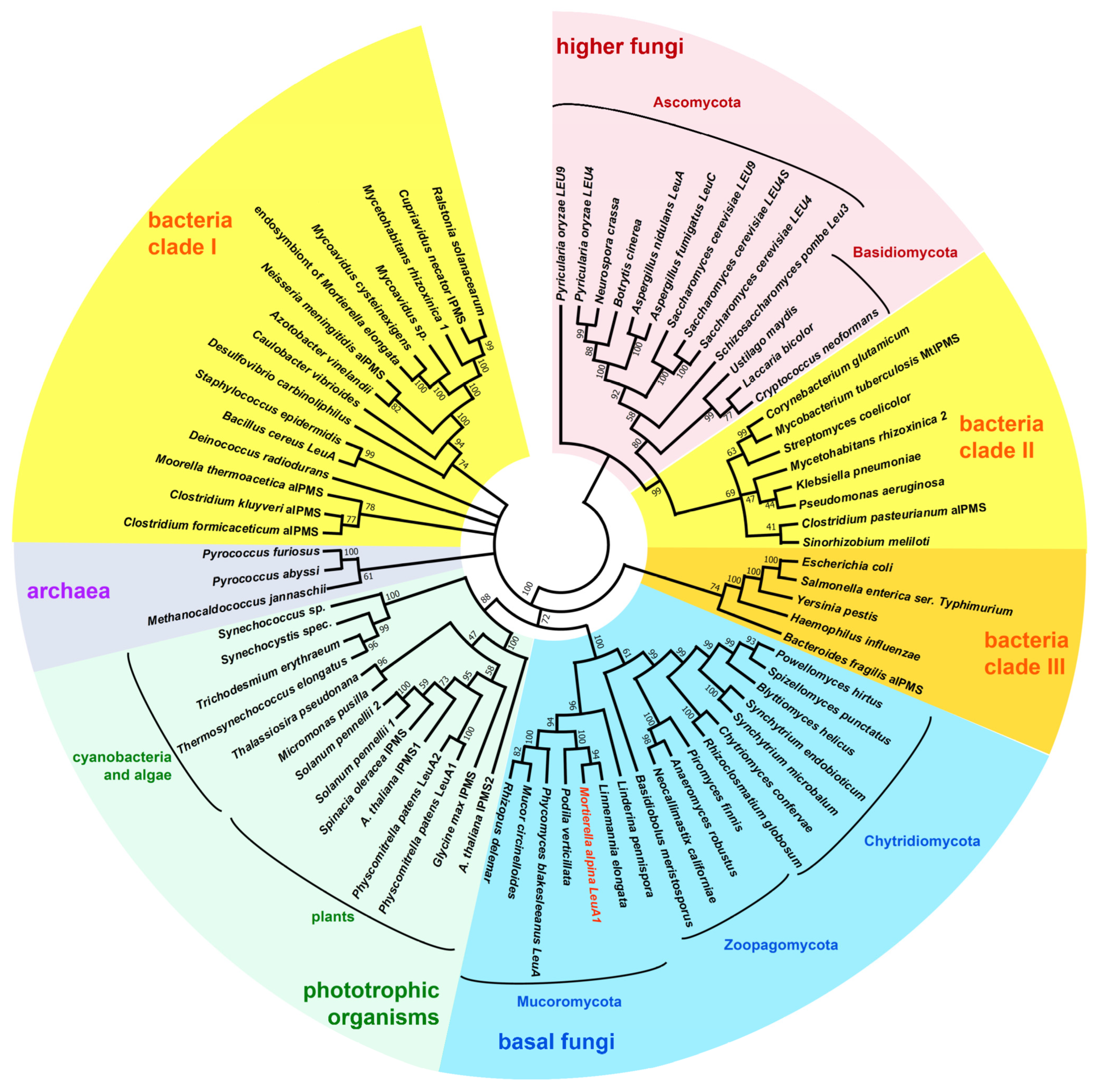
3.1. Identification of BCAA Biosynthetic Genes in M. alpina
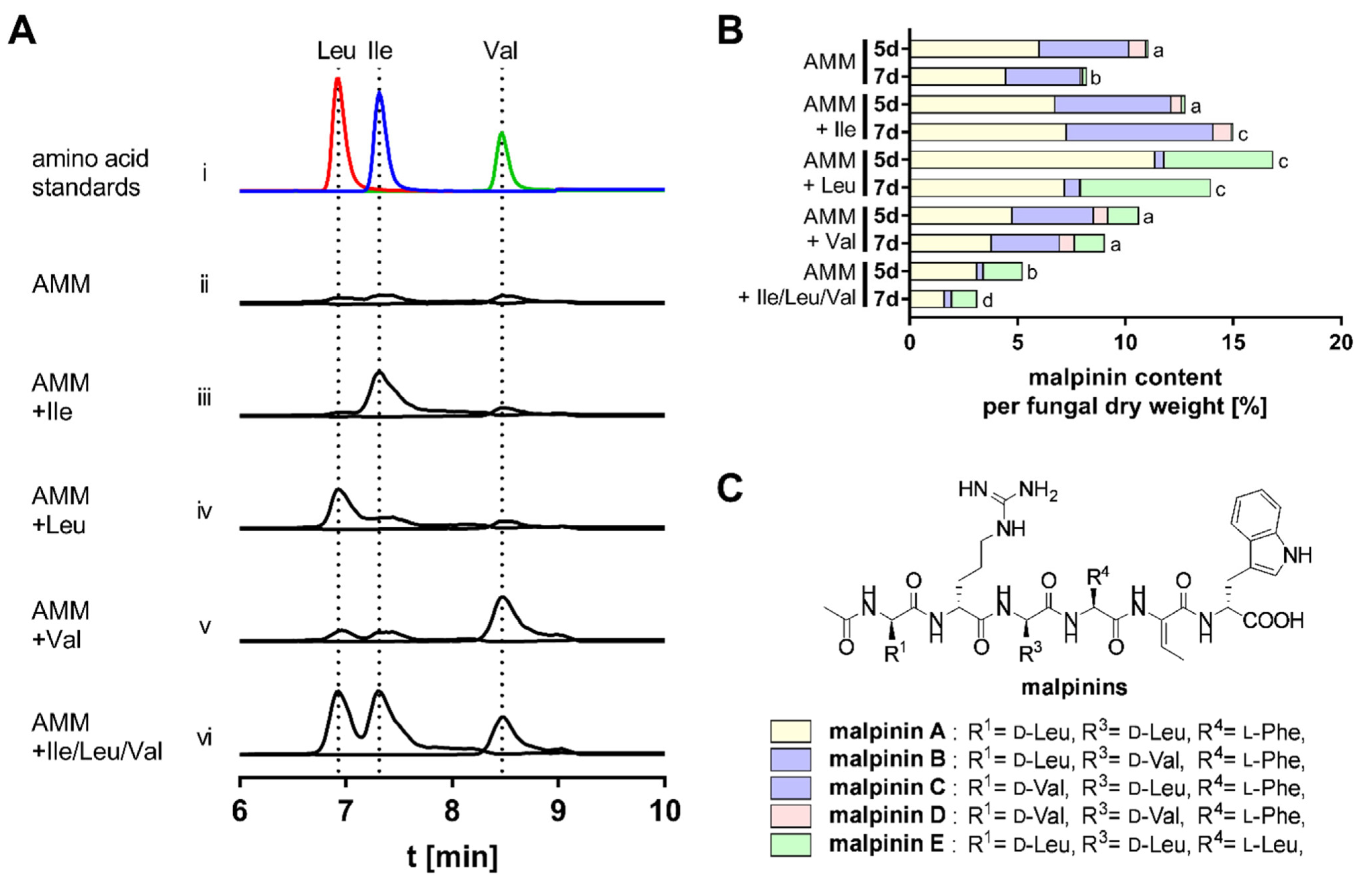
3.2. BCAA Uptake and Its Impact on Malpinin Biosynthesis
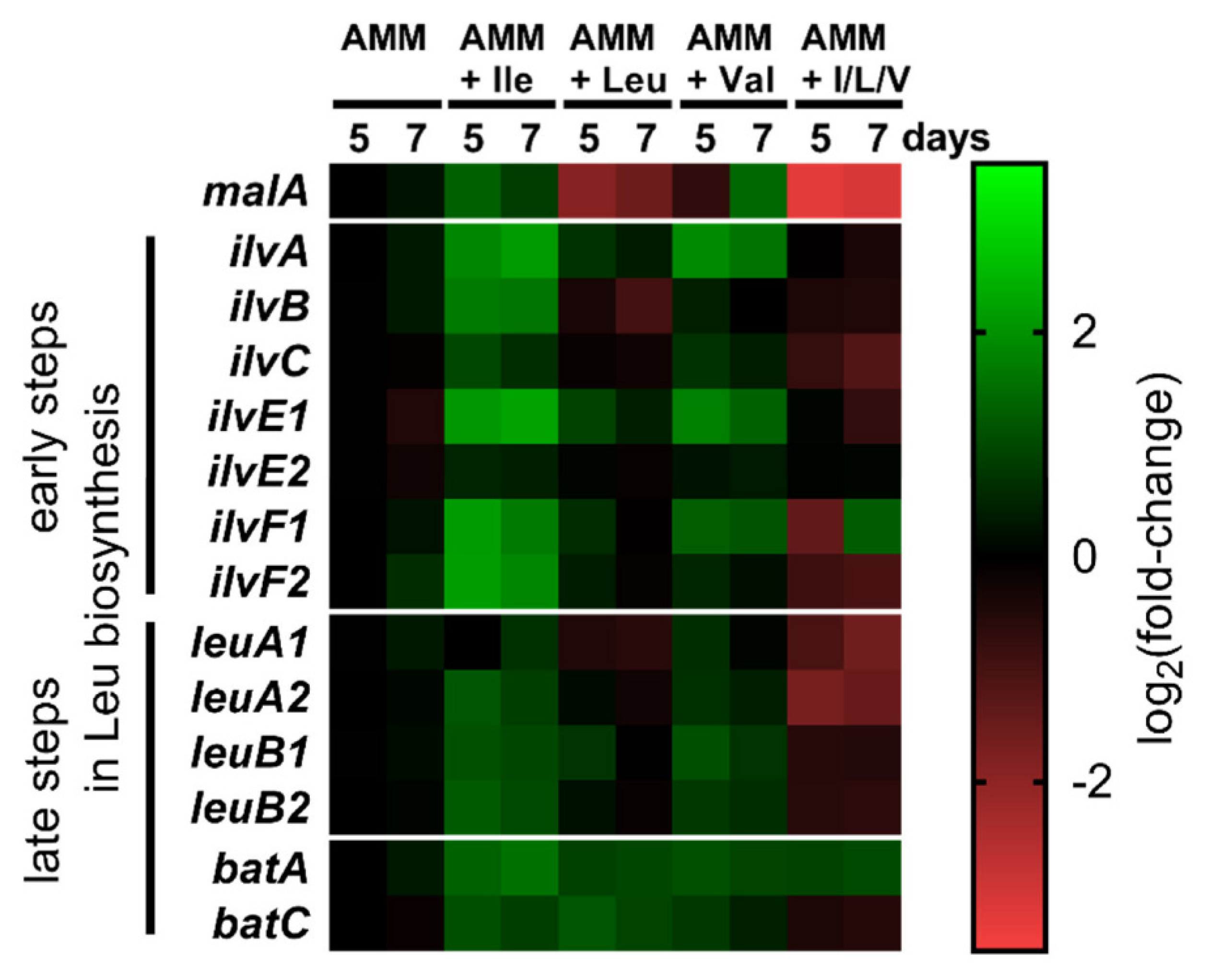
3.3. Transcriptional Regulation of BCAA Biosynthesis
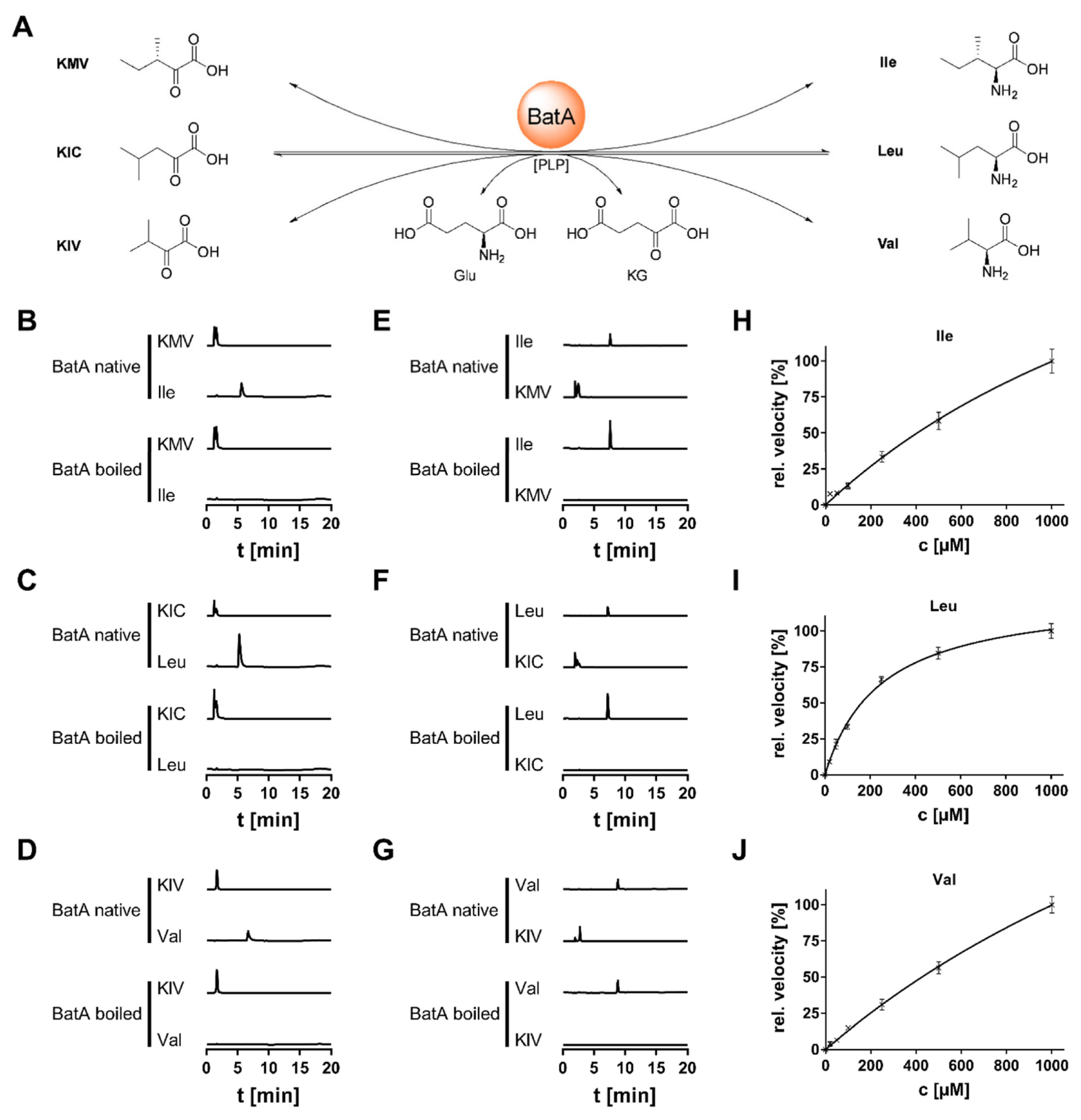
3.4. Biochemical Characterization of BatA
| Organism | Enzyme | Substrate | KM [µM] | kcat [s−1] | kcat/KM [µM−1 s−1] | Ref. |
|---|---|---|---|---|---|---|
| Mortierella alpina | BatA | Ile | 2091.0 ± 505.1 | 49.67 ± 8.79 | 0.024 | |
| Leu | 245.7 ± 18.6 | 17.73 ± 0.50 | 0.072 | |||
| Val | 2830.0 ± 435.5 | 84.87 ± 10.22 | 0.029 | |||
| Rattus norvegicus | BCATm | Ile | 1300 | n.d. | n.d. | [35] |
| Leu | 1600 | n.d. | n.d. | |||
| Val | 7700 | n.d. | n.d. | |||
| human | BCATm | Ile | 600 ± 20 | 80 ± 1 | 0.165 | [37] |
| Leu | 1210 ± 120 | 115 ± 11 | 0.088 | |||
| Val | 6100 ± 110 | 68 ± 2 | 0.011 | |||
| BCATc | Ile | 770 ± 20 | 172 ± 9 | 0.223 | ||
| Leu | 600 ± 40 | 132 ± 7 | 0.220 | |||
| Val | 2400 ± 90 | 122 ± 8 | 0.051 | |||
| Arabidopsis thaliana | BCAT3 | Ile | 140 ± 30 | n.d. | n.d. | [36] |
| Leu | 140 ± 40 | n.d. | n.d. | |||
| Val | 1380 ± 100 | n.d. | n.d. | |||
| Solanum lycopersicum | BCAT-1 | Ile | 670 ± 90 | 40.8 | 0.061 | [38] |
| mitochondrial | Leu | 560 ± 40 | 39.1 | 0.070 | ||
| Val | 1000 ± 100 | 50.5 | 0.050 | |||
| BCAT-5 | Ile | 3200 ± 200 | 163.0 | 0.051 | ||
| cytosolic | Leu | 1800 ± 100 | 118.0 | 0.066 | ||
| Val | 2600 ± 200 | 123.0 | 0.047 |
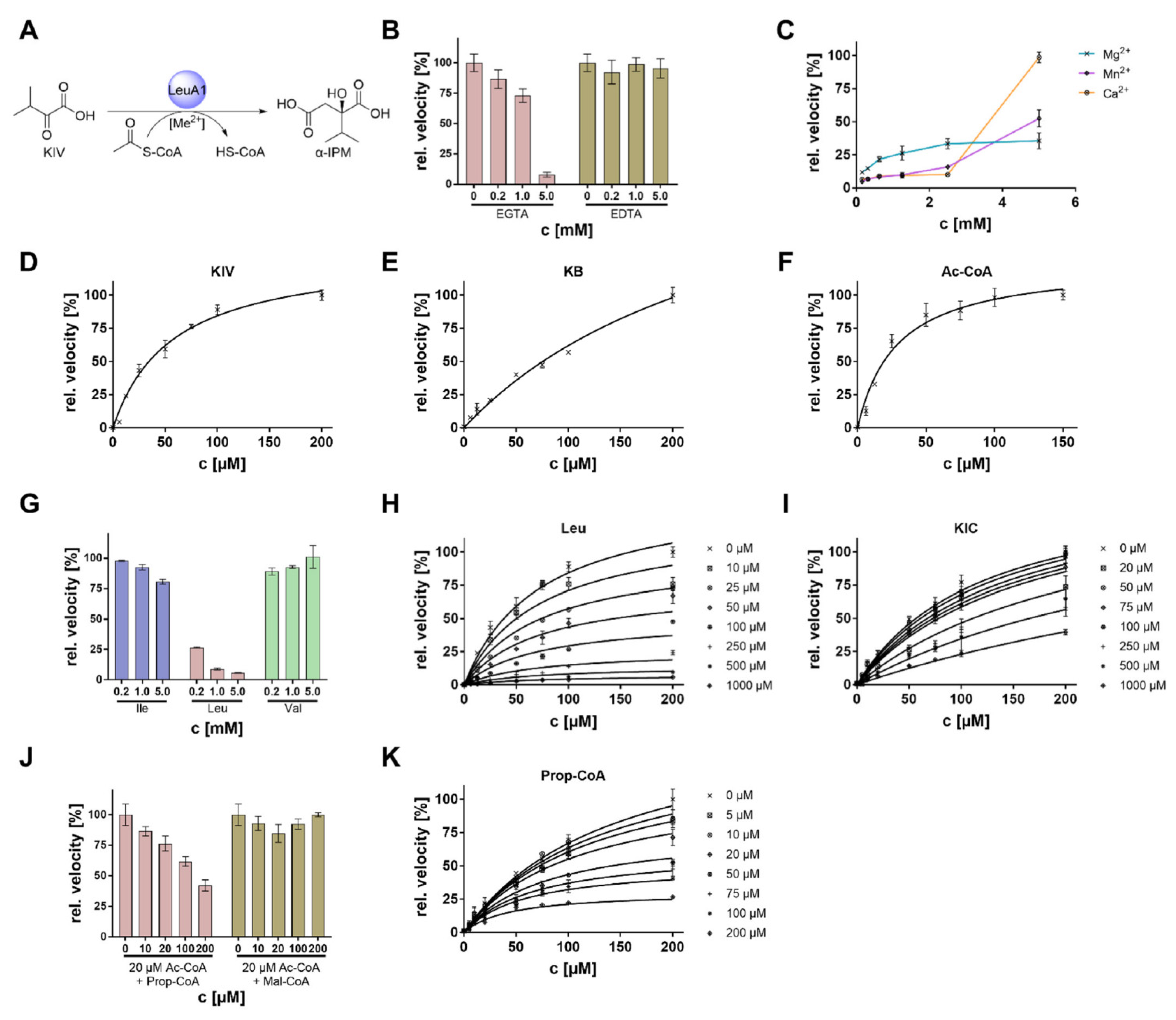
3.5. Biochemical Characterization of LeuA1
| Organism | Enzyme | Substrate | KM [µM] | kcat [s−1] | kcat/KM [µM−1 s−1] | Ref. |
|---|---|---|---|---|---|---|
| Mortierella alpina | LeuA1 | KIV | 56.9 ± 6.3 | 26.57 ± 1.20 | 0.467 | |
| KB | 280.8 ± 45.0 | 47.35 ± 5.41 | 0.169 | |||
| KG | Not a substrate | n.d. | n.d. | |||
| Pyr | Not a substrate | n.d. | n.d. | |||
| Ac-CoA | 28.6 ± 4.1 | 24.98 ± 1.16 | 0.873 | |||
| Prop-CoA | Not a substrate | n.d. | n.d. | |||
| Mal-CoA | Not a substrate | n.d. | n.d. | |||
| Mycobacterium tuberculosis | MtIPMS | KIV | 12 ± 1 | 3.50 ± 0.10 | 0.292 | [39] |
| KB | 410 ± 180 | 7.00 ± 1.30 | 0.012 | |||
| KG | n.d. | n.d. | n.d. | |||
| Pyr | 9500 ± 1680 | 6.10 ± 0.50 | 0.001 | |||
| Ac-CoA | 136 ± 5 | 2.10 ± 0.10 | 0.015 | |||
| Prop-CoA | 220 ± 40 | 0.04 ± 0.003 | 0.0002 | |||
| Mal-CoA | n.d. | n.d. | n.d. | |||
| Mycobacterium tuberculosis | IPMS-2CR | KIV | 261 | 1.17 | 0.005 | [41] |
| Ac-CoA | 568 | 2.22 | 0.004 | |||
| IPMS-14CR | KIV | 35 | 0.52 | 0.015 | ||
| Ac-CoA | 27 | 0.61 | 0.023 | |||
| Thermus thermophilus | TTC0849 | KIV | 54.0 ± 5.8 | 1.20 ± 0.03 | 0.022 | [9] |
