Turning Inside Out: Filamentous Fungal Secretion and Its Applications in Biotechnology, Agriculture, and the Clinic
Abstract
1. Fungal Secretion: The Global Bioeconomy and beyond
2. Polar Growth
2.1. From Dormancy to Growth: Establishing Polarity
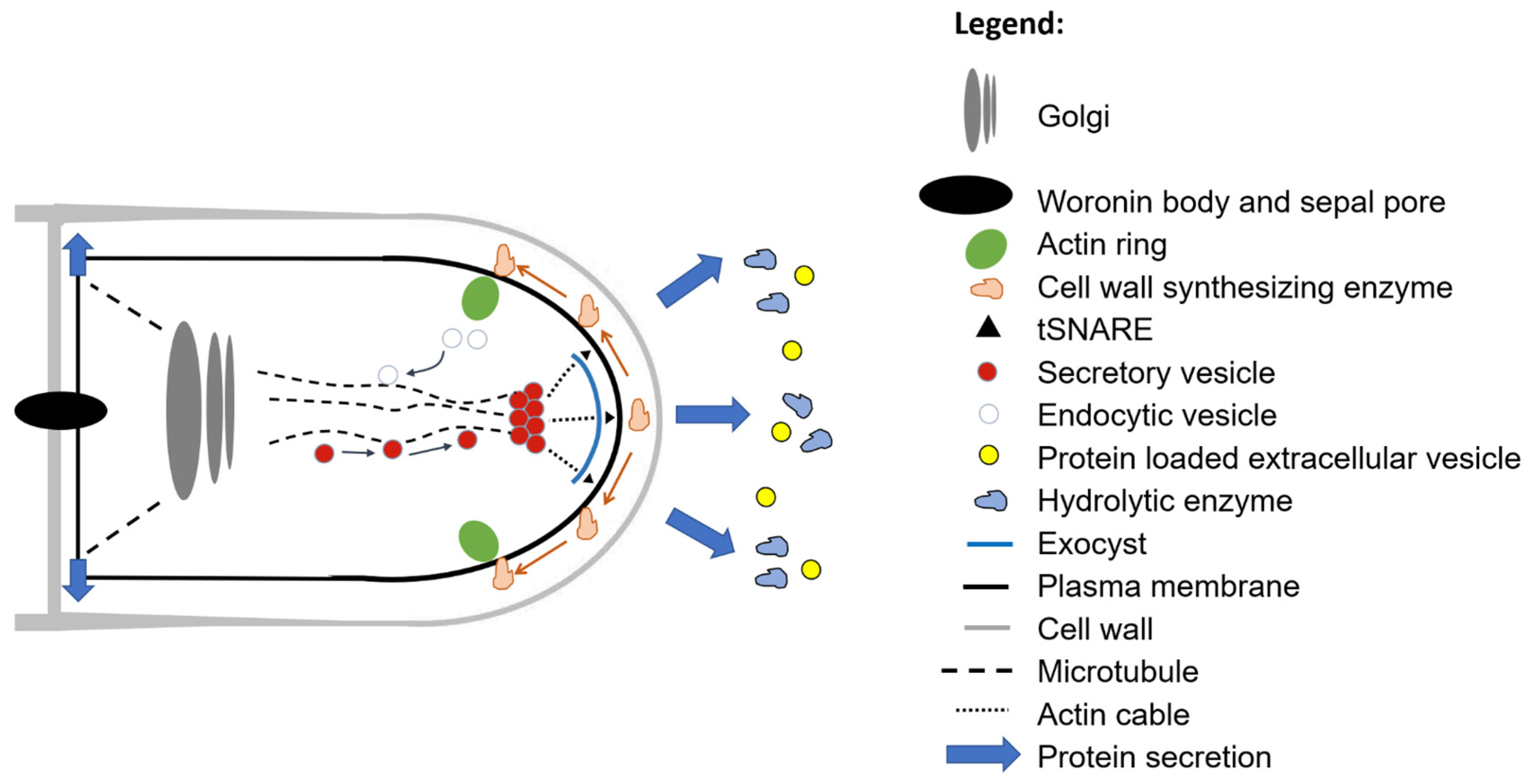
2.2. The Classical Secretion Route and Hyphal Polarity
2.3. Endocytosis and Branching
2.4. Repurposing Polar Growth and Classical Secretion in Biotechnology: A Case Study Using A. niger RacA and Glucoamylase
2.5. Controlling Filamentous Growth for Optimal Macromorphologies
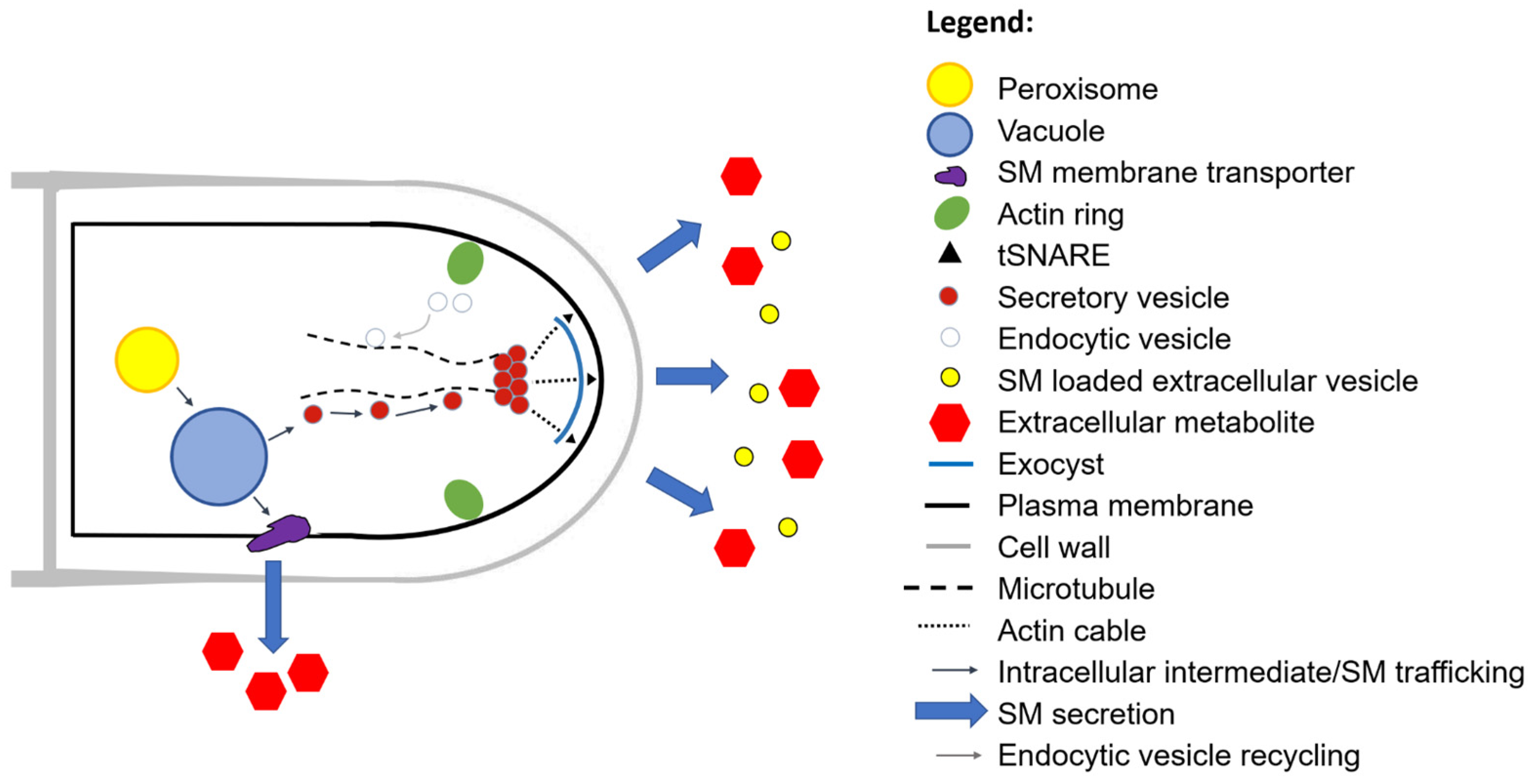
2.6. New Additions to the Classical Secretion Pathway: Extracellular Secretory Vesicles
2.7. Summary
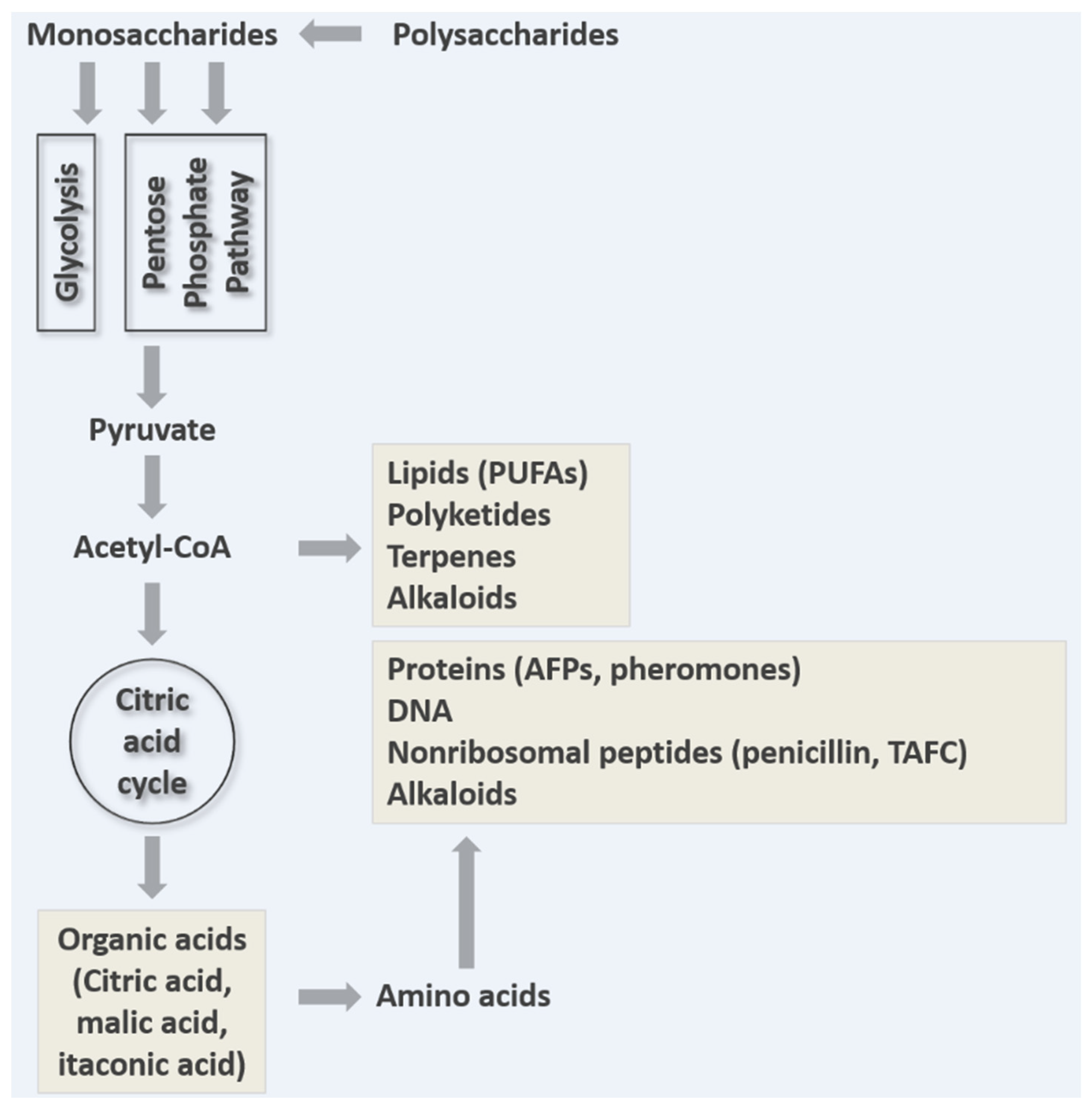
3. Breakdown of Macromolecules for Nutrient Acquisition
3.1. A Stomach Outside the Body: Digesting the Extracellular Space
3.2. Secreted Lipases in Industrial Biotechnology: An Exemplar Success Story
3.3. New Frontiers: Degrading Lignocellulose and Plastics
4. Secretion and Micronutrients
4.1. Siderophores and Iron Availability
4.2. Organic Acids May Liberate and Chelate Nutrients from the External Environment
4.3. Summary
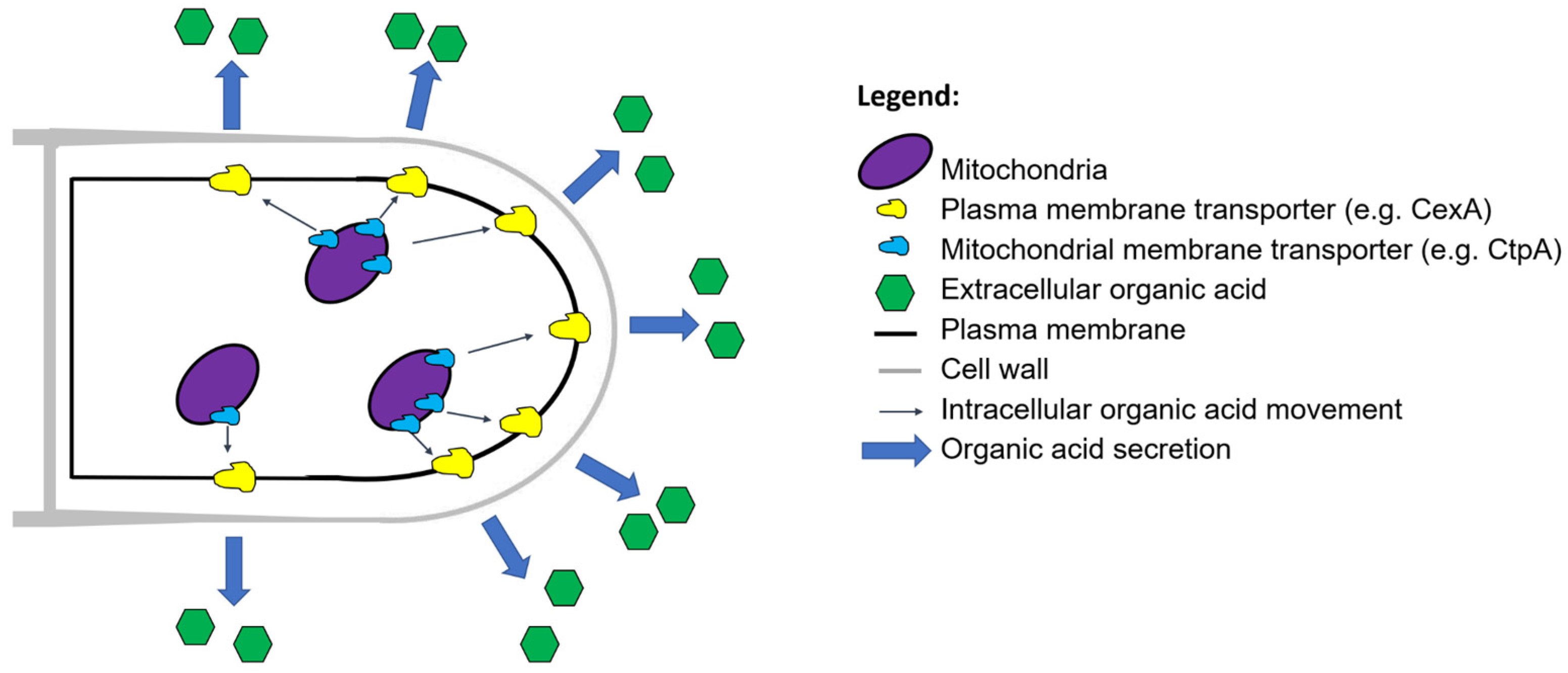
5. Removal of Overflow Metabolites
5.1. Organic Acids as Overflow Metabolites
5.2. Increasing Glycolytic Flux for Organic Acid Hypersecretors
5.3. Secretion of Organic Acids and Their Applications
6. Infection
6.1. Secreted Molecules and Immune Evasion during Plant or Human Infection
6.2. Direct Host Attack by Secreted Secondary Metabolites and Proteins
6.2.1. Plants
6.2.2. Humans and Other Mammals
6.3. Applications of Fungal Secretion and Extracellular Molecules
6.3.1. Purified PAMPs in Agriculture
6.3.2. Discovery and Cloning of Resistance Genes in Crop Genomes
6.3.3. Inhibiting Secretion for New Mode of Action Antifungals
6.3.4. Secreted Molecules and Vaccines for Fungal Infection
6.4. Summary
7. Interspecies Communication
7.1. Fungal Extracellular Molecules in Mutualist Interactions: Lichens
7.2. Arbuscular Mycorrhizae and Fungal Secretion
7.3. Summary
8. Intraspecies Communication
8.1. Sexual Reproduction and Secretion of Pheromones
8.2. Applications of Fungal Pheromones
8.3. Applications of Antifungal Proteins
8.4. Summary
9. Surviving in the Community
9.1. Harnessing Fungal Chemical Defence for New Bioactive Molecule Discovery: A Case Study Using Enniatin and Related Compounds
9.2. Secretion of Xenobiotics: Surviving Chemical Attack
9.3. Applications of Fungal Efflux Pumps
9.4. Revisiting Organic Acids: Are They Secreted to Inhibit Competing Neutrophiles?
9.5. Summary
10. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blackwell, M. The fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot. 2011, 98, 426–438. [Google Scholar] [CrossRef]
- Li, Y.; Steenwyk, J.L.; Chang, Y.; Wang, Y.; James, T.Y.; Stajich, J.E.; Spatafora, J.W.; Groenewald, M.; Dunn, C.W.; Hittinger, C.T.; et al. A genome-scale phylogeny of the kingdom Fungi. Curr. Biol. 2021, 31, 1653–1665.e5. [Google Scholar] [CrossRef]
- Cairns, T.C.; Zheng, X.; Zheng, P.; Sun, J.; Meyer, V. Moulding the mould: Understanding and reprogramming filamentous fungal growth and morphogenesis for next generation cell factories. Biotechnol. Biofuels 2019, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Cairns, T.C.; Nai, C.; Meyer, V. How a fungus shapes biotechnology: 100 years of Aspergillus niger research. Fungal Biol. Biotechnol. 2018, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Meyer, V.; Andersen, M.R.; Brakhage, A.A.; Braus, G.H.; Caddick, M.X.; Cairns, C.T.; de Vries, R.P.; Haarmann, T.; Hansen, K.; Hertz-Fowler, C.; et al. Current challenges of research on filamentous fungi in relation to human welfare and a sustainable bio-economy: A white paper. Fungal Biol. Biotechnol. 2016, 3, 1–17. [Google Scholar] [CrossRef]
- Meyer, V.; Basenko, E.Y.; Benz, J.P.; Braus, G.H.; Caddick, M.X.; Csukai, M.; de Vries, R.P.; Endy, D.; Frisvad, J.C.; Gunde-Cimerman, N.; et al. Growing a circular economy with fungal biotechnology: A white paper. Fungal Biol. Biotechnol. 2020, 7, 5. [Google Scholar] [CrossRef]
- Grimm, D.; Wösten, H.A.B. Mushroom cultivation in the circular economy. Appl. Microbiol. Biotechnol. 2018, 102, 7795–7803. [Google Scholar] [CrossRef]
- Kemona, A.; Piotrowska, M. Polyurethane recycling and disposal: Methods and prospects. Polymers 2020, 12, 1752. [Google Scholar] [CrossRef]
- Ostrov, N.; Jimenez, M.; Billerbeck, S.; Brisbois, J.; Matragrano, J.; Ager, A.; Cornish, V.W. A modular yeast biosensor for low-cost point-of-care pathogen detection. Sci. Adv. 2017, 3, e1603221. [Google Scholar] [CrossRef] [PubMed]
- Cortesão, M.; Schütze, T.; Marx, R.; Moeller, R.; Meyer, V. Fungal Biotechnology in Space: Why and How? In Grand Challenges in Fungal Biotechnology; Springer: Cham, Germany, 2020. [Google Scholar]
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef]
- Denning, D.W.; Bromley, M.J. How to bolster the antifungal pipeline. Science 2015, 347, 1414–1416. [Google Scholar] [CrossRef]
- Barnes, R.A.; Gow, N.A.; Denning, D.W.; May, R.C.; Haynes, K.; British Society of Medical Mycology. Antifungal resistance: More research needed. Lancet 2014, 384, 1427. [Google Scholar] [CrossRef]
- Meyer, V. Metabolic Engineering of Filamentous Fungi. In Metabolic Engineering: Concepts and Applications; Wiley-VCH GmbH: Weinheim, Germany, 2021. [Google Scholar]
- Fiedler, M.R.M.; Barthel, L.; Kubisch, C.; Nai, C.; Meyer, V. Construction of an improved Aspergillus niger platform for enhanced glucoamylase secretion. Microb. Cell Fact. 2018, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bischof, R.H.; Ramoni, J.; Seiboth, B. Cellulases and beyond: The first 70 years of the enzyme producer Trichoderma reesei. Microb. Cell Fact. 2016, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Contesini, F.J.; Davanço, M.G.; Borin, G.P.; Vanegas, K.G.; Cirino, J.P.G.; de Melo, R.R.; Mortensen, U.H.; Hildén, K.; Campos, D.R.; de Carvalho, P.O. Advances in recombinant lipases: Production, engineering, immobilization and application in the pharmaceutical industry. Catalysts 2020, 10, 1032. [Google Scholar] [CrossRef]
- Mehta, A.; Bodh, U.; Gupta, R. Fungal lipases: A review. J. Biotech. Res. 2017, 8, 58–77. [Google Scholar]
