Phylogenomic and Evolutionary Analyses Reveal Diversifications of SET-Domain Proteins in Fungi
Abstract
1. Introduction
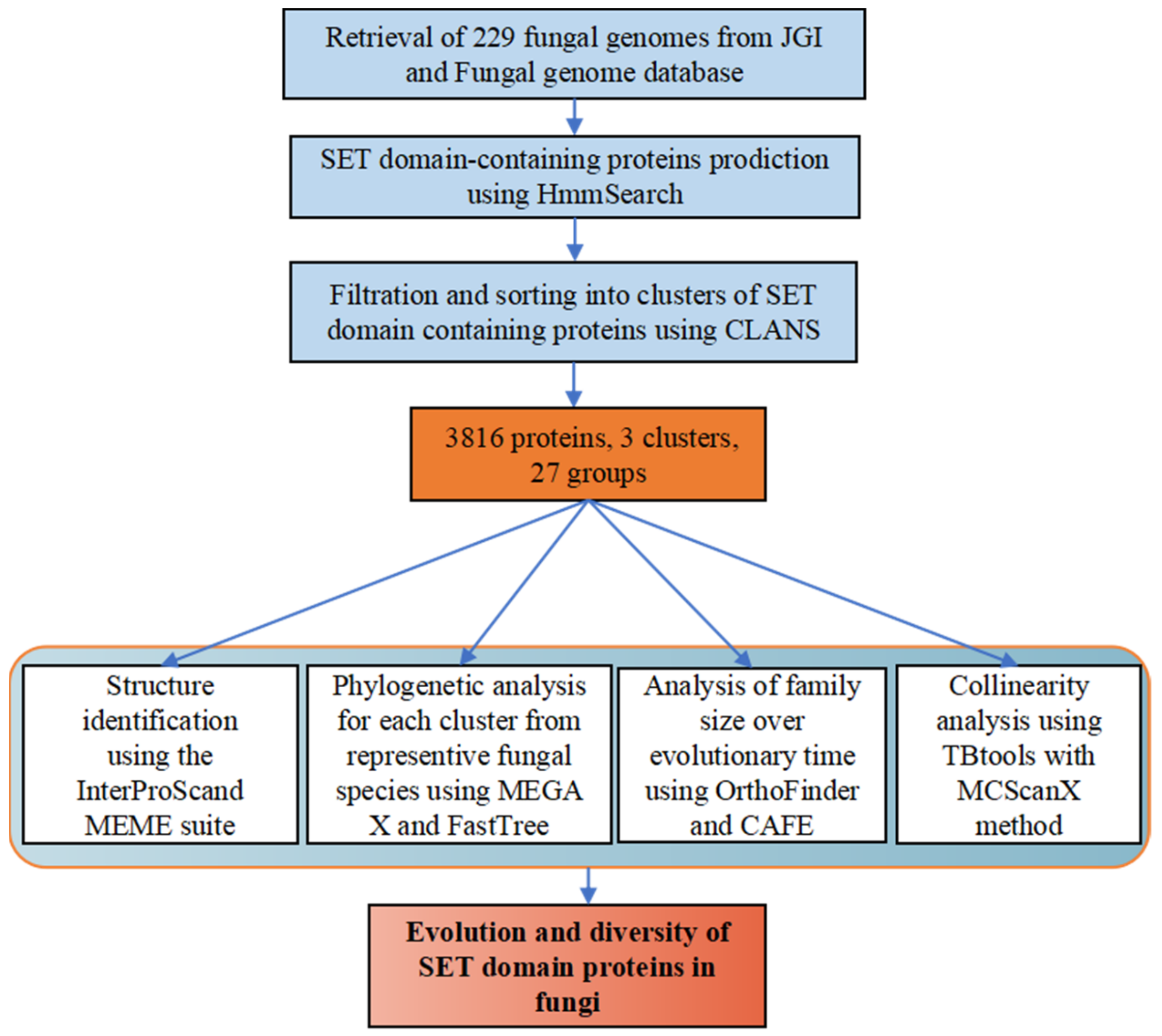
2. Materials and Methods
2.1. Identification and Homologous Searches of SET-Domain Containing Genes in Fungi
2.2. Distribution, Structure Identification of SET-Domain Containing Genes in Fungi
2.3. Phylogenetic Analysis of SET-Domain Containing Genes in Fungi
2.4. Analysis of Family Size over Evolutionary Time
2.5. Collinearity Analysis of SET-Domain Genes in Specific Fungal Species
3. Results and Discussions
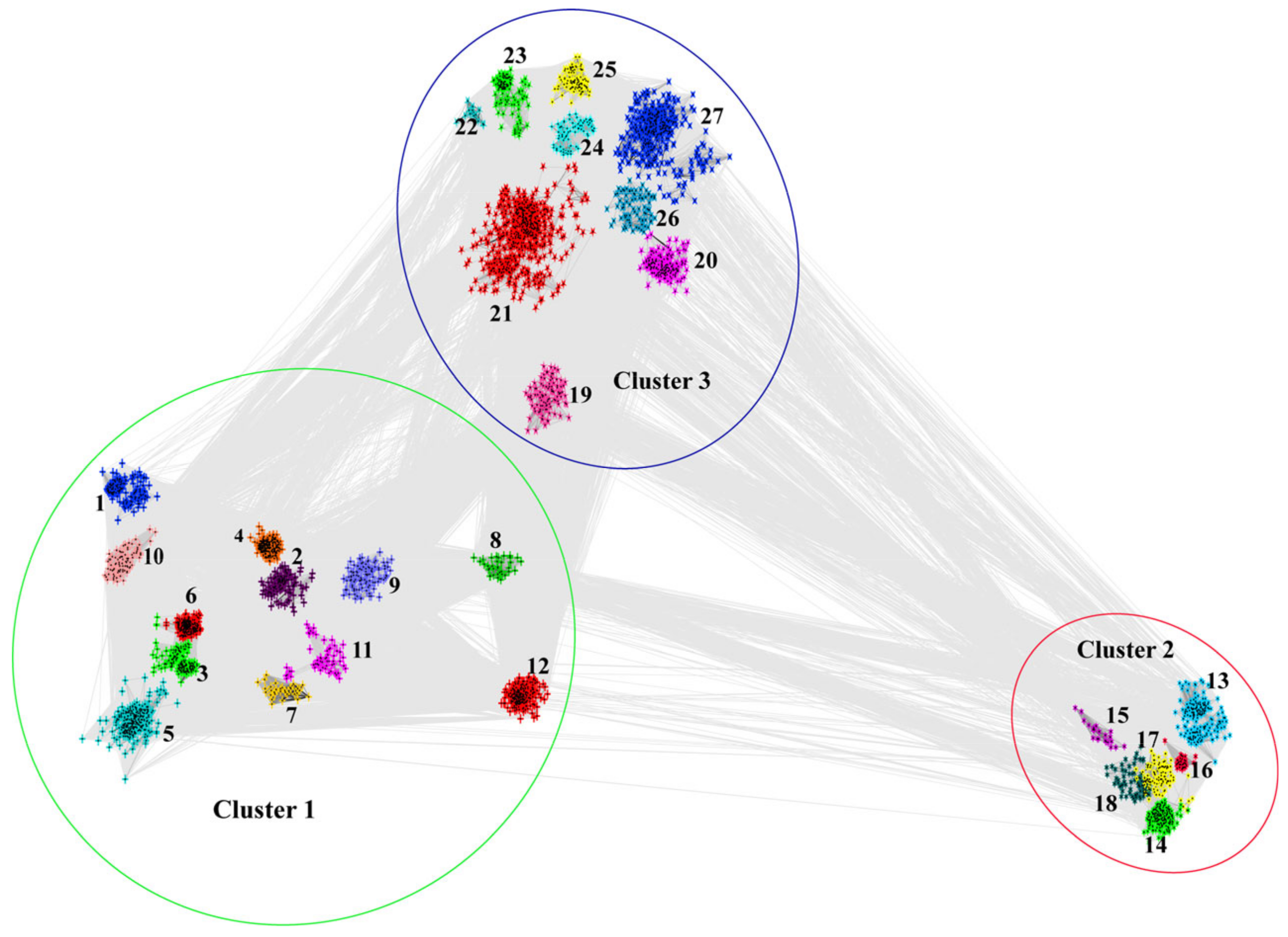
3.1. Distribution of SET-Domain-Containing Genes in Fungi
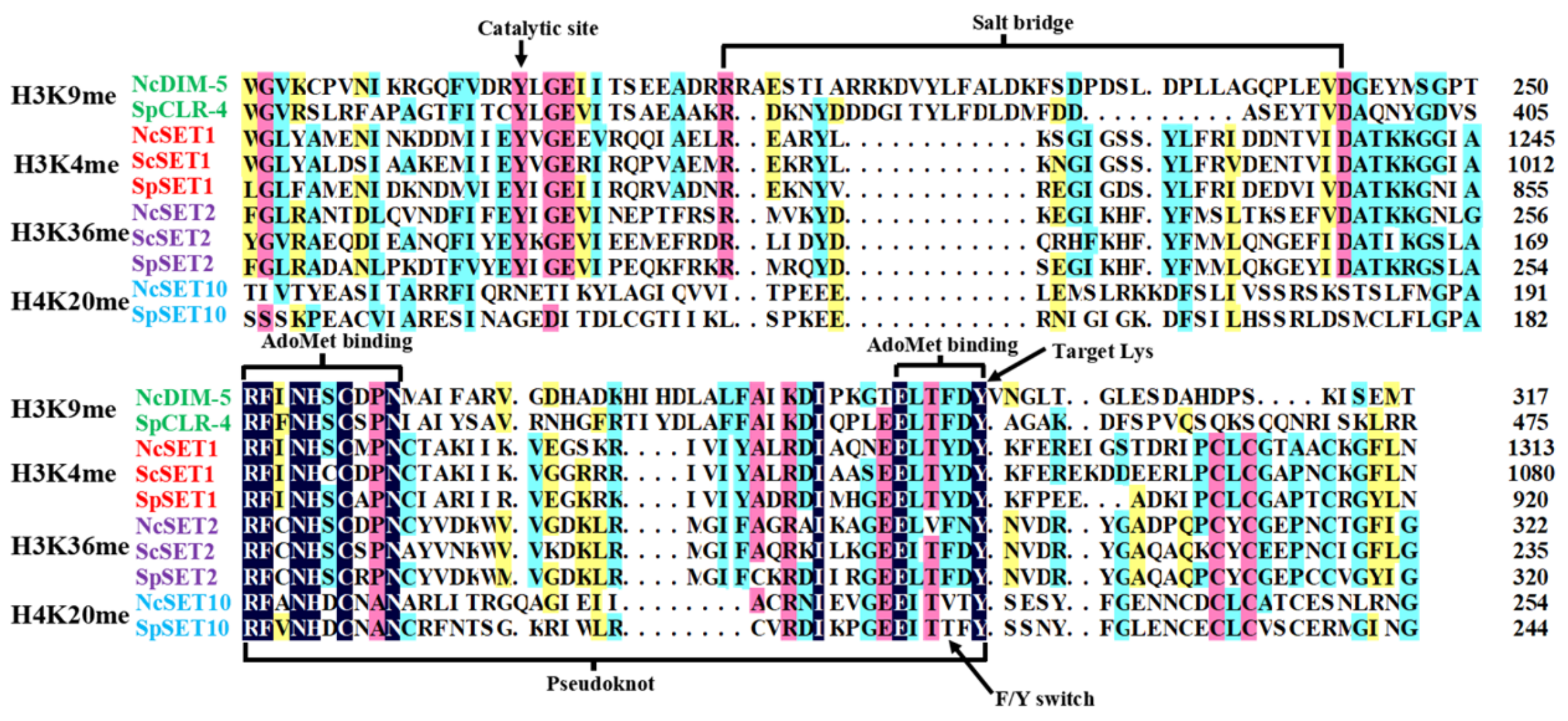
3.2. Conserved Motifs and Residues in SET Domain Proteins of Fungi
3.3. Collinearity Analysis of SET-Domain Genes in Specific Fungal Species
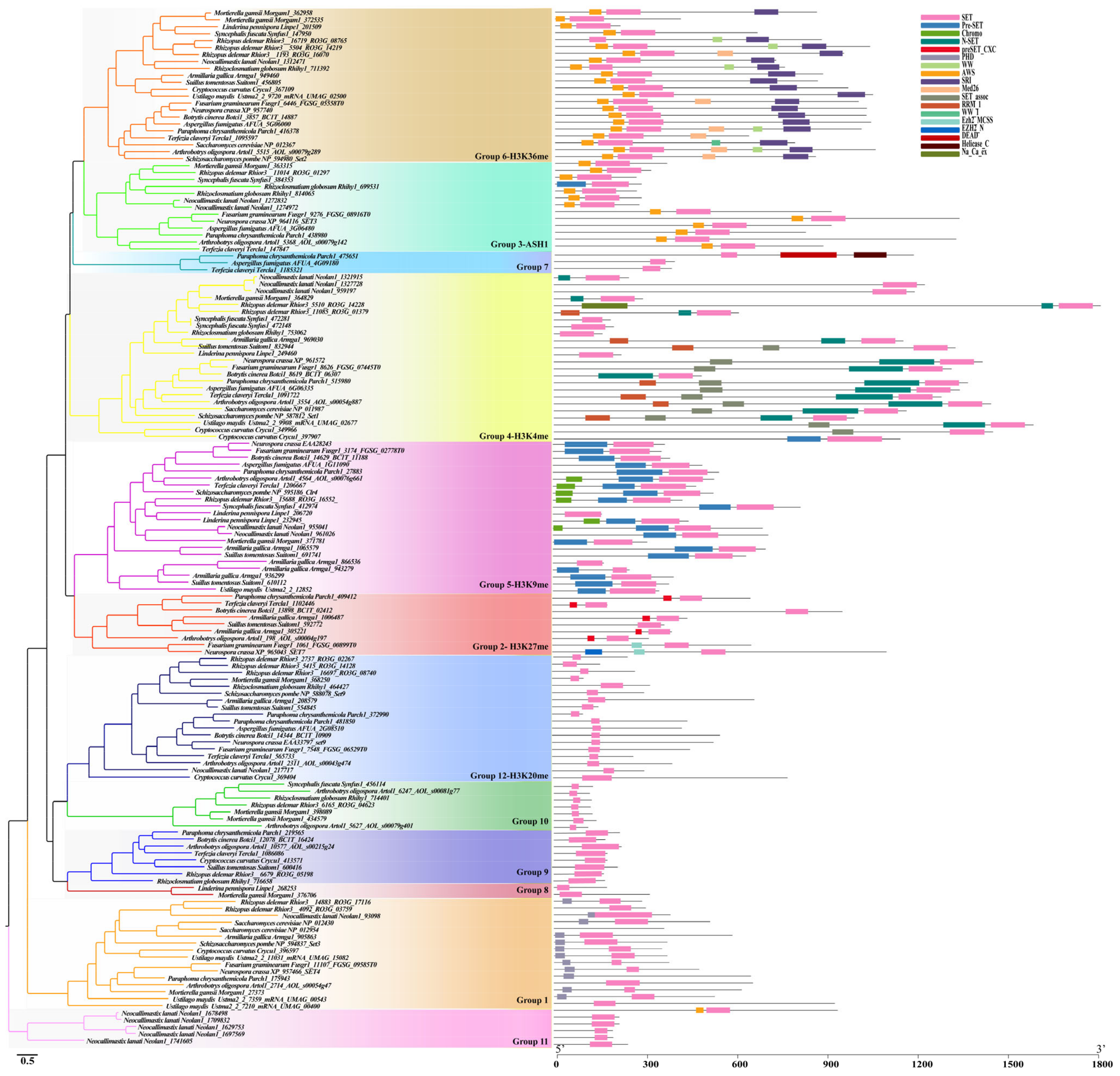
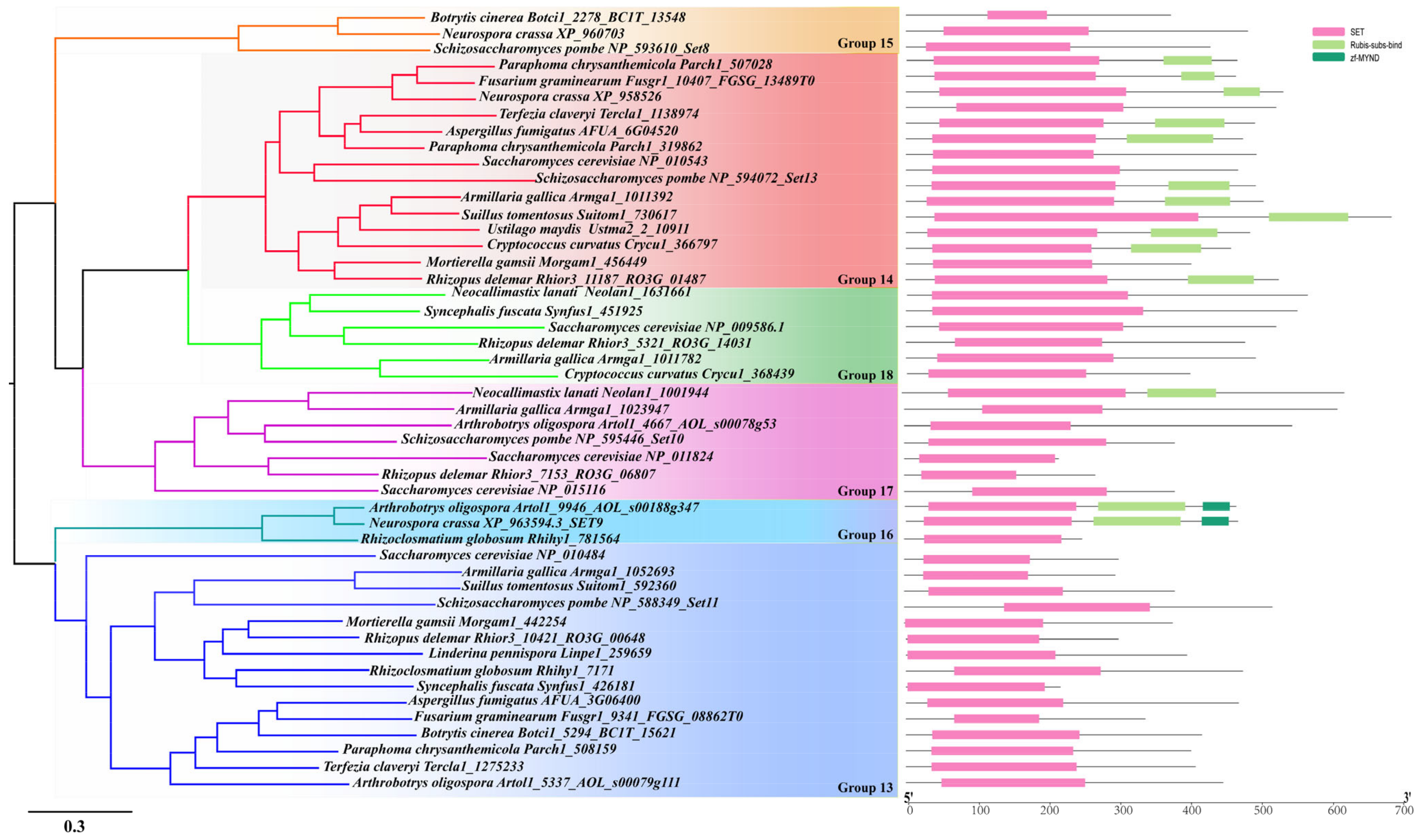
3.4. Characterization of Each Identified SET-Domain-Containing Gene in Fungi
3.4.1. Cluster 1
The SET3/4 Family (Group 1)
The Enhancer of Zeste (EZ) Family (Group 2)
The ASH1-Like Family (Group 3)
The SET1 Family (Group 4)
The SU(Var)3-9 Family (Group 5)
The SET2 Family (Group 6)
The Group 7-11 Family
The SU(Var)4-20 Family (Group 12)
3.4.2. Cluster 2
3.4.3. Cluster 3
3.5. Different Families/Groups Have Distinct Evolutionary Histories
| Cluster | Groups | Previous Names | Presence of Methylation | Neurospora crassa | Saccharomyces cerevisiae | Schizosaccharomyces pombe |
|---|---|---|---|---|---|---|
| Cluster 1 | Group 1 | SET3C family | XP_957466 (SET4) | NP_012430(SET4)/NP_012954 (SET3) | NP_594837 (SET3) | |
| Group 2 | Ez family | H3K27me | XP_965043 (SET7) | |||
| Group 3 | Ash1 family | XP_964116 (SET3) | ||||
| Group 4 | SET1 family | H3K4me | XP_961572 (SET1) | NP_011987 (SET1) | NP_587812 (SET1) | |