| Pyr | 130.0 ± 12.0 | 1.20 ± 0.03 | 0.009 | |||
| Ac-CoA | 23.0 ± 2.5 | 1.70 ± 0.07 | 0.073 | |||
| Arabidopsis thaliana | IPMS1 | KIV | 304.0 ± 68.0 | 2.4 ± 0.21 | 0.008 | [40] |
| Ac-CoA | 45.0 ± 10.0 | 2.2 ± 0.21 | 0.047 | |||
| IPMS2 | KIV | 279.0 ± 68.0 | 2.3 ± 0.17 | 0.008 | ||
| Ac-CoA | 16.0 ± 10.0 | 1.9 ± 0.11 | 0.120 |
| Organism | Enzyme | Inhibitor | KM [µM] | KI [µM] | α # | Comment |
|---|---|---|---|---|---|---|
| Mortierella alpina | LeuA1 | Ile | n.d. | n.d. | n.d. | slight inhibition |
| Leu | 72.6 ± 4.8 | 53.0 ± 2.5 | n.d. | non-competitive inhibition | ||
| Val | n.d. | n.d. | n.d. | no inhibition | ||
| KIC | 124.5 ± 13.1 | 293.3 ± 30.4 | n.d. | competitive inhibition | ||
| Prop-CoA | 145.2 ± 11.0 | 320.36 ± 121.17 | 0.14 | mixed type inhibition |
3.6. Complex Metabolic Regulation of the Key Enzyme LeuA1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kohlhaw, G.B. Leucine biosynthesis in fungi: Entering metabolism through the back door. Microbiol. Mol. Biol. Rev. 2003, 67, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Orasch, T.; Dietl, A.M.; Shadkchan, Y.; Binder, U.; Bauer, I.; Lass-Florl, C.; Osherov, N.; Haas, H. The leucine biosynthetic pathway is crucial for adaptation to iron starvation and virulence in Aspergillus fumigatus. Virulence 2019, 10, 925–934. [Google Scholar] [CrossRef]
- Que, Y.; Yue, X.; Yang, N.; Xu, Z.; Tang, S.; Wang, C.; Lv, W.; Xu, L.; Talbot, N.J.; Wang, Z. Leucine biosynthesis is required for infection-related morphogenesis and pathogenicity in the rice blast fungus Magnaporthe oryzae. Curr. Genet. 2020, 66, 155–171. [Google Scholar] [CrossRef]
- Larson, E.M.; Idnurm, A. Two origins for the gene encoding alpha-isopropylmalate synthase in fungi. PLoS ONE 2010, 5, e11605. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tang, X.; Chang, L.L.; Gu, S.J.; Zhang, H.; Chen, Y.Q.; Chen, H.Q.; Zhao, J.X.; Chen, W. Role of beta-isopropylmalate dehydrogenase in lipid biosynthesis of the oleaginous fungus Mortierella alpina. Fungal Genet. Biol. 2021, 152, 103572. [Google Scholar] [CrossRef] [PubMed]
- Colon, M.; Hernandez, F.; Lopez, K.; Quezada, H.; Gonzalez, J.; Lopez, G.; Aranda, C.; Gonzalez, A. Saccharomyces cerevisiae BAT1 and BAT2 aminotransferases have functionally diverged from the ancestral-like Kluyveromyces lactis orthologous enzyme. PLoS ONE 2011, 6, e16099. [Google Scholar] [CrossRef] [PubMed]
- Lopez, G.; Quezada, H.; Duhne, M.; Gonzalez, J.; Lezama, M.; El-Hafidi, M.; Colon, M.; Martinez de la Escalera, X.; Flores-Villegas, M.C.; Scazzocchio, C.; et al. Diversification of paralogous alpha-isopropylmalate synthases by modulation of feedback control and hetero-oligomerization in Saccharomyces cerevisiae. Eukaryot. Cell 2015, 14, 564–577. [Google Scholar] [CrossRef]