- Schmidt, C.S.; White, C.J.; Ibrahim, A.S.; Filler, S.G.; Fu, Y.; Yeaman, M.R.; Edwards, J.E.; Hennessey, J.P. NDV-3, a recombinant alum-adjuvanted vaccine for Candida and Staphylococcus aureus, is safe and immunogenic in healthy adults. Vaccine 2012, 30, 7594–7600. [Google Scholar] [CrossRef]
- Singh, S.; Uppuluri, P.; Mamouei, Z.; Alqarihi, A.; Elhassan, H.; French, S.; Lockhart, S.R.; Chiller, T.; Edwards, J.E.; Ibrahim, A.S. The NDV-3A vaccine protects mice from multidrug resistant Candida auris infection. PLoS Pathog. 2019, 15, e1007460. [Google Scholar] [CrossRef]
- Vilanova, M.; Teixeira, L.; Caramalho, Í.; Torrado, E.; Marques, A.; Madureira, P.; Ribeiro, A.; Ferreira, P.; Gama, M.; Demengeot, J. Protection against systemic candidiasis in mice immunized with secreted aspartic proteinase 2. Immunology 2004, 111, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Meyer, V.; Jung, S. Antifungal Peptides of the AFP Family Revisited: Are These Cannibal Toxins? Microorganisms 2018, 6, 50. [Google Scholar] [CrossRef]
- Garrigues, S.; Gandía, M.; Popa, C.; Borics, A.; Marx, F.; Coca, M.; Marcos, J.F.; Manzanares, P. Efficient production and characterization of the novel and highly active antifungal protein AfpB from Penicillium digitatum. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Giraldo, M.C.; Dagdas, Y.F.; Gupta, Y.K.; Mentlak, T.A.; Yi, M.; Martinez-Rocha, A.L.; Saitoh, H.; Terauchi, R.; Talbot, N.J.; Valent, B. Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat. Commun. 2013, 4, 1996. [Google Scholar] [CrossRef]
- Kanja, C.; Hammond-Kosack, K.E. Proteinaceous effector discovery and characterization in filamentous plant pathogens. Mol. Plant Pathol. 2020, 21, 1353–1376. [Google Scholar] [CrossRef]
- Liu, T.; Song, T.; Zhang, X.; Yuan, H.; Su, L.; Li, W.; Xu, J.; Liu, S.; Chen, L.; Chen, T.; et al. Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 2014, 5, 4686. [Google Scholar] [CrossRef]
- Schrettl, M.; Bignell, E.; Kragl, C.; Sabiha, Y.; Loss, O.; Eisendle, M.; Wallner, A.; Arst, H.N.; Haynes, K.; Haas, H. Distinct roles for intra- and extracellular siderophores during Aspergillus fumigatus infection. PLoS Pathog. 2007, 3, e128. [Google Scholar] [CrossRef]
- Haas, H. Fungal siderophore metabolism with a focus on Aspergillus fumigatus. Nat. Prod. Rep. 2014, 31, 1266–1276. [Google Scholar] [CrossRef]
- Martín, J.F. Transport systems, intracellular traffic of intermediates and secretion of β-lactam antibiotics in fungi. Fungal Biol. Biotechnol. 2020, 7, 1–15. [Google Scholar]
- Werner, S.; Schroeter, A.; Schimek, C.; Vlaic, S.; Wöstemeyer, J.; Schuster, S. Model of the synthesis of trisporic acid in Mucorales showing bistability. IET Syst. Biol. 2012, 6, 207–214. [Google Scholar] [CrossRef]
- Sy-Cordero, A.A.; Pearce, C.J.; Oberlies, N.H. Revisiting the enniatins: A review of their isolation, biosynthesis, structure determination and biological activities. J. Antibiot. 2012, 65, 541–549. [Google Scholar] [CrossRef]
- Tong, Z.; Zheng, X.; Tong, Y.; Shi, Y.-C.; Sun, J. Systems metabolic engineering for citric acid production by Aspergillus niger in the post-genomic era. Microb. Cell Fact. 2019, 18, 28. [Google Scholar] [CrossRef]
- Cao, W.; Yan, L.; Li, M.; Liu, X.; Xu, Y.; Xie, Z.; Liu, H. Identification and engineering a C4-dicarboxylate transporter for improvement of malic acid production in Aspergillus niger. Appl. Microbiol. Biotechnol. 2020, 104, 9773–9783. [Google Scholar] [CrossRef]
- Karaffa, L.; Kubicek, C.P. Citric acid and itaconic acid accumulation: Variations of the same story? Appl. Microbiol. Biotechnol. 2019, 103, 2889–2902. [Google Scholar] [CrossRef] [PubMed]
- Kiss, E.; Hegedüs, B.; Virágh, M.; Varga, T.; Merényi, Z.; Kószó, T.; Bálint, B.; Prasanna, A.N.; Krizsán, K.; Kocsubé, S.; et al. Comparative genomics reveals the origin of fungal hyphae and multicellularity. Nat. Commun. 2019, 10, 1–13. [Google Scholar]
- Odoni, D.I.; van Gaal, M.P.; Schonewille, T.; Tamayo-Ramos, J.A.; dos Santos, V.A.P.; Suarez-Diez, M.; Schaap, P.J. Aspergillus niger Secretes Citrate to Increase Iron Bioavailability. Front. Microbiol. 2017, 8, 1424. [Google Scholar] [CrossRef]
- Dolan, S.K.; Bock, T.; Hering, V.; Owens, R.A.; Jones, G.W.; Blankenfeldt, W.; Doyle, S. Structural, mechanistic and functional insight into gliotoxin bis-thiomethylation in Aspergillus fumigatus. Open Biol. 2017. [Google Scholar] [CrossRef]
- Golan, J.J.; Pringle, A. Long-Distance Dispersal of Fungi. Microbiol. Spectr. 2017. [Google Scholar] [CrossRef]
- Riquelme, M.; Munro, C.; Gow, N. The fungal cell wall: Biology, biosynthesis and biotechnology. Cell Surf. 2020, 6, 100037. [Google Scholar] [CrossRef]
- Gow, N.A.R.; Latgé, J.P.; Munro, C.A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. In The Fungal Kingdom; Heitman, J., Gow, N.A.R., Howlett, B., Crous, P., Stukenbroch, E., James, T., Eds.; ASM Press: Washington, DC, USA, 2017; pp. 267–292. [Google Scholar]
- Mélida, H.; Sain, D.; Stajich, J.E.; Bulone, V. Deciphering the uniqueness of Mucoromycotina cell walls by combining biochemical and phylogenomic approaches. Environ. Microbiol. 2015, 17, 1649–1662. [Google Scholar] [CrossRef]
- Lecointe, K.; Cornu, M.; Leroy, J.; Coulon, P.; Sendid, B. Polysaccharides cell wall architecture of mucorales. Front. Microbiol. 2019, 10, 469. [Google Scholar] [CrossRef]
- Ferguson, B.A.; Dreisbach, T.A.; Parks, C.G.; Filip, G.M.; Schmitt, C.L. Coarse-scale population structure of pathogenic Armillaria species in a mixed-conifer forest in the Blue Mountains of northeast Oregon. Can. J. For. Res. 2003, 33, 612–623. [Google Scholar] [CrossRef]
- Osherov, N.; May, G.S. The molecular mechanisms of conidial germination. FEMS Microbiol. Lett. 2001, 199, 153–160. [Google Scholar] [CrossRef]
- Si, H.; Rittenour, W.R.; Harris, S.D. Roles of Aspergillus nidulans Cdc42/Rho GTPase regulators in hyphal morphogenesis and development. Mycologia 2016, 108, 543–555. [Google Scholar] [CrossRef]
- Virag, A.; Lee, M.P.; Si, H.; Harris, S.D. Regulation of hyphal morphogenesis by cdc42 and rac1 homologues in Aspergillus nidulans. Mol. Microbiol. 2007, 66, 1579–1596. [Google Scholar] [CrossRef]