| Group 5 | SUV39 family | H3K9me | EAA28243 (DIM-5) | NP_595186 (Clr4) | ||
| Group 6 | SET2 family | H3K36me | XP_957740 (SET2) | NP_012367 (SET2) | NP_594980 (SET2) | |
| Group 7 | ||||||
| Group 8 | ||||||
| Group 9 | ||||||
| Group 10 | ||||||
| Group 11 | ||||||
| Group 12 | Suv4-20 family | H4K20me | EAA33797 (SET10) | NP_588078 (SET9) | ||
| Cluster 2 | Group 13 | NP_010484 | NP_588349 (SET11) | |||
| Group 14 | XP_958526 | NP_010543 | NP_594072 (SET13) | |||
| Group 15 | XP_960703 | NP_593610 (SET8) | ||||
| Group 16 | XP_963594 (SET9) | |||||
| Group 17 | NP_011824/NP_015116 | NP_595446 (SET10) | ||||
| Group 18 | NP_009586 | |||||
| Cluster 3 | Group 19 | NP_588361 (SET7) | ||||
| Group 20 | EAA35550/NCU00870 | |||||
| Group 21 | XP_957968(SET11)/XP_963161/NCU_00296 | NP_588413 (SET5) | ||||
| Group 22 | ||||||
| Group 23 | ||||||
| Group 24 | ||||||
| Group 25 | ||||||
| Group 26 | NP_015160(SET6) | |||||
| Group 27 | NCU_002962 | P38890(SET5) | NP_596514 (SET6) |
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Campos, E.I.; Reinberg, D. Histones: Annotating chromatin. Annu. Rev. Genet. 2009, 43, 559. [Google Scholar] [CrossRef] [PubMed]
- Carrozza, M.J.; Li, B.; Florens, L.; Suganuma, T.; Swanson, S.K.; Lee, K.K.; Shia, W.J.; Anderson, S.; Yates, J.; Washburn, M.P.; et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 2005, 123, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Briggs, S.D.; Bryk, M.; Strahl, B.D.; Cheung, W.L.; Davie, J.K.; Dent, S.Y.; Winston, F.; Allis, C.D. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001, 15, 3286–3295. [Google Scholar] [CrossRef]
- Freitag, M. Histone methylation by SET domain proteins in fungi. Annu. Rev. Microbiol. 2017, 71, 413–439. [Google Scholar] [CrossRef]
- Huck Hui, N.; Qin, F.; Hengbin, W.; Hediye, E.B.; Paul, T.; Yi, Z.; Kevin, S. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002, 16, 1518–1527. [Google Scholar]