- Friden, P.; Schimmel, P. LEU3 of Saccharomyces cerevisiae activates multiple genes for branched-chain amino acid biosynthesis by binding to a common decanucleotide core sequence. Mol. Cell. Biol. 1988, 8, 2690–2697. [Google Scholar]
- Yoshida, A.; Kosono, S.; Nishiyama, M. Characterization of two 2-isopropylmalate synthase homologs from Thermus thermophilus HB27. Biochem. Biophys. Res. Commun. 2018, 501, 465–470. [Google Scholar] [CrossRef]
- Shinmen, Y.; Shimizu, S.; Akimoto, K.; Kawashima, H.; Yamada, H. Production of arachidonic acid by Mortierella fungi. Appl. Microbiol. Biotechnol. 1989, 31, 11–16. [Google Scholar] [CrossRef]
- Dai, P.; Chen, H.Q.; Yang, B.; Wang, H.C.; Yang, Q.; Zhang, H.; Chen, W.; Chen, Y.Q. Mortierella alpina feed supplementation enriched hen eggs with DHA and AA. RSC Adv. 2016, 6, 1694–1699. [Google Scholar] [CrossRef]
- Slany, O.; Klempova, T.; Shapaval, V.; Zimmermann, B.; Kohler, A.; Certik, M. Biotransformation of animal fat-by products into ARA-enriched fermented bioproducts by solid-state fermentation of Mortierella alpina. J Fungi 2020, 6, 236. [Google Scholar] [CrossRef]
- Shimizu, S.; Kawashima, H.; Akimoto, K.; Shinmen, Y.; Yamada, H. Production of odd chain polyunsaturated fatty-acids by Mortierella fungi. J. Am. Oil Chem. Soc. 1991, 68, 254–258. [Google Scholar] [CrossRef]
- Chang, L.L.; Chen, H.Q.; Tang, X.; Zhao, J.X.; Zhang, H.; Chen, Y.Q.; Chen, W. Advances in improving the biotechnological application of oleaginous fungus Mortierella alpina. Appl. Microbiol. Biotechnol. 2021, 105, 6275–6289. [Google Scholar] [CrossRef] [PubMed]
- Downes, D.J.; Davis, M.A.; Kreutzberger, S.D.; Taig, B.L.; Todd, R.B. Regulation of the NADP-glutamate dehydrogenase gene gdhA in Aspergillus nidulans by the Zn(II)2Cys6 transcription factor LeuB. Microbiology 2013, 159, 2467–2480. [Google Scholar] [CrossRef][Green Version]
- Armenteros, J.J.A.; Salvatore, M.; Emanuelsson, O.; Winther, O.; von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef] [PubMed]
- Fukasawa, Y.; Tsuji, J.; Fu, S.C.; Tomii, K.; Horton, P.; Imai, K. Mitofates: Improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteom. 2015, 14, 1113–1126. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, T.; Itoh, M.; Izumikawa, M.; Sakata, N.; Tsuchida, T.; Shin-Ya, K. Novel arginine-containing peptides MBJ-0173 and MBJ-0174 from Mortierella alpina f28740. J. Antibiot. 2017, 70, 226–229. [Google Scholar] [CrossRef]
- Baldeweg, F.; Warncke, P.; Fischer, D.; Gressler, M. Fungal biosurfactants from Mortierella alpina. Org. Lett. 2019, 21, 1444–1448. [Google Scholar] [CrossRef]
