The Loss-of-Function Mutation aldA67 Leads to Enhanced α-L-Rhamnosidase Production by Aspergillus nidulans
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. RNA Isolation and RT-qPCR
2.3. α-L-Rhamnosidase Assays
3. Results
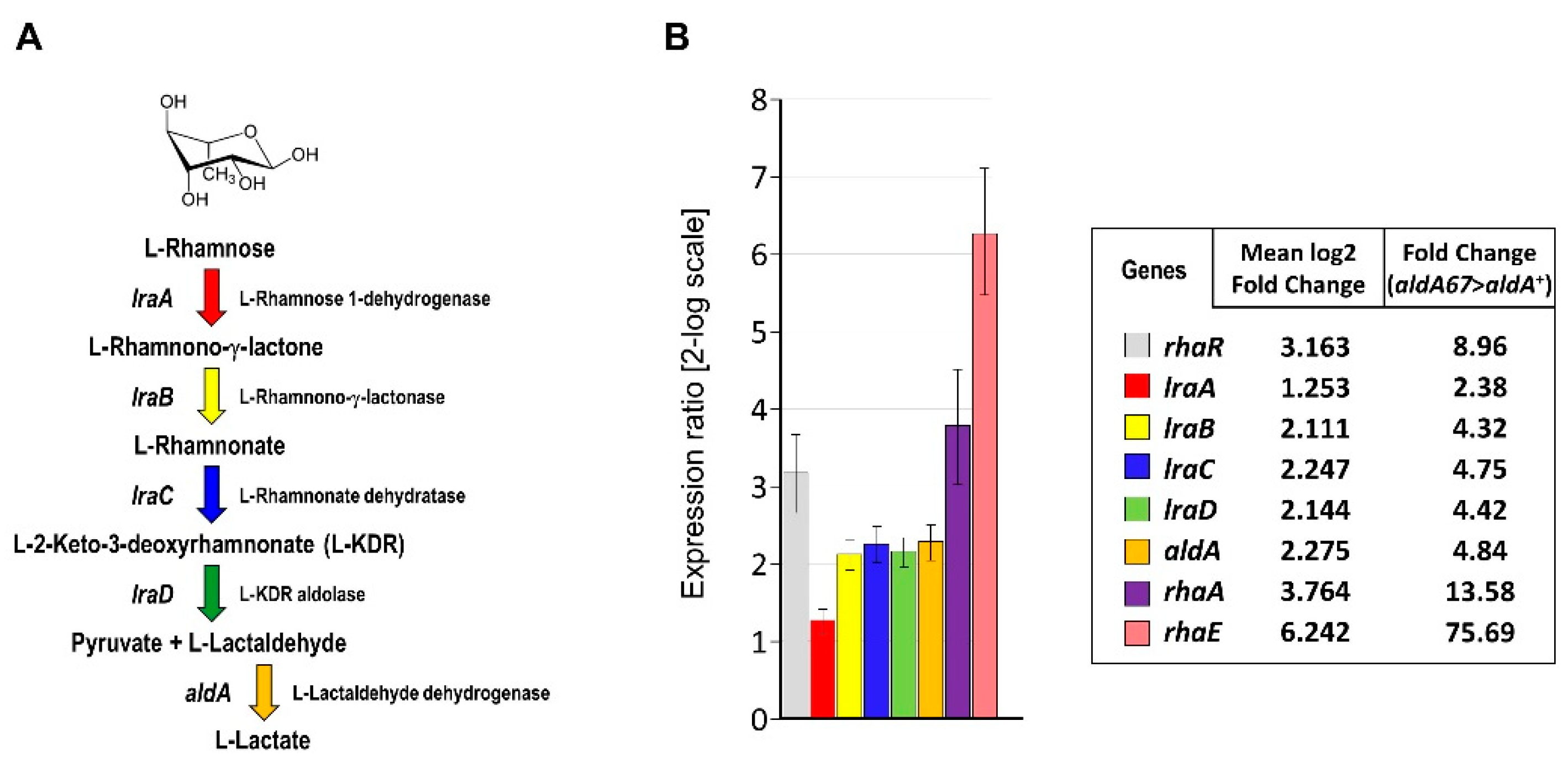
3.1. The aldA67 Loss-of-Function Mutation Affects the Expression of Genes Involved in L-Rhamnose Utilisation
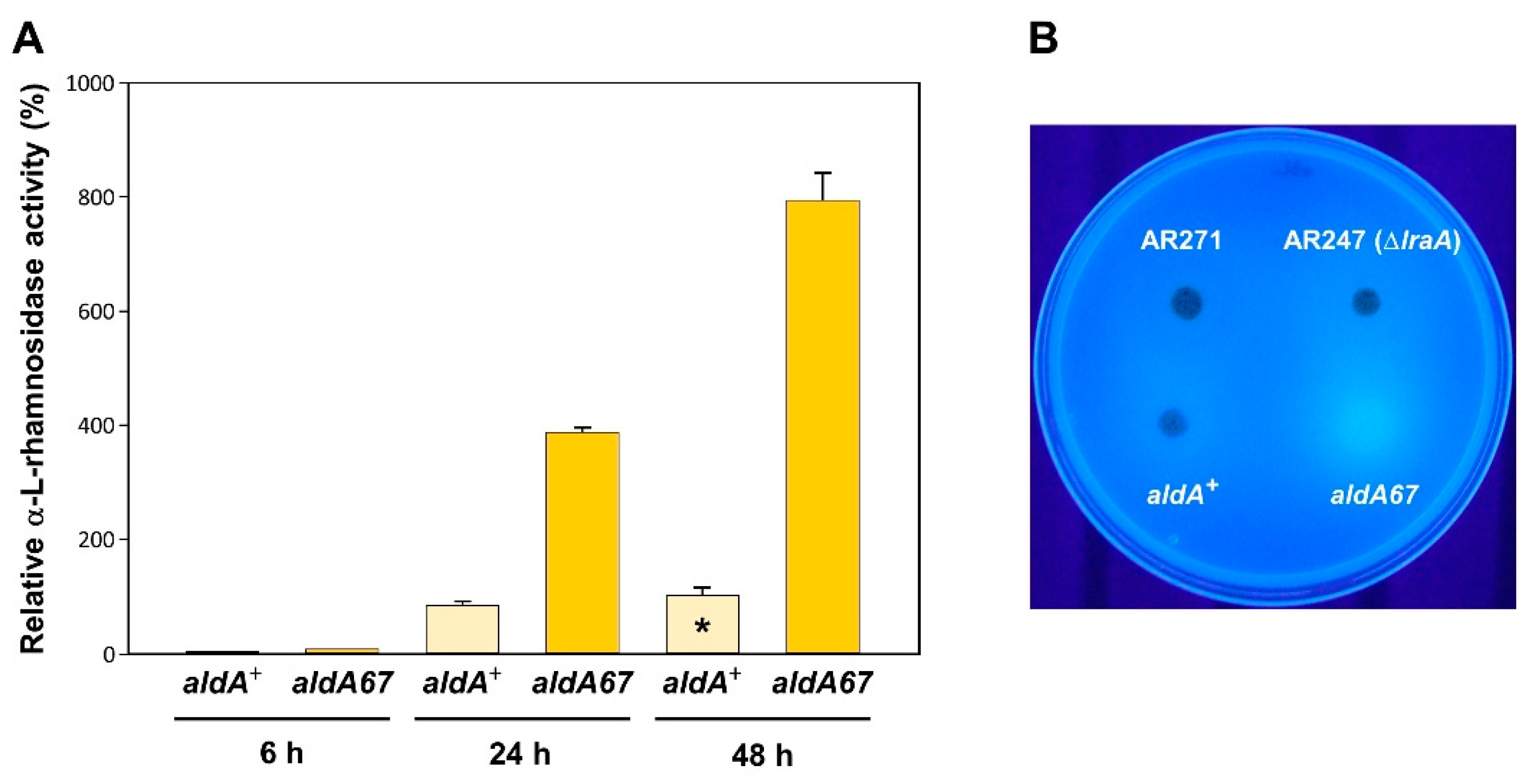
3.2. The aldA67 Mutation Results in Greater α-L-Rhamnosidase Production
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zeng, Y.; Himmel, M.E.; Ding, S.Y. Visualizing chemical functionality in plant cell walls. Biotechnol. Biofuels 2017, 10, 263. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Daniel, G. Heterogeneous distribution of pectin and hemicellulose epitopes in the phloem of four hardwood species. Trees 2018, 32, 393–414. [Google Scholar] [CrossRef]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, J.P.; Wubben, J.P.; Osbourn, A.E. Stagonospora avenae secretes multiple enzymes that hydrolyze oat leaf saponins. Mol. Plant Microbe Interact. 2000, 13, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- González-Barrio, R.; Trindade, L.M.; Manzanares, P.; de Graaff, L.H.; Tomás-Barberán, F.A.; Espín, J.C. Production of bioavailable flavonoid glucosides in fruit juices and green tea by use of fungal α-L-rhamnosidases. J. Agric. Food Chem. 2004, 52, 6136–6142. [Google Scholar] [CrossRef]
- Manzanares, P.; Vallés, S.; Ramón, D.; Orejas, M. α-L-Rhamnosidases: Old and new insights. In Industrial Enzymes: Structure, Function and Applications; Polaina, J., MacCabe, A.P., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 117–140. Available online: https://link.springer.com/content/pdf/10.1007/1-4020-5377-0_8.pdf (accessed on 4 October 2022).