- Qin, F.; Hengbin, W.; Huck Hui, N.; Hediye, E.B.; Paul, T.; Kevin, S.; Yi, Z. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002, 12, 1052–1058. [Google Scholar]
- Tschiersch, B.; Hofmann, A.; Krauss, V.; Dorn, R.; Korge, G.; Reuter, G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3–9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994, 13, 3822–3831. [Google Scholar] [CrossRef]
- Jones, R.S.; Gelbart, W.M. The Drosophila Polycomb-group gene Enhancer of zeste contains a region with sequence similarity to trithorax. Mol. Cell. Biol. 1993, 13, 6357. [Google Scholar] [PubMed]
- Dillon, S.C.; Zhang, X.; Trievel, R.C.; Cheng, X. The SET-domain protein superfamily: Protein lysine methyltransferases. Genome Biol. 2005, 6, 227. [Google Scholar] [CrossRef]
- Jenuwein, T.; Laible, G.; Dorn, R.; Reuter, G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell. Mol. Life Sci. CMLS 1997, 54, 80–93. [Google Scholar] [CrossRef]
- Black, J.C.; Van Rechem, C.; Whetstine, J.R. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell 2012, 48, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Fahrenkrog, B. Histone modifications as regulators of life and death in Saccharomyces cerevisiae. Microb. Cell 2015, 3, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Cooper, S.; Brockdorff, N. The interplay of histone modifications—Writers that read. EMBO Rep. 2015, 16, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Rea, S.; Eisenhaber, F.; O’Carroll, D.; Strahl, B.D.; Sun, Z.W.; Schmid, M.; Opravil, S.; Mechtler, K.; Ponting, C.P.; Allis, C.D.; et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 2000, 406, 593–599. [Google Scholar] [CrossRef]
- Nakayama, J.; Rice, J.C.; Strahl, B.D.; Allis, C.D.; Grewal, S.I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 2001, 292, 110–113. [Google Scholar] [CrossRef]