- Koyama, N.; Kojima, S.; Nonaka, K.; Masuma, R.; Matsumoto, M.; Omura, S.; Tomoda, H. Calpinactam, a new anti-mycobacterial agent, produced by Mortierella alpina FKI-4905. J. Antibiot. 2010, 63, 183–186. [Google Scholar] [CrossRef]
- Wurlitzer, J.M.; Stanisic, A.; Wasmuth, I.; Jungmann, S.; Fischer, D.; Kries, H.; Gressler, M. Bacterial-like nonribosomal peptide synthetases produce cyclopeptides in the zygomycetous fungus Mortierella alpina. Appl. Environ. Microbiol. 2021, 87, e02051-20. [Google Scholar] [CrossRef] [PubMed]
- Wurlitzer, J.M.; Stanišić, A.; Ziethe, S.; Jordan, P.M.; Günther, K.; Werz, O.; Kries, H.; Gressler, M. Macrophage-targeting oligopeptides from Mortierella alpina. 2022; submitted manuscript. [Google Scholar]
- Tabima, J.F.; Trautman, I.A.; Chang, Y.; Wang, Y.; Mondo, S.; Kuo, A.; Salamov, A.; Grigoriev, I.V.; Stajich, J.E.; Spatafora, J.W. Phylogenomic analyses of non-dikarya fungi supports horizontal gene transfer driving diversification of secondary metabolism in the amphibian gastrointestinal symbiont, Basidiobolus. G3 Genes Genomes Genet. 2020, 10, 3417–3433. [Google Scholar] [CrossRef] [PubMed]
- Gressler, M.; Meyer, F.; Heine, D.; Hortschansky, P.; Hertweck, C.; Brock, M. Phytotoxin production in Aspergillus terreus is regulated by independent environmental signals. eLife 2015, 4, e07861. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Steyer, J.T.; Downes, D.J.; Hunter, C.C.; Migeon, P.A.; Todd, R.B. Duplication and functional divergence of branched-chain amino acid biosynthesis genes in Aspergillus nidulans. mBio 2021, 12, e0076821. [Google Scholar] [CrossRef]
- Gonzalez, J.; Lopez, G.; Argueta, S.; Escalera-Fanjul, X.; El Hafidi, M.; Campero-Basaldua, C.; Strauss, J.; Riego-Ruiz, L.; Gonzalez, A. Diversification of transcriptional regulation determines subfunctionalization of paralogous branched chain aminotransferases in the yeast Saccharomyces cerevisiae. Genetics 2017, 207, 975–991. [Google Scholar] [CrossRef][Green Version]
- Long, N.; Orasch, T.; Zhang, S.; Gao, L.; Xu, X.; Hortschansky, P.; Ye, J.; Zhang, F.; Xu, K.; Gsaller, F.; et al. The Zn2Cys6-type transcription factor LeuB cross-links regulation of leucine biosynthesis and iron acquisition in Aspergillus fumigatus. PLoS Genet. 2018, 14, e1007762. [Google Scholar] [CrossRef]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, S.; Yin, X.; Yu, D.; Xie, X.; Huang, B. Functional analysis of keto-acid reductoisomerase IlvC in the entomopathogenic fungus Metarhizium robertsii. J. Fungi 2021, 7, 737. [Google Scholar] [CrossRef]