- Kwon, M.J.; Arentshorst, M.; Roos, E.D.; Van Den Hondel, C.A.M.J.J.; Meyer, V.; Ram, A.F.J. Functional characterization of Rho GTPases in Aspergillus niger uncovers conserved and diverged roles of Rho proteins within filamentous fungi. Mol. Microbiol. 2011, 79, 1151–1167. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.D.; Read, N.D.; Roberson, R.W.; Shaw, B.; Seiler, S.; Plamann, M.; Momany, M. Polarisome meets Spitzenkörper: Microscopy, genetics, and genomics converge. Eukaryot. Cell 2005, 4, 225–229. [Google Scholar] [CrossRef]
- Schuster, M.; Martin-Urdiroz, M.; Higuchi, Y.; Hacker, C.; Kilaru, S.; Gurr, S.J.; Steinberg, G. Co-Delivery of Cell-Wall-Forming enzymes in the same vesicle for coordinated fungal cell wall formation. Nat. Microbiol. 2016, 1, 16149. [Google Scholar] [CrossRef] [PubMed]
- Schuster, M.; Guiu-Aragones, C.; Steinberg, G. Class V chitin synthase and β(1,3)-glucan synthase co-travel in the same vesicle in Zymoseptoria tritici. Fungal Genet. Biol. 2020, 135, 103286. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, N.; Fischer, R. On the role of microtubules, cell end markers, and septal microtubule organizing centres on site selection for polar growth in Aspergillus nidulans. Fungal Biol. 2011, 115, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, N.; Mania, D.; Herrero, S.; Ishitsuka, Y.; Nienhaus, G.U.; Podolski, M.; Howard, J.; Fischer, R. The cell-end marker TeaA and the microtubule polymerase AlpA contribute to microtubule guidance at the hyphal tip cortex of Aspergillus nidulans to provide polarity maintenance. J. Cell Sci. 2013, 126, 5400–5411. [Google Scholar] [CrossRef] [PubMed]
- Ishitsuka, Y.; Savage, N.; Li, Y.; Bergs, A.; Grün, N.; Kohler, D.; Donnelly, R.; Nienhaus, G.U.; Fischer, R.; Takeshita, N. Superresolution microscopy reveals a dynamic picture of cell polarity maintenance during directional growth. Sci. Adv. 2015, 1, e1500947. [Google Scholar] [CrossRef]
- Steinberg, G.; Peñalva, M.A.; Riquelme, M.; Wösten, H.A.; Harris, S.D. Cell Biology of Hyphal Growth. In The Fungal Kingdom; ASM Press: Washington, DC, USA, 2017. [Google Scholar]
- Meyer, V.; Arentshorst, M.; van den Hondel, C.A.M.J.J.; Ram, A.F.J. The polarisome component SpaA localises to hyphal tips of Aspergillus niger and is important for polar growth. Fungal Genet. Biol. 2008, 45, 152–164. [Google Scholar] [CrossRef]
- Zheng, P.; Nguyen, T.A.; Wong, J.Y.; Lee, M.; Nguyen, T.A.; Fan, J.S.; Yang, D.; Jedd, G. Spitzenkörper assembly mechanisms reveal conserved features of fungal and metazoan polarity scaffolds. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Fitz, E.; Gamauf, C.; Seiboth, B.; Wanka, F. Deletion of the small GTPase rac1 in Trichoderma reesei provokes hyperbranching and impacts growth and cellulase production. Fungal Biol. Biotechnol. 2019, 6, 1–11. [Google Scholar] [CrossRef]
- Kwon, M.J.; Nitsche, B.M.; Arentshorst, M.; Jorgensen, T.R.; Ram, A.F.; Meyer, V. The transcriptomic signature of RacA activation and inactivation provides new insights into the morphogenetic network of Aspergillus niger. PLoS ONE 2013, 8, e68946. [Google Scholar] [CrossRef]
- Takeshita, N.; Higashitsuji, Y.; Konzack, S.; Fischer, R. Apical sterol-rich membranes are essential for localizing cell end markers that determine growth directionality in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 2008, 19, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, M.; Bredeweg, E.L.; Callejas-Negrete, O.; Roberson, R.W.; Ludwig, S.; Beltrán-Aguilar, A.; Seiler, S.; Novick, P.; Freitag, M. The Neurospora crassa exocyst complex tethers Spitzenkörper vesicles to the apical plasma membrane during polarized growth. Mol. Biol. Cell 2014, 25, 1312–1326. [Google Scholar] [CrossRef] [PubMed]
- De Assis, L.J.; Ries, L.N.A.; Savoldi, M.; Dos Reis, T.F.; Brown, N.A.; Goldman, G.H. Aspergillus nidulans protein kinase A plays an important role in cellulase production. Biotechnol. Biofuels 2015, 8, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, M.; Aguirre, J.; Bartnicki-García, S.; Braus, G.H.; Feldbrügge, M.; Fleig, U.; Hansberg, W.; Herrera-Estrella, A.; Kämper, J.; Kück, U.; et al. Fungal Morphogenesis, from the Polarized Growth of Hyphae to Complex Reproduction and Infection Structures. Microbiol. Mol. Biol. Rev. 2018, 82, e00068-17. [Google Scholar] [CrossRef]
- Takeshita, N.; Evangelinos, M.; Zhou, L.; Serizawa, T.; Somera-Fajardo, R.A.; Lu, L.; Takaya, N.; Nienhaus, G.U.; Fischer, R. Pulses of Ca2+ coordinate actin assembly and exocytosis for stepwise cell extension. Proc. Natl. Acad. Sci. USA 2017, 114, 5701–5706. [Google Scholar] [CrossRef]
- Takeshita, N. Coordinated process of polarized growth in filamentous fungi. Biosci. Biotechnol. Biochem. 2016, 80, 1693–1699. [Google Scholar] [CrossRef]
- Steinberg, G. Hyphal growth: A tale of motors, lipids, and the spitzenkörper. Eukaryot. Cell 2007, 6, 351–360. [Google Scholar] [CrossRef]
- Fiedler, M.R.M.; Cairns, T.C.; Koch, O.; Kubisch, C.; Meyer, V. Conditional Expression of the Small GTPase ArfA Impacts Secretion, Morphology, Growth, and Actin Ring Position in Aspergillus niger. Front. Microbiol. 2018, 9, 878. [Google Scholar] [CrossRef]
- Caballero-Lima, D.; Kaneva, I.N.; Watton, S.P.; Sudbery, P.E.; Craven, C.J. The spatial distribution of the exocyst and actin cortical patches is sufficient to organize hyphal tip growth. Eukaryot. Cell 2013, 12, 998–1008. [Google Scholar] [CrossRef] [PubMed]
- Taheri-Talesh, N.; Horio, T.; Araujo-Bazán, L.; Dou, X.; Espeso, E.A.; Peñalva, M.A.; Osmani, S.A.; Oakley, B.R. The tip growth apparatus of Aspergillus nidulans. Mol. Biol. Cell 2008, 19, 1439–1449. [Google Scholar] [CrossRef] [PubMed]
- Cairns, T.C.; Feurstein, C.; Zheng, X.; Zheng, P.; Sun, J.; Meyer, V. A quantitative image analysis pipeline for the characterization of filamentous fungal morphologies as a tool to uncover targets for morphology engineering: A case study using aplD in Aspergillus niger. Biotechnol. Biofuels 2019, 12, 149. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.D. Hyphal branching in filamentous fungi. Dev. Biol. 2019, 451, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://marketersmedia.com/glucoamylase-industry-2017-global-market-demand-growth-trends-and-2022-forecast-report/259805 (accessed on 1 January 2018).