- Grewal, S.I.; Moazed, D. Heterochromatin and epigenetic control of gene expression. Science 2003, 301, 798–802. [Google Scholar] [CrossRef]
- Sims, R.J., 3rd; Reinberg, D. Histone H3 Lys 4 methylation: Caught in a bind? Genes Dev. 2006, 20, 2779–2786. [Google Scholar] [CrossRef]
- Bilokapic, S.; Halic, M. Nucleosome and ubiquitin position Set2 to methylate H3K36. Nat. Commun. 2019, 10, 3795. [Google Scholar] [CrossRef]
- Jamieson, K.; Rountree, M.R.; Lewis, Z.A.; Stajich, J.E.; Selker, E.U. Regional control of histone H3 lysine 27 methylation in Neurospora. Proc. Natl. Acad. Sci. USA 2013, 110, 6027–6032. [Google Scholar] [CrossRef]
- Connolly, L.R.; Smith, K.M.; Freitag, M. The Fusarium graminearum histone H3 K27 methyltransferase KMT6 regulates development and expression of secondary metabolite gene clusters. PLoS Genet. 2013, 9, e1003916. [Google Scholar] [CrossRef]
- Ridenour, J.B.; Möller, M.; Freitag, M. Polycomb Repression without Bristles: Facultative Heterochromatin and Genome Stability in Fungi. Genes 2020, 11, 638. [Google Scholar] [CrossRef] [PubMed]
- Tamaru, H.; Selker, E.U. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 2001, 414, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Adhvaryu, K.K.; Morris, S.A.; Strahl, B.D.; Selker, E.U. Methylation of histone H3 lysine 36 is required for normal development in Neurospora crassa. Eukaryot. Cell 2005, 4, 1455–1464. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.; Jethmalani, Y.; Jaiswal, D.; Green, E.M. Set4 is a chromatin-associated protein, promotes survival during oxidative stress, and regulates stress response genes in yeast. J. Biol. Chem. 2018, 293, 14429–14443. [Google Scholar] [CrossRef]
- Pham, K.T.; Inoue, Y.; Vu, B.V.; Nguyen, H.H.; Nakayashiki, T.; Ikeda, K.; Nakayashiki, H. MoSET1 (histone H3K4 methyltransferase in Magnaporthe oryzae) regulates global gene expression during infection-related morphogenesis. PLoS Genet. 2015, 11, e1005385. [Google Scholar] [CrossRef]