- Orij, R.; Postmus, J.; Ter Beek, A.; Brul, S.; Smits, G.J. In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology 2009, 155, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Kotogán, A.; Zambrano, C.; Kecskeméti, A.; Varga, M.; Szekeres, A.; Papp, T.; Vágvölgyi, C.; Takó, M. An organic solvent-tolerant lipase with both hydrolytic and synthetic activities from the oleaginous fungus Mortierella echinosphaera. Int. J. Mol. Sci. 2018, 19, 1129. [Google Scholar] [CrossRef] [PubMed]
- Bezsudnova, E.Y.; Boyko, K.M.; Popov, V.O. Properties of bacterial and archaeal branched-chain amino acid aminotransferases. Biochemistry 2017, 82, 1572–1591. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Nautiyal, M.; Wynn, R.M.; Mobley, J.A.; Chuang, D.T.; Hutson, S.M. Branched-chain amino acid metabolon: Interaction of glutamate dehydrogenase with the mitochondrial branched-chain aminotransferase (BCATm). J. Biol. Chem. 2010, 285, 265–276. [Google Scholar] [CrossRef]
- Knill, T.; Schuster, J.; Reichelt, M.; Gershenzon, J.; Binder, S. Arabidopsis branched-chain aminotransferase 3 functions in both amino acid and glucosinolate biosynthesis. Plant Physiol. 2008, 146, 1028–1039. [Google Scholar] [CrossRef]
- Davoodi, J.; Drown, P.M.; Bledsoe, R.K.; Wallin, R.; Reinhart, G.D.; Hutson, S.M. Overexpression and characterization of the human mitochondrial and cytosolic branched-chain aminotransferases. J. Biol. Chem. 1998, 273, 4982–4989. [Google Scholar] [CrossRef]
- Maloney, G.S.; Kochevenko, A.; Tieman, D.M.; Tohge, T.; Krieger, U.; Zamir, D.; Taylor, M.G.; Fernie, A.R.; Klee, H.J. Characterization of the branched-chain amino acid aminotransferase enzyme family in tomato. Plant Physiol. 2010, 153, 925–936. [Google Scholar] [CrossRef]
- Casey, A.K.; Hicks, M.A.; Johnson, J.L.; Babbitt, P.C.; Frantom, P.A. Mechanistic and bioinformatic investigation of a conserved active site helix in alpha-isopropylmalate synthase from Mycobacterium tuberculosis, a member of the DRE-TIM metallolyase superfamily. Biochemistry 2014, 53, 2915–2925. [Google Scholar] [CrossRef]
- De Kraker, J.W.; Luck, K.; Textor, S.; Tokuhisa, J.G.; Gershenzon, J. Two Arabidopsis genes (IPMS1 and IMPS2) encode isopropylmalate synthase, the branchpoint step in the biosynthesis of leucine. Plant Physiol. 2007, 143, 970–986. [Google Scholar] [CrossRef]
- Yindeeyoungyeon, W.; Likitvivatanavong, S.; Palittapongarnpim, P. Characterization of alpha-isopropylmalate synthases containing different copy numbers of tandem repeats in Mycobacterium tuberculosis. BMC Microbiol. 2009, 9, 122. [Google Scholar] [CrossRef]
- De Carvalho, L.P.; Blanchard, J.S. Kinetic analysis of the effects of monovalent cations and divalent metals on the activity of Mycobacterium tuberculosis alpha-isopropylmalate synthase. Arch. Biochem. Biophys. 2006, 451, 141–148. [Google Scholar] [CrossRef]