- Meyer, V.; Wanka, F.; van Gent, J.; Arentshorst, M.; van den Hondel, C.A.M.J.J.; Ram, A.F.J. Fungal gene expression on demand: An inducible, tunable, and metabolism-independent expression system for Aspergillus niger. Appl. Environ. Microbiol. 2011, 77, 2975–2983. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, J. The filamentous fungal pellet and forces driving its formation. Crit. Rev. Biotechnol. 2016, 36, 1066–1077. [Google Scholar] [CrossRef]
- Wucherpfennig, T.; Lakowitz, A.; Driouch, H.; Krull, R.; Wittmann, C. Customization of Aspergillus niger Morphology through Addition of Talc Micro Particles. J. Vis. Exp. 2012. [Google Scholar] [CrossRef] [PubMed]
- Schmideder, S.; Barthel, L.; Müller, H.; Meyer, V.; Briesen, H. From three-dimensional morphology to effective diffusivity in filamentous fungal pellets. Biotechnol. Bioeng. 2019, 116, 3360–3371. [Google Scholar] [CrossRef]
- Te Biesebeke, R.; Record, E.; Van Biezen, N.; Heerikhuisen, M.; Franken, A.; Punt, P.J.; Van Den Hondel, C.A.M.J.J. Branching mutants of Aspergillus oryzae with improved amylase and protease production on solid substrates. Appl. Microbiol. Biotechnol. 2005, 69, 44–50. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Li, C.; Ma, L.; Zhang, D.; Chen, S. Effect of highly branched hyphal morphology on the enhanced production of cellulase in Trichoderma reesei DES-15. 3 Biotech 2016, 6, 214. [Google Scholar] [CrossRef][Green Version]
- Yin, X.; Shin, H.D.; Li, J.; Du, G.; Liu, L.; Chen, J. Comparative genomics and transcriptome analysis of Aspergillus niger and metabolic engineering for citrate production. Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Papagianni, M.; Mattey, M.; Kristiansen, B. Morphology and citric acid production of Aspergillus niger PM1. Biotechnol. Lett. 1994, 16, 929–934. [Google Scholar] [CrossRef]
- Gonciarz, J.; Bizukojc, M. Adding talc microparticles to Aspergillus terreus ATCC 20542 preculture decreases fungal pellet size and improves lovastatin production. Eng. Life Sci. 2014, 14, 190–200. [Google Scholar] [CrossRef]
- Cairns, T.C.; Feurstein, C.; Zheng, X.; Zhang, L.H.; Zheng, P.; Sun, J.; Meyer, V. Functional exploration of co-expression networks identifies a nexus for modulating protein and citric acid titres in Aspergillus niger submerged culture. Fungal Biol. Biotechnol. 2019, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Schmideder, S.; Barthel, L.; Friedrich, T.; Thalhammer, M.; Kovačević, T.; Niessen, L.; Meyer, V.; Briesen, H. An X-ray microtomography-based method for detailed analysis of the three-dimensional morphology of fungal pellets. Biotechnol. Bioeng. 2019, 116, 1355–1365. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nimrichter, L.; Oliveira, D.L.; Frases, S.; Miranda, K.; Zaragoza, O.; Alvarez, M.; Nakouzi, A.; Feldmesser, M.; Casadevall, A. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell 2007, 6, 48–59. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nakayasu, E.S.; Oliveira, D.L.; Nimrichter, L.; Nosanchuk, J.D.; Almeida, I.C.; Casadevall, A. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot. Cell 2008, 7, 58–67. [Google Scholar] [CrossRef]
- Zhao, K.; Bleackley, M.; Chisanga, D.; Gangoda, L.; Fonseka, P.; Liem, M.; Kalra, H.; Al Saffar, H.; Keerthikumar, S.; Ang, C.-S.; et al. Extracellular vesicles secreted by Saccharomyces cerevisiae are involved in cell wall remodelling. Commun. Biol. 2019, 2, 305. [Google Scholar] [CrossRef]
- Oliveira, D.L.; Nakayasu, E.S.; Joffe, L.S.; Guimarães, A.J.; Sobreira, T.J.P.; Nosanchuk, J.D.; Cordero, R.J.B.; Frases, S.; Casadevall, A.; Almeida, I.C.; et al. Characterization of yeast extracellular vesicles: Evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS ONE 2010, 5, 1–13. [Google Scholar] [CrossRef]
- Hill, E.H.; Solomon, P.S. Extracellular vesicles from the apoplastic fungal wheat pathogen Zymoseptoria tritici. Fungal Biol. Biotechnol. 2020, 7, 1–14. [Google Scholar] [CrossRef]
- Bleackley, M.R.; Samuel, M.; Garcia-Ceron, D.; McKenna, J.A.; Lowe, R.G.T.; Pathan, M.; Zhao, K.; Ang, C.S.; Mathivanan, S.; Anderson, M.A. Extracellular Vesicles from the Cotton Pathogen Fusarium oxysporum f. sp. vasinfectum Induce a Phytotoxic Response in Plants. Front. Plant Sci. 2020, 10, 1610. [Google Scholar] [CrossRef] [PubMed]
- Bielska, E.; May, R.C. Extracellular vesicles of human pathogenic fungi. Curr. Opin. Microbiol. 2019, 52, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Vallejo, M.C.; Nakayasu, E.S.; Matsuo, A.L.; Sobreira, T.J.P.; Longo, L.V.G.; Ganiko, L.; Almeida, I.C.; Puccia, R. Vesicle and vesicle-free extracellular proteome of Paracoccidioides brasiliensis: Comparative analysis with other pathogenic fungi. J. Proteome Res. 2012, 11, 1676–1685. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, R.P.; Puccia, R.; Rodrigues, M.L.; Oliveira, D.L.; Joffe, L.S.; César, G.V.; Nimrichter, L.; Goldenberg, S.; Alves, L.R. Extracellular vesicle-mediated export of fungal RNA. Sci. Rep. 2015, 5, 7763. [Google Scholar] [CrossRef]
- Vallejo, M.C.; Nakayasu, E.S.; Longo, L.V.G.; Ganiko, L.; Lopes, F.G.; Matsuo, A.L.; Almeida, I.C.; Puccia, R. Lipidomic analysis of extracellular vesicles from the pathogenic phase of Paracoccidioides brasiliensis. PLoS ONE 2012, 7, e39463. [Google Scholar]
- da Silva, R.P.; Heiss, C.; Black, I.; Azadi, P.; Gerlach, J.Q.; Travassos, L.R.; Joshi, L.; Kilcoyne, M.; Puccia, R. Extracellular vesicles from Paracoccidioides pathogenic species transport polysaccharide and expose ligands for DC-SIGN receptors. Sci. Rep. 2015, 5, 14213. [Google Scholar] [CrossRef] [PubMed]
- Bleackley, M.R.; Dawson, C.S.; Anderson, M.A. Fungal Extracellular Vesicles with a Focus on Proteomic Analysis. Proteomics 2019, 19, e1800232. [Google Scholar] [CrossRef]
- Kwon, M.J.; Schütze, T.; Spohner, S.; Haefner, S.; Meyer, V. Practical guidance for the implementation of the CRISPR genome editing tool in filamentous fungi. Fungal Biol. Biotechnol. 2019, 6, 15. [Google Scholar] [CrossRef]
- Dadachova, E.; Bryan, R.A.; Huang, X.; Moadel, T.; Schweitzer, A.D.; Aisen, P.; Nosanchuk, J.D.; Casadevall, A. Ionizing radiation changes the electronic properties of melanin and enhances the growth of melanized fungi. PLoS ONE 2007, 2, e457. [Google Scholar] [CrossRef] [PubMed]
- Casadeval, A.; Cordero, R.J.B.; Bryan, R.; Nosanchuk, J.; Dadachova, E. Melanin, Radiation, and Energy Transduction in Fungi. In The Fungal Kingdom; ASM Press: Washington, DC, USA, 2017. [Google Scholar] [CrossRef]
- Burggraaf, A.M.; Punt, P.J.; Ram, A.F.J. The unconventional secretion of PepN is independent of a functional autophagy machinery in the filamentous fungus Aspergillus niger. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed]
- Richards, T.A.; Talbot, N.J. Osmotrophy. Curr. Biol. 2018, 28, R1179–R1180. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.; Swinnen, S.; Thevelein, J.M.; Nevoigt, E. Glycerol metabolism and transport in yeast and fungi: Established knowledge and ambiguities. Environ. Microbiol. 2017, 19, 878–893. [Google Scholar] [CrossRef]
- Stajich, J.E.; Harris, T.; Brunk, B.P.; Brestelli, J.; Fischer, S.; Harb, O.S.; Kissinger, J.C.; Li, W.; Nayak, V.; Pinney, D.F.; et al. FungiDB: An integrated functional genomics database for fungi. Nucleic Acids Res. 2012, 40, D675–D681. [Google Scholar] [CrossRef] [PubMed]
- Martinez, C.A.; Hu, S.; Dumond, Y.; Tao, J.; Kelleher, P.; Tully, L. Development of a chemoenzymatic manufacturing process for Pregabalin. Org. Process. Res. Dev. 2008, 12, 392–398. [Google Scholar] [CrossRef]
- Gopinath, S.C.B.; Anbu, P.; Lakshmipriya, T.; Hilda, A. Strategies to Characterize Fungal Lipases for Applications in Medicine and Dairy Industry. Biomed. Res. Int. 2013, 2013, 154549. [Google Scholar] [CrossRef]
- Glass, N.L.; Schmoll, M.; Cate, J.H.D.; Coradetti, S. Plant cell wall deconstruction by ascomycete fungi. Annu. Rev. Microbiol. 2013, 67, 477–498. [Google Scholar] [CrossRef]
- Andlar, M.; Rezić, T.; Marđetko, N.; Kracher, D.; Ludwig, R.; Šantek, B. Lignocellulose degradation: An overview of fungi and fungal enzymes involved in lignocellulose degradation. Eng. Life Sci. 2018, 18, 768–778. [Google Scholar] [CrossRef]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 2013, 6, 41. [Google Scholar] [CrossRef]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef]
- Kameshwar, A.K.S.; Ramos, L.P.; Qin, W. CAZymes-based ranking of fungi (CBRF): An interactive web database for identifying fungi with extrinsic plant biomass degrading abilities. Bioresour. Bioprocess. 2019, 6, 1–10. [Google Scholar] [CrossRef]
- Durand, H.; Clanet, M.; Tiraby, G. Genetic improvement of Trichoderma reesei for large scale cellulase production. Enzym. Microb. Technol. 1988, 10, 341–346. [Google Scholar] [CrossRef]