- Gu, Q.; Tahir, H.A.; Zhang, H.; Huang, H.; Ji, T.; Sun, X.; Wu, L.; Wu, H.; Gao, X. Involvement of FvSet1 in Fumonisin B1 biosynthesis, vegetative growth, fungal virulence, and environmental stress responses in Fusarium verticillioides. Toxins 2017, 9, 43. [Google Scholar] [CrossRef]
- Fingerman, I.M.; Wu, C.L.; Wilson, B.D.; Briggs, S.D. Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 28761–28765. [Google Scholar] [CrossRef]
- McDaniel, S.L.; Hepperla, A.J.; Huang, J.; Dronamraju, R.; Adams, A.T.; Kulkarni, V.G.; Davis, I.J.; Strahl, B.D. H3K36 Methylation Regulates Nutrient Stress Response in Saccharomyces cerevisiae by Enforcing Transcriptional Fidelity. Cell Rep. 2017, 19, 2371–2382. [Google Scholar] [CrossRef]
- Allshire, R.C.; Ekwall, K. Epigenetic Regulation of Chromatin States in Schizosaccharomyces pombe. Cold Spring Harb. Perspect. Biol. 2015, 7, a018770. [Google Scholar] [CrossRef]
- Veerappan, C.S.; Avramova, Z.; Moriyama, E.N. Evolution of SET-domain protein families in the unicellular and multicellular Ascomycota fungi. BMC Evol. Biol. 2008, 8, 190. [Google Scholar] [CrossRef]
- Frickey, T.; Lupas, A. CLANS: A Java application for visualizing protein families based on pairwise similarity. Bioinformatics 2004, 20, 3702–3704. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 2005, 21, 2104–2105. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- De Bie, T.; Cristianini, N.; Demuth, J.P.; Hahn, M.W. CAFE: A computational tool for the study of gene family evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef]