- Crea, F.; De Stefano, C.; Gianguzza, A.; Piazzese, D.; Sammartano, S. Speciation of poly-amino carboxylic compounds in seawater. Chem. Speciat. Bioavailab. 2003, 15, 75–86. [Google Scholar] [CrossRef]
- Koon, N.; Squire, C.J.; Baker, E.N. Crystal structure of LeuA from Mycobacterium tuberculosis, a key enzyme in leucine biosynthesis. Proc. Natl. Acad. Sci. USA 2004, 101, 8295–8300. [Google Scholar] [CrossRef] [PubMed]
- Wiegel, J.; Schlegel, H.G. Alpha-isopropylmalate synthase from Alcaligenes eutrophus H16. 2. Substrate-specificity and kinetics. Arch. Microbiol. 1977, 112, 247–254. [Google Scholar] [CrossRef] [PubMed]
- De Castro, P.A.; Colabardini, A.C.; Moraes, M.; Horta, M.A.C.; Knowles, S.L.; Raja, H.A.; Oberlies, N.H.; Koyama, Y.; Ogawa, M.; Gomi, K.; et al. Regulation of gliotoxin biosynthesis and protection in Aspergillus species. PLoS Genet. 2022, 18, e1009965. [Google Scholar] [CrossRef]
- Gressler, M.; Zaehle, C.; Scherlach, K.; Hertweck, C.; Brock, M. Multifactorial induction of an orphan PKS-NRPS gene cluster in Aspergillus terreus. Chem. Biol. 2011, 18, 198–209. [Google Scholar] [CrossRef]
- Wang, B.; Han, X.; Bai, Y.; Lin, Z.; Qiu, M.; Nie, X.; Wang, S.; Zhang, F.; Zhuang, Z.; Yuan, J.; et al. Effects of nitrogen metabolism on growth and aflatoxin biosynthesis in Aspergillus flavus. J. Hazard. Mater. 2017, 324, 691–700. [Google Scholar] [CrossRef]
- Subramaniam, R.; Narayanan, S.; Walkowiak, S.; Wang, L.; Joshi, M.; Rocheleau, H.; Ouellet, T.; Harris, L.J. Leucine metabolism regulates tri6 expression and affects deoxynivalenol production and virulence in Fusarium graminearum. Mol. Microbiol. 2015, 98, 760–769. [Google Scholar] [CrossRef]
- García-Estrada, C.; Domínguez-Santos, R.; Kosalková, K.; Martín, J. Transcription factors controlling primary and secondary metabolism in filamentous fungi: The beta-lactam paradigm. Fermentation 2018, 4, 47. [Google Scholar] [CrossRef]
- Shelest, E. Transcription factors in fungi: TFome dynamics, three major families, and dual-specificity TFs. Front. Genet. 2017, 8, 53. [Google Scholar] [CrossRef]
- Zhou, K.M.; Bai, Y.L.; Kohlhaw, G.B. Yeast regulatory protein LEU3: A structure-function analysis. Nucleic Acids Res. 1990, 18, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Cooper, T.G.; Kohlhaw, G.B. The Saccharomyces cerevisiae LEU3 protein activates expression of gdh1, a key gene in nitrogen assimilation. Mol. Cell. Biol. 1995, 15, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Prohl, C.; Kispal, G.; Lill, R. Branched-chain-amino-acid transaminases of yeast Saccharomyces cerevisiae. Methods Enzymol. 2000, 324, 365–375. [Google Scholar] [PubMed]