- Blumer-Schuette, S.E.; Brown, S.D.; Sander, K.B.; Bayer, E.A.; Kataeva, I.; Zurawski, J.V.; Conway, J.M.; Adams, M.W.W.; Kelly, R.M. Thermophilic lignocellulose deconstruction. FEMS Microbiol. Rev. 2014, 38, 393–448. [Google Scholar] [CrossRef] [PubMed]
- Brenelli, L.B.; Persinoti, G.F.; Cairo, J.P.L.F.; Liberato, M.V.; Gonçalves, T.A.; Otero, I.V.R.; Mainardi, P.H.; Felby, C.; Sette, L.D.; Squina, F.M. Novel redox-active enzymes for ligninolytic applications revealed from multiomics analyses of Peniophora sp. CBMAI 1063, a laccase hyper-producer strain. Sci. Rep. 2019, 9, 1–15. [Google Scholar]
- Willems, T.; De Mol, M.L.; De Bruycker, A.; De Maeseneire, S.L.; Soetaert, W.K. Alkaloids from marine fungi: Promising antimicrobials. Antibiotics 2020, 9, 340. [Google Scholar] [CrossRef]
- Russell, J.R.; Huang, J.; Anand, P.; Kucera, K.; Sandoval, A.G.; Dantzler, K.W.; Hickman, D.S.; Jee, J.; Kimovec, F.M.; Koppstein, D.; et al. Biodegradation of polyester polyurethane by endophytic fungi. Appl. Environ. Microbiol. 2011, 77, 6076–6084. [Google Scholar] [CrossRef]
- Brunner, I.; Fischer, M.; Rüthi, J.; Stierli, B.; Frey, B. Ability of fungi isolated from plastic debris floating in the shoreline of a lake to degrade plastics. PLoS ONE 2018, 13, e0202047. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Barragán, J.; Domínguez-Malfavón, L.; Vargas-Suárez, M.; González-Hernández, R.; Aguilar-Osorio, G.; Loza-Tavera, H. Biodegradative activities of selected environmental fungi on a polyester polyurethane varnish and polyether polyurethane foams. Appl. Environ. Microbiol. 2016, 82, 5225–5235. [Google Scholar] [CrossRef]
- Smith, A.D.; Logeman, B.L.; Thiele, D.J. Copper Acquisition and Utilization in Fungi. Annu. Rev. Microbiol. 2017, 71, 597–623. [Google Scholar] [CrossRef]
- Philpott, C.C. Iron uptake in fungi: A system for every source. Biochim. Biophys. Acta Mol. Cell Res. 2006, 1763, 636–645. [Google Scholar] [CrossRef] [PubMed]
- Yasmin, S.; Alcazar-Fuoli, L.; Grundlinger, M.; Puempel, T.; Cairns, T.; Blatzer, M.; Lopez, J.F.; Grimalt, J.O.; Bignell, E.; Haas, H. Mevalonate governs interdependency of ergosterol and siderophore biosyntheses in the fungal pathogen Aspergillus fumigatus. Proc. Natl. Acad. Sci. USA 2012, 109, E497–E504. [Google Scholar] [CrossRef]
- Ahmed, E.; Holmström, S.J.M. Siderophores in environmental research: Roles and applications. Microb. Biotechnol. 2014, 7, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Renshaw, J.C.; Robson, G.D.; Trinci, A.P.J.; Wiebe, M.G.; Livens, F.R.; Collison, D.; Taylor, R.J. Fungal siderophores: Structures, functions and applications. Mycol. Res. 2002, 106, 1123–1142. [Google Scholar] [CrossRef]
- Andersen, M.R.; Lehmann, L.; Nielsen, J. Systemic analysis of the response of Aspergillus niger to ambient pH. Genome Biol. 2009, 10, R47. [Google Scholar] [CrossRef]
- Dutton, M.V.; Evans, C.S. Oxalate production by fungi: Its role in pathogenicity and ecology in the soil environment. Can. J. Microbiol. 1996, 42, 881–895. [Google Scholar] [CrossRef]
- Landeweert, R.; Hoffland, E.; Finlay, R.D.; Kuyper, T.W.; Van Breemen, N. Linking plants to rocks: Ectomycorrhizal fungi mobilize nutrients from minerals. Trends Ecol. Evol. 2001, 16, 248–254. [Google Scholar] [CrossRef]
- Upton, D.J.; McQueen-Mason, S.J.; Wood, A.J. An accurate description of Aspergillus niger organic acid batch fermentation through dynamic metabolic modelling. Biotechnol. Biofuels 2017, 10, 258. [Google Scholar] [CrossRef] [PubMed]
- Li, R.-X.; Cai, F.; Pang, G.; Shen, Q.R.; Li, R.; Chen, W. Solubilisation of phosphate and micronutrients by Trichoderma harzianum and its relationship with the promotion of tomato plant growth. PLoS ONE 2015, 10, e0130081. [Google Scholar] [CrossRef]
- Guerinot, M.L.; Meidl, E.J.; Plessner, O. Citrate as a siderophore in Bradyrhizobium japonicum. J. Bacteriol. 1990, 172, 3298–3303. [Google Scholar] [CrossRef] [PubMed]
- Legiša, M.; Mattey, M. Changes in primary metabolism leading to citric acid overflow in Aspergillus niger. Biotechnol. Lett. 2007, 29, 181–190. [Google Scholar] [CrossRef]
- Zhang, L.; Zheng, X.; Cairns, T.C.; Zhang, Z.; Wang, D.; Zheng, P.; Sun, J. Disruption or reduced expression of the orotidine-5′-decarboxylase gene pyrG increases citric acid production: A new discovery during recyclable genome editing in Aspergillus niger. Microb. Cell Fact. 2020, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kirimura, K.; Kobayashi, K.; Ueda, Y.; Hattori, T. Phenotypes of gene disruptants in relation to a putative mitochondrial malate-citrate shuttle protein in citric acid-producing Aspergillus niger. Biosci. Biotechnol. Biochem. 2016, 80, 1737–1746. [Google Scholar] [CrossRef] [PubMed][Green Version]
- van der Straat, L.; Vernooij, M.; Lammers, M.; van den Berg, W.; Schonewille, T.; Cordewener, J.; van der Meer, I.; Koops, A.; De Graaff, L.H. Expression of the Aspergillus terreus itaconic acid biosynthesis cluster in Aspergillus niger. Microb. Cell Fact. 2014, 13, 11. [Google Scholar] [CrossRef] [PubMed]
- Wierckx, N.; Agrimi, G.; Lübeck, P.S.; Steiger, M.G.; Mira, N.P.; Punt, P.J. Metabolic specialization in itaconic acid production: A tale of two fungi. Curr. Opin. Biotechnol. 2020, 62, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Steiger, M.G.; Rassinger, A.; Mattanovich, D.; Sauer, M. Engineering of the citrate exporter protein enables high citric acid production in Aspergillus niger. Metab. Eng. 2019, 52, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Linde, T.; Hossain, A.H.; Lübeck, M.; Punt, P.J.; Lübeck, P.S. Disruption of a putative mitochondrial oxaloacetate shuttle protein in Aspergillus carbonarius results in secretion of malic acid at the expense of citric acid production. BMC Biotechnol. 2019, 19, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Meyer, V.; Fiedler, M.; Nitsche, B.; King, R. The Cell Factory Aspergillus Enters the Big Data Era: Opportunities and Challenges for Optimising Product Formation. Adv. Biochem. Eng. Biotechnol. 2015, 149, 91–132. [Google Scholar] [PubMed]
- Fischer, G.J.; Keller, N.P. Production of cross-kingdom oxylipins by pathogenic fungi: An update on their role in development and pathogenicity. J. Microbiol. 2016, 54, 254–264. [Google Scholar] [CrossRef]
- Lu, T.; Yao, B.; Zhang, C. DFVF: Database of fungal virulence factors. Database 2012, 2012, bas032. [Google Scholar] [CrossRef]
- Kouzai, Y.; Nakajima, K.; Hayafune, M.; Ozawa, K.; Kaku, H.; Shibuya, N.; Minami, E.; Nishizawa, Y. CEBiP is the major chitin oligomer-binding protein in rice and plays a main role in the perception of chitin oligomers. Plant Mol. Biol. 2014, 84, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Mentlak, T.A.; Kombrink, A.; Shinya, T.; Ryder, L.S.; Otomo, I.; Saitoh, H.; Terauchi, R.; Nishizawa, Y.; Shibuya, N.; Thomma, B.P.H.J.; et al. Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 2012, 24, 322–335. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.Q.; Wang, F.Z.; Li, J.F. Hide-and-Seek: Chitin-Triggered Plant Immunity and Fungal Counterstrategies. Trends Plant Sci. 2020, 25, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Cen, K.; Li, B.; Lu, Y.; Zhang, S.; Wang, C. Divergent LysM effectors contribute to the virulence of Beauveria bassiana by evasion of insect immune defenses. PLoS Pathog. 2017, 13, e1006604. [Google Scholar] [CrossRef]
- Hardison, S.E.; Brown, G.D. C-type lectin receptors orchestrate antifungal immunity. Nat. Immunol. 2012, 13, 817–822. [Google Scholar] [CrossRef]
- Childers, D.S.; Avelar, G.M.; Bain, J.M.; Pradhan, A.; Larcombe, D.E.; Netea, M.G.; Erwig, L.P.; Gow, N.A.R.; Brown, A.J.P. Epitope shaving promotes fungal immune evasion. MBio 2020. [Google Scholar] [CrossRef]
- Behnsen, J.; Lessing, F.; Schindler, S.; Wartenberg, D.; Jacobsen, I.D.; Thoen, M.; Zipfel, P.F.; Brakhage, A.A. Secreted Aspergillus fumigatus protease Alp1 degrades human complement proteins C3, C4, and C5. Infect. Immun. 2010, 78, 3585–3594. [Google Scholar] [CrossRef]
- Shende, R.; Wong, S.S.W.; Rapole, S.; Beau, R.; Ibrahim-Granet, O.; Monod, M.; Gührs, K.H.; Pal, J.K.; Latgé, J.P.; Madan, T.; et al. Aspergillus fumigatus conidial metalloprotease Mep1p cleaves host complement proteins. J. Biol. Chem. 2018, 293, 15538–15555. [Google Scholar] [CrossRef] [PubMed]
- Gropp, K.; Schild, L.; Schindler, S.; Hube, B.; Zipfel, P.F.; Skerka, C. The yeast Candida albicans evades human complement attack by secretion of aspartic proteases. Mol. Immunol. 2009, 47, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, E.; Schneider, A.E.; Sándor, N.; Lermann, U.; Staib, P.; Kremlitzka, M.; Bajtay, Z.; Barz, D.; Erdei, A.; Józsi, M. Secreted aspartic protease 2 of Candida albicans inactivates factor H and the macrophage factor H-receptors CR3 (CD11b/CD18) and CR4 (CD11c/CD18). Immunol. Lett. 2015, 168, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Fontaine, T.; Latgé, J.P. Galactomannan produced by Aspergillus fumigatus: An update on the structure, biosynthesis and biological functions of an emblematic fungal biomarker. J. Fungi 2020, 6, 283. [Google Scholar] [CrossRef]