- Mendes, F.K.; Vanderpool, D.; Fulton, B.; Hahn, M.W. CAFE 5 models variation in evolutionary rates among gene families. Bioinformatics 2020, 36, 5516–5518. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Sanderson, M.J. r8s: Inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 2003, 19, 301–302. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Accari, S.L.; Fisher, P.R. Emerging Roles of JmjC Domain-Containing Proteins. Int. Rev. Cell Mol. Biol. 2015, 319, 165–220. [Google Scholar] [CrossRef]
- Catania, S.; Dumesic, P.A.; Pimentel, H.; Nasif, A.; Stoddard, C.I.; Burke, J.E.; Diedrich, J.K.; Cook, S.; Shea, T.; Geinger, E.; et al. Evolutionary Persistence of DNA Methylation for Millions of Years after Ancient Loss of a De Novo Methyltransferase. Cell 2020, 180, 263–277.e220. [Google Scholar] [CrossRef]
- Du, J.; Johnson, L.M.; Jacobsen, S.E.; Patel, D.J. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015, 16, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Buratowski, S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5’ transcribed regions. Cell 2009, 137, 259–272. [Google Scholar] [CrossRef]
- Pijnappel, W.W.; Schaft, D.; Roguev, A.; Shevchenko, A.; Tekotte, H.; Wilm, M.; Rigaut, G.; Séraphin, B.; Aasland, R.; Stewart, A.F. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001, 15, 2991–3004. [Google Scholar] [CrossRef]
- Nobile, C.J.; Fox, E.P.; Hartooni, N.; Mitchell, K.F.; Hnisz, D.; Andes, D.R.; Kuchler, K.; Johnson, A.D. A histone deacetylase complex mediates biofilm dispersal and drug resistance in Candida albicans. mBio 2014, 5, e01201–e01214. [Google Scholar] [CrossRef]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef]
- Beisel, C.; Imhof, A.; Greene, J.; Kremmer, E.; Sauer, F. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 2002, 419, 857–862. [Google Scholar] [CrossRef]
- Dorafshan, E.; Kahn, T.G.; Glotov, A.; Savitsky, M.; Walther, M.; Reuter, G.; Schwartz, Y.B. Ash1 counteracts Polycomb repression independent of histone H3 lysine 36 methylation. EMBO Rep. 2019, 20, e46762. [Google Scholar] [CrossRef] [PubMed]