- Yennawar, N.; Dunbar, J.; Conway, M.; Hutson, S.; Farber, G. The structure of human mitochondrial branched-chain aminotransferase. Acta Crystallogr. D Biol. Crystallogr. 2001, 57, 506–515. [Google Scholar] [CrossRef]
- Shafei, M.A.; Forshaw, T.; Davis, J.; Flemban, A.; Qualtrough, D.; Dean, S.; Perks, C.; Dong, M.; Newman, R.; Conway, M.E. BCATc modulates crosstalk between the PI3K/Akt and the Ras/ERK pathway regulating proliferation in triple negative breast cancer. OncoTarget 2020, 11, 1971–1987. [Google Scholar] [CrossRef]
- Gakh, O.; Cavadini, P.; Isaya, G. Mitochondrial processing peptidases. Biochim. Biophys. Acta 2002, 1592, 63–77. [Google Scholar] [CrossRef]
- Cavalieri, D.; Casalone, E.; Bendoni, B.; Fia, G.; Polsinelli, M.; Barberio, C. Trifluoroleucine resistance and regulation of alpha-isopropyl malate synthase in Saccharomyces cerevisiae. Mol. Gen. Genet. 1999, 261, 152–160. [Google Scholar] [CrossRef]
- Hewton, K.G.; Johal, A.S.; Parker, S.J. Transporters at the interface between cytosolic and mitochondrial amino acid metabolism. Metabolites 2021, 11, 112. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Brock, M.; Keller, N.P. Connection of propionyl-CoA metabolism to polyketide biosynthesis in Aspergillus nidulans. Genetics 2004, 168, 785–794. [Google Scholar] [CrossRef]
- Brock, M.B.W. On the mechanism of action of the antifungal agent propionate: Propionyl-CoA inhibits glucose metabolism in Aspergillus nidulans. Eur. J. Biochem. 2004, 271, 3227–3241. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Bartnik, E.; Bonnet-Bidaud, A.; Kowalska, I.; Pieniazek, N.J. New leucine auxotrophs of Aspergillus nidulans. Acta Microbiol. Pol. 1980, 29, 29–33. [Google Scholar] [PubMed]
- Wiegel, J. Alpha-isopropylmalate synthase as a marker for the leucine biosynthetic pathway in several Clostridia and in Bacteroides fragilis. Arch. Microbiol. 1981, 130, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Wiegel, J.; Schlegel, H.G. Alpha-isopropylmalate synthase from Alcaligenes eutrophus H 16. I. Purification and general properties. Arch. Microbiol. 1977, 112, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Nouwen, N.; Arrighi, J.F.; Cartieaux, F.; Chaintreuil, C.; Gully, D.; Klopp, C.; Giraud, E. The role of rhizobial (NifV) and plant (Fen1) homocitrate synthases in Aeschynomene/photosynthetic Bradyrhizobium symbiosis. Sci. Rep. 2017, 7, e448. [Google Scholar] [CrossRef]
- Huisman, F.H.; Hunter, M.F.; Devenish, S.R.; Gerrard, J.A.; Parker, E.J. The C-terminal regulatory domain is required for catalysis by Neisseria meningitidis alpha-isopropylmalate synthase. Biochem. Biophys. Res. Commun. 2010, 393, 168–173. [Google Scholar] [CrossRef]