- Speth, C.; Rambach, G.; Lass-Flörl, C.; Howell, P.L.; Sheppard, D.C. Galactosaminogalactan (GAG) and its multiple roles in Aspergillus pathogenesis. Virulence 2019, 10, 976–983. [Google Scholar] [CrossRef] [PubMed]
- Spassieva, S.D.; Markham, J.E.; Hille, J. The plant disease resistance gene Asc-1 prevents disruption of sphingolipid metabolism during AAL-toxin-induced programmed cell death. Plant J. 2002, 32, 561–572. [Google Scholar] [CrossRef] [PubMed]
- Meiss, E.; Konno, H.; Groth, G.; Hisabori, T. Molecular processes of inhibition and stimulation of ATP synthase caused by the phytotoxin tentoxin. J. Biol. Chem. 2008, 283, 24594–24599. [Google Scholar] [CrossRef] [PubMed]
- Berestetskiy, A.; Dmitriev, A.; Mitina, G.; Lisker, I.; Andolfi, A.; Evidente, A. Nonenolides and cytochalasins with phytotoxic activity against Cirsium arvense and Sonchus arvensis: A structure-activity relationships study. Phytochemistry 2008, 69, 953–960. [Google Scholar] [CrossRef] [PubMed]
- Malerba, M.; Contran, N.; Tonelli, M.; Crosti, P.; Cerana, R. Role of nitric oxide in actin depolymerization and programmed cell death induced by fusicoccin in sycamore (Acer pseudoplatanus) cultured cells. Physiol. Plant. 2008, 133, 449–457. [Google Scholar] [CrossRef]
- Reineke, G.; Heinze, B.; Schirawski, J.; Buettner, H.; Kahmann, R.; Basse, C.W. Indole-3-acetic acid (IAA) biosynthesis in the smut fungus Ustilago maydis and its relevance for increased IAA levels in infected tissue and host tumour formation. Mol. Plant Pathol. 2008, 9, 339–355. [Google Scholar] [CrossRef]
- Möbius, N.; Hertweck, C. Fungal phytotoxins as mediators of virulence. Curr. Opin. Plant Biol. 2009, 12, 390–398. [Google Scholar] [CrossRef]
- Pusztahelyi, T.; Holb, I.J.; PÃ3csi, I. Secondary metabolites in fungus-plant interactions. Front. Plant Sci. 2015, 6, 573. [Google Scholar] [CrossRef]
- Bradshaw, M.J.; Bartholomew, H.P.; Fonseca, J.M.; Gaskins, V.L.; Prusky, D.; Jurick, W.M. Delivering the goods: Fungal secretion modulates virulence during host–pathogen interactions. Fungal Biol. Rev. 2021, 36, 76–86. [Google Scholar] [CrossRef]
- Kim, S.; Kim, C.-Y.; Park, S.-Y.; Kim, K.-T.; Jeon, J.; Chung, H.; Choi, G.; Kwon, S.; Choi, J.; Jeon, J.; et al. Two nuclear effectors of the rice blast fungus modulate host immunity via transcriptional reprogramming. Nat. Commun. 2020, 11, 5845. [Google Scholar] [CrossRef]
- McDonagh, A.; Fedorova, N.D.; Crabtree, J.; Yu, Y.; Kim, S.; Chen, D.; Loss, O.; Cairns, T.; Goldman, G.; Armstrong-James, D.; et al. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog. 2008, 4, e1000154. [Google Scholar] [CrossRef]
- Choi, H.S.; Shim, J.S.; Kim, J.-A.; Kang, S.W.; Kwon, H.J. Discovery of gliotoxin as a new small molecule targeting thioredoxin redox system. Biochem. Biophys. Res. Commun. 2007, 359, 523–528. [Google Scholar] [CrossRef] [PubMed]
- Bok, J.W.; Chung, D.; Balajee, S.A.; Marr, K.A.; Andes, D.; Nielsen, K.F.; Frisvad, J.C.; Kirby, K.A.; Keller, N.P. GliZ, a transcriptional regulator of gliotoxin biosynthesis, contributes to Aspergillus fumigatus virulence. Infect. Immun. 2006, 74, 6761–6768. [Google Scholar] [CrossRef] [PubMed]
- Spikes, S.; Xu, R.; Nguyen, C.K.; Chamilos, G.; Kontoyiannis, D.P.; Jacobson, R.H.; Ejzykowicz, D.E.; Chiang, L.Y.; Filler, S.G.; May, G.S. Gliotoxin production in Aspergillus fumigatus contributes to host-specific differences in virulence. J. Infect. Dis. 2008, 197, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Naglik, J.R.; Challacombe, S.J.; Hube, B. Candida albicans Secreted Aspartyl Proteinases in Virulence and Pathogenesis. Microbiol. Mol. Biol. Rev. 2003, 67, 400–428. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, T.; Cairns, T.C.; Olbermann, P.; Morschhauser, J.; Bignell, E.M.; Krappmann, S. Oligopeptide transport and regulation of extracellular proteolysis are required for growth of Aspergillus fumigatus on complex substrates but not for virulence. Mol. Microbiol. 2011, 82, 917–935. [Google Scholar] [CrossRef]
- Moyes, D.L.; Wilson, D.; Richardson, J.P.; Mogavero, S.; Tang, S.X.; Wernecke, J.; Höfs, S.; Gratacap, R.L.; Robbins, J.; Runglall, M.; et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 2016, 532, 64–68. [Google Scholar] [CrossRef] [PubMed]
- De Miccolis Angelini, R.M.; Rotolo, C.; Gerin, D.; Abate, D.; Pollastro, S.; Faretra, F. Global transcriptome analysis and differentially expressed genes in grapevine after application of the yeast-derived defense inducer cerevisane. Pest. Manag. Sci. 2019, 75, 2020–2033. [Google Scholar] [CrossRef]
- Becker, S.; Tebben, J.; Coffinet, S.; Wiltshire, K.; Iversen, M.H.; Harder, T.; Hinrichs, K.U.; Hehemann, J.H. Laminarin is a major molecule in the marine carbon cycle. Proc. Natl. Acad. Sci. USA 2020, 117, 6599–6607. [Google Scholar] [CrossRef]
- Jia, Y.; McAdams, S.A.; Bryan, G.T.; Hershey, H.P.; Valent, B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 2000, 19, 4004–4014. [Google Scholar] [CrossRef] [PubMed]
- Rooney, H.C.E.; Van’t Klooster, J.W.; van der Hoorn, R.A.L.; Joosten, M.H.A.J.; Jones, J.D.G.; de Wit, P.J.G.M. Cladosporium Avr2 Inhibits Tomato Rcr3 Protease Required for Cf-2-Dependent Disease Resistance. Science 2005, 308, 1783–1786. [Google Scholar] [CrossRef]
- Saintenac, C.; Lee, W.-S.; Cambon, F.; Rudd, J.J.; King, R.C.; Marande, W.; Powers, S.J.; Bergès, H.; Phillips, A.L.; Uauy, C.; et al. Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat. Genet. 2018, 50, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, C.G.; Guimarães, G.A.; Nogueira, S.R.; MacLean, D.; Cook, D.R.; Steuernagel, B.; Baek, J.; Bouyioukos, C.; Melo, B.D.V.A.; Tristão, G.; et al. A pigeonpea gene confers resistance to Asian soybean rust in soybean. Nat. Biotechnol. 2016, 34, 661–665. [Google Scholar] [CrossRef]
- Ghislain, M.; Byarugaba, A.A.; Magembe, E.; Njoroge, A.; Rivera, C.; Román, M.L.; Tovar, J.C.; Gamboa, S.; Forbes, G.A.; Kreuze, J.F.; et al. Stacking three late blight resistance genes from wild species directly into African highland potato varieties confers complete field resistance to local blight races. Plant Biotechnol. J. 2019, 17, 1119–1129. [Google Scholar] [CrossRef]
- Luo, M.; Xie, L.; Chakraborty, S.; Wang, A.; Matny, O.; Jugovich, M.; Kolmer, J.A.; Richardson, T.; Bhatt, D.; Hoque, M.; et al. A five-transgene cassette confers broad-spectrum resistance to a fungal rust pathogen in wheat. Nat. Biotechnol. 2021, 39, 561–566. [Google Scholar] [CrossRef]
- Mor, V.; Rella, A.; Farnou, A.M.; Singh, A.; Munshi, M.; Bryan, A.; Naseem, S.; Konopka, J.B.; Ojima, I.; Bullesbach, E.; et al. Identification of a new class of antifungals targeting the synthesis of fungal sphingolipids. MBio 2015, 6, e00647-15. [Google Scholar] [CrossRef]
- Lazzarini, C.; Haranahalli, K.; Rieger, R.; Ananthula, H.K.; Desai, P.B.; Ashbaugh, A.; Linke, M.J.; Cushion, M.T.; Ruzsicska, B.; Haley, J.; et al. Acylhydrazones as antifungal agents targeting the synthesis of fungal sphingolipids. Antimicrob. Agents Chemother. 2018. [Google Scholar] [CrossRef] [PubMed]
- Pierce, S.E.; Davis, R.W.; Nislow, C.; Giaever, G. Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat. Protoc. 2007, 2, 2958–2974. [Google Scholar] [CrossRef]
- Zhang, F.; Zhao, M.; Braun, D.R.; Ericksen, S.S.; Piotrowski, J.S.; Nelson, J.; Peng, J.; Ananiev, G.E.; Chanana, S.; Barns, K.; et al. A marine microbiome antifungal targets urgent-threat drug-resistant fungi. Science 2020, 370, 974–978. [Google Scholar] [CrossRef] [PubMed]
- Bankaitis, V.A.; Aitken, J.R.; Cleves, A.E.; Dowhan, W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature 1990, 347, 561–562. [Google Scholar] [CrossRef]
- Vossen, J.H.; Müller, W.H.; Lipke, P.N.; Klis, F.M. Restrictive glycosylphosphatidylinositol anchor synthesis in cwh6/gpi3 yeast cells causes aberrant biogenesis of cell wall proteins. J. Bacteriol. 1997, 179, 2202–2209. [Google Scholar] [CrossRef]
- McLellan, C.A.; Whitesell, L.; King, O.D.; Lancaster, A.K.; Mazitschek, R.; Lindquist, S. Inhibiting GPI anchor biosynthesis in fungi stresses the endoplasmic reticulum and enhances immunogenicity. ACS Chem. Biol. 2012, 7, 1520–1528. [Google Scholar] [CrossRef]