- Schlichter, A.; Cairns, B.R. Histone trimethylation by Set1 is coordinated by the RRM, autoinhibitory, and catalytic domains. EMBO J. 2005, 24, 1222–1231. [Google Scholar] [CrossRef] [PubMed]
- Thon, G.; Hansen, K.R.; Altes, S.P.; Sidhu, D.; Singh, G.; Verhein-Hansen, J.; Bonaduce, M.J.; Klar, A.J. The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics 2005, 171, 1583–1595. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, Z.; Khan, S.I.; Horton, J.R.; Tamaru, H.; Selker, E.U.; Cheng, X. Structural basis for the product specificity of histone lysine methyltransferases. Mol. Cell 2003, 12, 177–185. [Google Scholar] [CrossRef]
- Li, J.; Moazed, D.; Gygi, S.P. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 2002, 277, 49383–49388. [Google Scholar] [CrossRef]
- Du, L.L.; Nakamura, T.M.; Russell, P. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev. 2006, 20, 1583–1596. [Google Scholar] [CrossRef]
- Strahl, B.D.; Grant, P.A.; Briggs, S.D.; Sun, Z.W.; Bone, J.R.; Caldwell, J.A.; Mollah, S.; Cook, R.G.; Shabanowitz, J.; Hunt, D.F.; et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 2002, 22, 1298–1306. [Google Scholar] [CrossRef]
- Schotta, G.; Lachner, M.; Sarma, K.; Ebert, A.; Sengupta, R.; Reuter, G.; Reinberg, D.; Jenuwein, T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004, 18, 1251–1262. [Google Scholar] [CrossRef]
- Hamamoto, R.; Furukawa, Y.; Morita, M.; Iimura, Y.; Silva, F.P.; Li, M.; Yagyu, R.; Nakamura, Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell Biol. 2004, 6, 731–740. [Google Scholar] [CrossRef]
- Brown, M.A.; Sims, R.J., 3rd; Gottlieb, P.D.; Tucker, P.W. Identification and characterization of Smyd2: A split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol. Cancer 2006, 5, 26. [Google Scholar] [CrossRef]