- Yadav, V.; Yadav, P.K.; Yadav, S.; Yadav, K.D.S. α-L-Rhamnosidase: A review. Process Biochem. 2010, 45, 1226–1235. [Google Scholar] [CrossRef]
- Robert, L.; Labat-Robert, J.; Robert, A.-M. Physiology of skin aging. Clin. Plast. Surg. 2012, 39, 1–8. [Google Scholar] [CrossRef]
- Mueller, M.; Zartl, B.; Schleritzko, A. Rhamnosidase activity of selected probiotics and their ability to hydrolyse flavonoid rhamnoglucosides. Bioprocess Biosyst. Eng. 2018, 41, 221–228. [Google Scholar] [CrossRef]
- Westrick, N.M.; Smith, D.L.; Kabbage, M. Disarming the host: Detoxification of plant defense compounds during fungal necrotrophy. Front. Plant Sci. 2021, 12, 651716. [Google Scholar] [CrossRef]
- Monti, D.; Pišvejcová, A.; Křen, V.; Lama, M.; Riva, S. Generation of an α-L-rhamnosidase library and its application for the selective derhamnosylation of natural products. Biotechnol. Bioeng. 2004, 87, 763–771. [Google Scholar] [CrossRef]
- Watanabe, S.; Makino, K. Novel modified version of nonphosphorylated sugar metabolism—An alternative L-rhamnose pathway of Sphingomonas sp. FEBS J. 2009, 276, 1554–1567. [Google Scholar] [CrossRef] [PubMed]
- MacCabe, A.P.; Ninou, E.I.; Pardo, E.; Orejas, M. Catabolism of L-rhamnose in A. nidulans proceeds via the non-phosphorylated pathway and is glucose repressed by a CreA-independent mechanism. Microb. Cell Fact. 2020, 19, 188. [Google Scholar] [CrossRef] [PubMed]
- MacCabe, A.; Sanmartín, G.; Orejas, M. Identification of the genes encoding the catalytic steps corresponding to LRA4 (L-2-keto-3-deoxyrhamnonate aldolase) and L-lactaldehyde dehydrogenase in Aspergillus nidulans: Evidence for involvement of the loci AN9425/lraD and AN0544/aldA in the L-rhamnose catabolic pathway. Environ. Microbiol. 2021, 23, 2420–2432. [Google Scholar] [CrossRef] [PubMed]
- Orejas, M.; Ibáñez, E.; Ramón, D. The filamentous fungus Aspergillus nidulans produces an α-L-rhamnosidase of potential oenological interest. Lett. Appl. Microbiol. 1999, 28, 383–388. [Google Scholar] [CrossRef]
- Manzanares, P.; Orejas, M.; Ibáñez, E.; Vallés, S.; Ramón, D. Purification and characterization of an α-L-rhamnosidase from Aspergillus nidulans. Lett. Appl. Microbiol. 2000, 31, 198–202. [Google Scholar] [CrossRef]
- Tamayo-Ramos, J.A.; Flipphi, M.; Pardo, E.; Manzanares, P.; Orejas, M. L-rhamnose induction of Aspergillus nidulans α-L-rhamnosidase genes is glucose repressed via a CreA-independent mechanism acting at the level of inducer uptake. Microb. Cell Fact. 2012, 21, 11–26. [Google Scholar] [CrossRef]
- Pardo, E.; Orejas, M. The Aspergillus nidulans Zn(II)2Cys6 transcription factor AN5673/RhaR mediates L-rhamnose utilization and the production of α-L-rhamnosidases. Microb. Cell Fact. 2014, 13, 161. [Google Scholar] [CrossRef]
- Tamayo, E.N.; Villanueva, A.; Hasper, A.A.; de Graaff, L.H.; Ramón, D.; Orejas, M. CreA mediates repression of the regulatory gene xlnR which controls the production of xylanolytic enzymes in Aspergillus nidulans. Fungal Genet. Biol. 2008, 45, 984–993. [Google Scholar] [CrossRef]