- Fu, Y.; Estoppey, D.; Roggo, S.; Pistorius, D.; Fuchs, F.; Studer, C.; Ibrahim, A.S.; Aust, T.; Grandjean, F.; Mihalic, M.; et al. Jawsamycin exhibits in vivo antifungal properties by inhibiting Spt14/Gpi3-mediated biosynthesis of glycosylphosphatidylinositol. Nat. Commun. 2020, 11, 3387. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, L.L.; Cota, E. Candida albicans agglutinin-like sequence (Als) family vignettes: A review of als protein structure and function. Front. Microbiol. 2016, 7, 280. [Google Scholar] [CrossRef]
- Richard, M.L.; Plaine, A. Comprehensive analysis of glycosylphosphatidylinositol-anchored proteins in Candida albicans. Eukaryot. Cell 2007, 6, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Schaller, M.; Bein, M.; Korting, H.C.; Baur, S.; Hamm, G.; Monod, M.; Beinhauer, S.; Hube, B. The secreted aspartyl proteinases Sap1 and Sap2 cause tissue damage in an in vitro model of vaginal candidiasis based on reconstituted human vaginal epithelium. Infect. Immun. 2003, 71, 3227–3234. [Google Scholar] [CrossRef] [PubMed]
- De Bernardis, F.; Amacker, M.; Arancia, S.; Sandini, S.; Gremion, C.; Zurbriggen, R.; Moser, C.; Cassone, A. A virosomal vaccine against candidal vaginitis: Immunogenicity, efficacy and safety profile in animal models. Vaccine 2012, 30, 4490–4498. [Google Scholar] [CrossRef] [PubMed]
- Wan Tso, G.H.; Reales-Calderon, J.A.; Pavelka, N. The Elusive Anti-Candida Vaccine: Lessons from the past and opportunities for the future. Front. Immunol. 2018, 9, 897. [Google Scholar]
- Nami, S.; Mohammadi, R.; Vakili, M.; Khezripour, K.; Mirzaei, H.; Morovati, H. Fungal vaccines, mechanism of actions and immunology: A comprehensive review. Biomed. Pharmacother. 2019, 109, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Asplund, J.; Wardle, D.A. How lichens impact on terrestrial community and ecosystem properties. Biol. Rev. 2017, 92, 1720–1738. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Liu, B.; Zhang, X.Y.; Zhou, Q.M.; Zhang, T.; Li, H.; Yu, Y.F.; Zhang, X.L.; Hao, X.Y.; Wang, M.; et al. Genome characteristics reveal the impact of lichenization on lichen-forming fungus Endocarpon pusillum Hedwig (Verrucariales, Ascomycota). BMC Genom. 2014, 15, 34. [Google Scholar] [CrossRef]
- Spribille, T.; Tuovinen, V.; Resl, P.; Vanderpool, D.; Wolinski, H.; Aime, M.C.; Schneider, K.; Stabentheiner, E.; Toome-Heller, M.; Thor, G.; et al. Basidiomycete yeasts in the cortex of ascomycete macrolichens. Science 2016, 353, 488–492. [Google Scholar] [CrossRef]
- Goga, M.; Elečko, J.; Marcinčinová, M.; Ručová, D.; Bačkorová, M.; Bačkor, M. Lichen Metabolites: An Overview of Some Secondary Metabolites and Their Biological Potential. In Co-Evolution of Secondary Metabolites; Reference Series in Phytochemistry; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Armaleo, D.; Müller, O.; Lutzoni, F.; Andrésson, Ó.S.; Blanc, G.; Bode, H.B.; Collart, F.R.; Dal Grande, F.; Dietrich, F.; Grigoriev, I.V.; et al. The lichen symbiosis re-viewed through the genomes of Cladonia grayi and its algal partner Asterochloris glomerata. BMC Genom. 2019, 20, 605. [Google Scholar] [CrossRef]
- Tagirdzhanova, G.; Saary, P.; Tingley, J.P.; Díaz-Escandón, D.; Abbott, D.W.; Finn, R.D.; Spribille, T. Predicted Input of Uncultured Fungal Symbionts to a Lichen Symbiosis from Metagenome-Assembled Genomes. Genome Biol. Evol. 2021, 13, evab047. [Google Scholar] [CrossRef] [PubMed]
- Zambare, V.P.; Christopher, L.P. Biopharmaceutical potential of lichens. Pharm. Biol. 2012. [Google Scholar] [CrossRef]
- Ulus, G. Antiangiogenic properties of lichen secondary metabolites. Phyther. Res. 2021, 35, 3046–3058. [Google Scholar] [CrossRef] [PubMed]
- Schuetze, T.; Meyer, V. Polycistronic gene expression in Aspergillus niger. Microb. Cell Fact. 2017, 16, 162. [Google Scholar] [CrossRef]
- Luginbuehl, L.H.; Menard, G.N.; Kurup, S.; Van Erp, H.; Radhakrishnan, G.V.; Breakspear, A.; Oldroyd, G.E.D.; Eastmond, P.J. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 2017, 356, 1175–1178. [Google Scholar] [CrossRef]
- Martin-Urdiroz, M.; Oses-Ruiz, M.; Ryder, L.S.; Talbot, N.J. Investigating the biology of plant infection by the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 2016, 90, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Parniske, M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat. Rev. Microbiol. 2008, 6, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Toro, K.S.; Brachmann, A. The effector candidate repertoire of the arbuscular mycorrhizal fungus Rhizophagus clarus. BMC Genom. 2016, 17, 101. [Google Scholar]
- Zeng, T.; Rodriguez-Moreno, L.; Mansurkhodzaev, A.; Wang, P.; van den Berg, W.; Gasciolli, V.; Cottaz, S.; Fort, S.; Thomma, B.P.H.J.; Bono, J.J.; et al. A lysin motif effector subverts chitin-triggered immunity to facilitate arbuscular mycorrhizal symbiosis. New Phytol. 2019, 225, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Voß, S.; Betz, R.; Heidt, S.; Corradi, N.; Requena, N. RiCRN1, a crinkler effector from the arbuscular mycorrhizal fungus rhizophagus irregularis, functions in arbuscule development. Front. Microbiol. 2018, 9, 2068. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Pieterse, C.M.J.; Bakker, P.A.H.M.; Berendsen, R.L. Beneficial microbes going underground of root immunity. Plant. Cell Environ. 2019, 42, 2860–2870. [Google Scholar] [CrossRef] [PubMed]
- Pozo, M.J.; Zabalgogeazcoa, I.; Vazquez de Aldana, B.R.; Martinez-Medina, A. Untapping the potential of plant mycobiomes for applications in agriculture. Curr. Opin. Plant. Biol. 2021, 60, 102034. [Google Scholar] [CrossRef]
- Amend, A.; Burgaud, G.; Cunliffe, M.; Edgcomb, V.P.; Ettinger, C.L.; Gutiérrez, M.H.; Heitman, J.; Hom, E.F.Y.; Ianiri, G.; Jones, A.C.; et al. Fungi in the marine environment: Open questions and unsolved problems. MBio 2019, 10, e01189-18. [Google Scholar] [CrossRef]
- Santus, W.; Devlin, J.R.; Behnsen, J. Crossing kingdoms: How the mycobiota and fungal-bacterial interactions impact host health and disease. Infect. Immun. 2021. [Google Scholar] [CrossRef]
- Leeder, A.C.; Palma-Guerrero, J.; Glass, N.L. The social network: Deciphering fungal language. Nat. Rev. Microbiol. 2011, 9, 440–451. [Google Scholar] [CrossRef]
- Fraser, J.A.; Heitman, J. Sex, MAT, and the Evolution of Fungal Virulence. In Molecular Principles of Fungal Pathogenesis; ASM Press: Washington, DC, USA, 2014. [Google Scholar]
- Nieuwenhuis, B.P.S.; James, T.Y. The frequency of sex in fungi. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150540. [Google Scholar] [CrossRef]
- O’Gorman, C.M.; Fuller, H.T.; Dyer, P.S. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 2009, 457, 471–474. [Google Scholar] [CrossRef]
- Jones, S.K.; Bennett, R.J. Fungal mating pheromones: Choreographing the dating game. Fungal Genet. Biol. 2011, 48, 668–676. [Google Scholar] [CrossRef]
- Chou, S.; Lane, S.; Liu, H. Regulation of Mating and Filamentation Genes by Two Distinct Ste12 Complexes in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006, 26, 4794–4805. [Google Scholar] [CrossRef] [PubMed]
- Frawley, D.; Stroe, M.C.; Oakley, B.R.; Heinekamp, T.; Straßburger, M.; Fleming, A.B.; Brakhage, A.A.; Bayram, Ö. The Pheromone Module SteC-MkkB-MpkB-SteD-HamE Regulates Development, Stress Responses and Secondary Metabolism in Aspergillus fumigatus. Front. Microbiol. 2020, 11, 811. [Google Scholar] [CrossRef]
- Frawley, D.; Greco, C.; Oakley, B.; Alhussain, M.M.; Fleming, A.B.; Keller, N.P.; Bayram, Ö. The tetrameric pheromone module SteC-MkkB-MpkB-SteD regulates asexual sporulation, sclerotia formation and aflatoxin production in Aspergillus flavus. Cell Microbiol. 2020, 11, 811. [Google Scholar] [CrossRef] [PubMed]
- Bayram, Ö.; Krappmann, S.; Ni, M.; Jin, W.B.; Helmstaedt, K.; Valerius, O.; Braus-Stromeyer, S.; Kwon, N.J.; Keller, N.P.; Yu, J.H.; et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 2008, 320, 1504–1506. [Google Scholar] [CrossRef] [PubMed]
- Frawley, D.; Bayram, Ö. The pheromone response module, a mitogen-activated protein kinase pathway implicated in the regulation of fungal development, secondary metabolism and pathogenicity. Fungal Genet. Biol. 2020, 144, 103469. [Google Scholar] [CrossRef]
- Nanou, K.; Roukas, T. Oxidative stress response of Blakeslea trispora induced by iron ions during carotene production in shake flask culture. Appl. Biochem. Biotechnol. 2020, 144, 103469. [Google Scholar] [CrossRef]
- Virágh, M.; Marton, A.; Vizler, C.; Tóth, L.; Vágvölgyi, C.; Marx, F.; Galgóczy, L. Insight into the antifungal mechanism of Neosartorya fischeri antifungal protein. Protein Cell 2015, 6, 518–528. [Google Scholar] [CrossRef]
- Bugeda, A.; Garrigues, S.; Gandía, M.; Manzanares, P.; Marcos, J.F.; Coca, M. The Antifungal Protein AfpB Induces Regulated Cell Death in Its Parental Fungus Penicillium digitatum. mSphere 2020. [Google Scholar] [CrossRef]