- Green, E.M.; Mas, G.; Young, N.L.; Garcia, B.A.; Gozani, O. Methylation of H4 lysines 5, 8 and 12 by yeast Set5 calibrates chromatin stress responses. Nat. Struct. Mol. Biol. 2012, 19, 361–363. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.S.; Xu, Z.; Clauder-Münster, S.; Steinmetz, L.; Buratowski, S. Set3 HDAC Mediates Effects of Overlapping Noncoding Transcription on Gene Induction Kinetics. Cell 2012, 150, 1158–1169. [Google Scholar] [CrossRef] [PubMed]
- Van Werven, F.J.; Neuert, G.; Hendrick, N.; Lardenois, A.; Buratowski, S.; van Oudenaarden, A.; Primig, M.; Amon, A. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell 2012, 150, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Tamaru, H.; Zhang, X.; McMillen, D.; Singh, P.B.; Nakayama, J.; Grewal, S.I.; Allis, C.D.; Cheng, X.; Selker, E.U. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 2003, 34, 75–79. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, G.; Shang, L.; Zhou, W.; Lu, S.; Zhou, Z.; Huang, X.; Li, J. Phylogenomic and Evolutionary Analyses Reveal Diversifications of SET-Domain Proteins in Fungi. J. Fungi 2022, 8, 1159. https://doi.org/10.3390/jof8111159
Ding G, Shang L, Zhou W, Lu S, Zhou Z, Huang X, Li J. Phylogenomic and Evolutionary Analyses Reveal Diversifications of SET-Domain Proteins in Fungi. Journal of Fungi. 2022; 8(11):1159. https://doi.org/10.3390/jof8111159
Chicago/Turabian StyleDing, Guoqing, Liqiu Shang, Wenliang Zhou, Siyi Lu, Zong Zhou, Xinyi Huang, and Juan Li. 2022. "Phylogenomic and Evolutionary Analyses Reveal Diversifications of SET-Domain Proteins in Fungi" Journal of Fungi 8, no. 11: 1159. https://doi.org/10.3390/jof8111159
APA StyleDing, G., Shang, L., Zhou, W., Lu, S., Zhou, Z., Huang, X., & Li, J. (2022). Phylogenomic and Evolutionary Analyses Reveal Diversifications of SET-Domain Proteins in Fungi. Journal of Fungi, 8(11), 1159. https://doi.org/10.3390/jof8111159