- Beltzer, J.P.; Morris, S.R.; Kohlhaw, G.B. Yeast Leu4 encodes mitochondrial and nonmitochondrial forms of alpha-isopropylmalate synthase. J. Biol. Chem. 1988, 263, 368–374. [Google Scholar] [CrossRef]
- Casalone, E.; Barberio, C.; Cavalieri, D.; Polsinelli, M. Identification by functional analysis of the gene encoding alpha-isopropylmalate synthase II (Leu9) in Saccharomyces cerevisiae. Yeast 2000, 16, 539–545. [Google Scholar] [CrossRef]
- Diebold, R.; Schuster, J.; Daschner, K.; Binder, S. The branched-chain amino acid transaminase gene family in Arabidopsis encodes plastid and mitochondrial proteins. Plant Physiol. 2002, 129, 540–550. [Google Scholar] [CrossRef]
- Berger, B.J.; English, S.; Chan, G.; Knodel, M.H. Methionine regeneration and aminotransferases in Bacillus subtilis, Bacillus cereus, and Bacillus anthracis. J. Bacteriol. 2003, 185, 2418–2431. [Google Scholar] [CrossRef]
- Chun, C.D.; Brown, J.C.S.; Madhani, H.D. A major role for capsule-independent phagocytosis-inhibitory mechanisms in mammalian infection by Cryptococcus neoformans. Cell Host Microbe 2011, 9, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Merchant, S.S.; Prochnik, S.E.; Vallon, O.; Harris, E.H.; Karpowicz, S.J.; Witman, G.B.; Terry, A.; Salamov, A.; Fritz-Laylin, L.K.; Marechal-Drouard, L.; et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 2007, 318, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Somers, J.M.; Amzallag, A.; Middleton, R.B. Genetic fine structure of the leucine operon of Escherichia coli k-12. J. Bacteriol. 1973, 113, 1268–1272. [Google Scholar] [CrossRef] [PubMed]
- Khoei, M.A.; Karimi, M.; Karamian, R.; Amini, S.; Soorni, A. Identification of the complex interplay between nematode-related lncRNAs and their target genes in Glycine max l. Front Plant Sci 2021, 12, e779597. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sonnabend, R.; Seiler, L.; Gressler, M. Regulation of the Leucine Metabolism in Mortierella alpina. J. Fungi 2022, 8, 196. https://doi.org/10.3390/jof8020196
Sonnabend R, Seiler L, Gressler M. Regulation of the Leucine Metabolism in Mortierella alpina. Journal of Fungi. 2022; 8(2):196. https://doi.org/10.3390/jof8020196
Chicago/Turabian StyleSonnabend, Robin, Lucas Seiler, and Markus Gressler. 2022. "Regulation of the Leucine Metabolism in Mortierella alpina" Journal of Fungi 8, no. 2: 196. https://doi.org/10.3390/jof8020196
APA StyleSonnabend, R., Seiler, L., & Gressler, M. (2022). Regulation of the Leucine Metabolism in Mortierella alpina. Journal of Fungi, 8(2), 196. https://doi.org/10.3390/jof8020196