- Chroumpi, T.; Aguilar-Pontes, M.V.; Peng, M.; Wang, M.; Lipzen, A.; Ng, V.; Grigoriev, I.V.; Mäkelä, M.R.; de Vries, R.P. Identification of a gene encoding the last step of the L-rhamnose catabolic pathway in Aspergillus niger revealed the inducer of the pathway regulator. Microbiol. Res. 2020, 234, 126426. [Google Scholar] [CrossRef]
- Kuivanen, J.; Richard, P. Engineering a filamentous fungus for L-rhamnose extraction. AMB Expr. 2016, 6, 27. [Google Scholar] [CrossRef]
- Linhardt, R.J.; Bakhit, R.; Daniels, L.; Mayerl, F.; Pickenhagen, W. Microbially produced rhamnolipid as a source of rhamnose. Biotechnol. Bioeng. 1989, 33, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Felenbok, B.; Flipphi, M.; Nikolaev, I. Ethanol catabolism in Aspergillus nidulans: A model system for studying gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 2001, 69, 149–204. [Google Scholar] [CrossRef] [PubMed]
- Pateman, J.A.; Doy, C.H.; Olsen, J.E.; Norris, U.; Creaser, E.H.; Hynes, M. Regulation of alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (AldDH) in Aspergillus nidulans. Proc. R. Soc. Lond. B Biol. Sci. 1983, 217, 243–264. [Google Scholar] [CrossRef] [PubMed]
- Flipphi, M.; Mathieu, M.; Cirpus, I.; Panozzo, C.; Felenbok, B. Regulation of the aldehyde dehydrogenase gene (aldA) and its role in the control of the coinducer level necessary for induction of the ethanol utilization pathway in Aspergillus nidulans. J. Biol. Chem. 2001, 276, 6950–6958. [Google Scholar] [CrossRef] [PubMed]
- Cove, D.J. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 1966, 113, 51–56. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
- McIlvaine, T.C. A buffer solution for colorimetric comparison. J. Biol. Chem. 1921, 49, 183–186. [Google Scholar] [CrossRef]
- Creaser, E.H.; Porter, R.L.; Pateman, J.A. Purification and properties of aldehyde dehydrogenase from Aspergillus nidulans. Int. J. Biochem. 1987, 19, 1009–1012. [Google Scholar] [CrossRef]
- Watanabe, S.; Saimura, M.; Makino, K. Eukaryotic and bacterial gene clusters related to an alternative pathway of nonphosphorylated L-rhamnose metabolism. J. Biol. Chem. 2008, 283, 20372–20382. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orejas, M.; MacCabe, A.P. The Loss-of-Function Mutation aldA67 Leads to Enhanced α-L-Rhamnosidase Production by Aspergillus nidulans. J. Fungi 2022, 8, 1181. https://doi.org/10.3390/jof8111181
Orejas M, MacCabe AP. The Loss-of-Function Mutation aldA67 Leads to Enhanced α-L-Rhamnosidase Production by Aspergillus nidulans. Journal of Fungi. 2022; 8(11):1181. https://doi.org/10.3390/jof8111181
Chicago/Turabian StyleOrejas, Margarita, and Andrew P. MacCabe. 2022. "The Loss-of-Function Mutation aldA67 Leads to Enhanced α-L-Rhamnosidase Production by Aspergillus nidulans" Journal of Fungi 8, no. 11: 1181. https://doi.org/10.3390/jof8111181
APA StyleOrejas, M., & MacCabe, A. P. (2022). The Loss-of-Function Mutation aldA67 Leads to Enhanced α-L-Rhamnosidase Production by Aspergillus nidulans. Journal of Fungi, 8(11), 1181. https://doi.org/10.3390/jof8111181