- Hegedüs, N.; Sigl, C.; Zadra, I.; Pócsi, I.; Marx, F. The paf gene product modulates asexual development in Penicillium chrysogenum. J. Basic Microbiol. 2011, 51, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Garrigues, S.; Gandía, M.; Marcos, J.F. Occurrence and function of fungal antifungal proteins: A case study of the citrus postharvest pathogen Penicillium digitatum. Appl. Microbiol. Biotechnol. 2016, 100, 2243–2256. [Google Scholar] [CrossRef]
- Schäpe, P.; Kwon, M.J.; Baumann, B.; Gutschmann, B.; Jung, S.; Lenz, S.; Nitsche, B.; Paege, N.; Schütze, T.; Cairns, T.C.; et al. Updating genome annotation for the microbial cell factory Aspergillus niger using gene co-expression networks. Nucleic Acids Res. 2019, 47, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Paege, N.; Jung, S.; Schäpe, P.; Müller-Hagen, D.; Ouedraogo, J.P.; Heiderich, C.; Jedamzick, J.; Nitsche, B.M.; Van Den Hondel, C.A.; Ram, A.F.; et al. A transcriptome meta-Analysis proposes novel biological roles for the antifungal protein anafp in Aspergillus niger. PLoS ONE 2016, 11, e0165755. [Google Scholar] [CrossRef] [PubMed]
- Meyer, V. A small protein that fights fungi: AFP as a new promising antifungal agent of biotechnological value. Appl. Microbiol. Biotechnol. 2008, 78, 17–28. [Google Scholar] [CrossRef]
- Coca, M.; Bortolotti, C.; Rufat, M.; Peñas, G.; Eritja, R.; Tharreau, D.; Martinez Del Pozo, A.; Messeguer, J.; San Segundo, B. Transgenic rice plants expressing the antifungal AFP protein from Aspergillus giganteus show enhanced resistance to the rice blast fungus Magnaporthe grisea. Plant Mol. Biol. 2004, 54, 245–259. [Google Scholar] [CrossRef] [PubMed]
- Künzler, M. How fungi defend themselves against microbial competitors and animal predators. PLoS Pathog. 2018, 14, e1007184. [Google Scholar] [CrossRef]
- Nai, C.; Meyer, V. From Axenic to Mixed Cultures: Technological Advances Accelerating a Paradigm Shift in Microbiology. Trends Microbiol. 2017, 26, 538–554. [Google Scholar] [CrossRef]
- Guo, Z.; Doll, K.; Dastjerdi, R.; Karlovsky, P.; Dehne, H.W.; Altincicek, B. Effect of fungal colonization of wheat grains with Fusarium spp. on food choice, weight gain and mortality of meal beetle larvae (Tenebrio molitor). PLoS ONE 2014, 9, e100112. [Google Scholar] [CrossRef]
- German-Fattal, M. Fusafungine, an antimicrobial with anti-inflammatory properties in respiratory tract infections. Clin. Drug Investig. 2001, 21, 653–670. [Google Scholar] [CrossRef]
- Richter, L.; Wanka, F.; Boecker, S.; Storm, D.; Kurt, T.; Vural, Ö.; Süßmuth, R.; Meyer, V. Engineering of Aspergillus niger for the production of secondary metabolites. Fungal Biol. Biotechnol. 2014, 1, 4. [Google Scholar] [CrossRef] [PubMed]
- Boecker, S.; Grätz, S.; Kerwat, D.; Adam, L.; Schirmer, D.; Richter, L.; Schütze, T.; Petras, D.; Süssmuth, R.D.; Meyer, V. Aspergillus niger is a superior expression host for the production of bioactive fungal cyclodepsipeptides. Fungal Biol. Biotechnol. 2018, 5, 4. [Google Scholar] [CrossRef]
- Steiniger, C.; Hoffmann, S.; Mainz, A.; Kaiser, M.; Voigt, K.; Meyer, V.; Süssmuth, R. Harnessing Fungal Nonribosomal Cyclodepsipeptide Synthetases for Mechanistic Insights and Tailored Engineering. Chem. Sci. 2017, 8, 7834–7843. [Google Scholar] [CrossRef]
- de Vries, R.P.; Riley, R.; Wiebenga, A.; Aguilar-Osorio, G.; Amillis, S.; Uchima, C.A.; Anderluh, G.; Asadollahi, M.; Askin, M.; Barry, K.; et al. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017, 18, 1–45. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A.; Schroeckh, V. Fungal secondary metabolites—Strategies to activate silent gene clusters. Fungal Genet. Biol. 2011, 48, 15–22. [Google Scholar] [CrossRef]
- Kovalchuk, A.; Driessen, A.J.M. Phylogenetic analysis of fungal ABC transporters. BMC Genom. 2010, 11, 177. [Google Scholar] [CrossRef]
- Coleman, J.J.; Mylonakis, E. Efflux in Fungi: La Pièce de Résistance. PLoS Pathog. 2009, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cairns, T.; Meyer, V. In silico prediction and characterization of secondary metabolite biosynthetic gene clusters in the wheat pathogen Zymoseptoria tritici. BMC Genom. 2017, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhou, Y.; Cao, W.; Liu, H. Improved Production of Malic Acid in Aspergillus niger by Abolishing Citric Acid Accumulation and Enhancing Glycolytic Flux. ACS Synth. Biol. 2020, 9, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-N.; Toyotome, T.; Muraosa, Y.; Watanabe, A.; Wuren, T.; Bunsupa, S.; Aoyagi, K.; Yamazaki, M.; Takino, M.; Kamei, K. GliA in Aspergillus fumigatus is required for its tolerance to gliotoxin and affects the amount of extracellular and intracellular gliotoxin. Med. Mycol. 2014, 52, 506. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.K.; Yu, J.; Yu, J.H. aflT, a MFS transporter-encoding gene located in the aflatoxin gene cluster, does not have a significant role in aflatoxin secretion. Fungal Genet. Biol. 2004, 41, 911–920. [Google Scholar] [CrossRef]
- Chowdhary, A.; Kathuria, S.; Xu, J.; Meis, J.F. Emergence of azole-resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. Plos Pathog. 2013, 9, e1003633. [Google Scholar] [CrossRef]
- Esquivel, B.D.; Rybak, J.M.; Barker, K.S.; Fortwendel, J.R.; Rogers, P.D.; White, T.C. Characterization of the efflux capability and substrate specificity of Aspergillus fumigatus PDR5-like ABC transporters expressed in Saccharomyces cerevisiae. MBio 2020. [Google Scholar] [CrossRef] [PubMed]
- Redhu, A.K.; Banerjee, A.; Shah, A.H.; Moreno, A.; Rawal, M.K.; Nair, R.; Falson, P.; Prasad, R. Molecular Basis of Substrate Polyspecificity of the Candida albicans Mdr1p Multidrug/H+ Antiporter. J. Mol. Biol. 2018, 430, 682–694. [Google Scholar] [CrossRef]
- Holmes, A.R.; Cardno, T.S.; Strouse, J.J.; Ivnitski-Steele, I.; Keniya, M.V.; Lackovic, K.; Monk, B.C.; Sklar, L.A.; Cannon, R.D. Targeting efflux pumps to overcome antifungal drug resistance. Future Med. Chem. 2016, 8, 1485–1501. [Google Scholar] [CrossRef] [PubMed]
- Lamping, E.; Monk, B.C.; Niimi, K.; Holmes, A.R.; Tsao, S.; Tanabe, K.; Niimi, M.; Uehara, Y.; Cannon, R.D. Characterization of three classes of membrane proteins involved in fungal azole resistance by functional hyperexpression in Saccharomyces cerevisiae. Eukaryot. Cell 2007, 6, 1150–1165. [Google Scholar] [CrossRef]
- Lee, M.D.; Galazzo, J.L.; Staley, A.L.; Lee, J.C.; Warren, M.S.; Fuernkranz, H.; Chamberland, S.; Lomovskaya, O.; Miller, G.H. Microbial fermentation-derived inhibitors of efflux-pump-mediated drug resistance. Farmaco 2001, 56, 81–85. [Google Scholar] [CrossRef]
- Holmes, A.R.; Keniya, M.V.; Ivnitski-Steele, I.; Monk, B.C.; Lamping, E.; Sklar, L.A.; Cannon, R.D. The monoamine oxidase A inhibitor clorgyline is a broad-spectrum inhibitor of fungal ABC and MFS transporter efflux pump activities which reverses the azole resistance of Candida albicans and Candida glabrata clinical isolates. Antimicrob. Agents Chemother. 2012, 56, 1508–1515. [Google Scholar] [CrossRef]
- Niimi, K.; Harding, D.R.K.; Holmes, A.R.; Lamping, E.; Niimi, M.; Tyndall, J.D.A.; Cannon, R.D.; Monk, B.C. Specific interactions between the Candida albicans ABC transporter Cdr1p ectodomain and a D-octapeptide derivative inhibitor. Mol. Microbiol. 2012, 85, 747–767. [Google Scholar] [CrossRef] [PubMed]
- Liaud, N.; Giniés, C.; Navarro, D.; Fabre, N.; Crapart, S.; Gimbert, I.H.-; Levasseur, A.; Raouche, S.; Sigoillot, J.-C. Exploring fungal biodiversity: Organic acid production by 66 strains of filamentous fungi. Fungal Biol. Biotechnol. 2014, 1, 1–10. [Google Scholar] [CrossRef]
- Strelko, C.L.; Lu, W.; Dufort, F.J.; Seyfried, T.N.; Chiles, T.C.; Rabinowitz, J.D.; Roberts, M.F. Itaconic acid is a mammalian metabolite induced during macrophage activation. J. Am. Chem. Soc. 2011, 133, 16386–16389. [Google Scholar] [CrossRef] [PubMed]
- Sasikaran, J.; Ziemski, M.; Zadora, P.K.; Fleig, A.; Berg, I.A. Bacterial itaconate degradation promotes pathogenicity. Nat. Chem. Biol. 2014, 10, 371–377. [Google Scholar] [CrossRef] [PubMed]

| Product Class | Product | Exemplar Producing Fungal Species | Secretion Route and/or Plasma Membrane Transporter | Postulated or Confirmed Function(s) in the Natural Niche | Current or Prospective Application(s) | References |
|---|---|---|---|---|---|---|
| Enzyme/proteins | Glucoamylase GlaA | A. niger | Classical secretion route | Nutrient acquisition | Food and beverage industries processing starch to glucose | [4,15] |
| Cellulases | T. reesei, T. thermophila | Textile, food, and other industries | [16] | |||
| Lipases | T. lanuginosus A. oryzae | Applications in biodiesel, dairy, textile, detergent, paper, pharmaceutical, leather and other industries | [17,18] | |||
| Agglutinin-like sequence protein Als3 | Candida spp. | Adhesion of fungal cells to host and abiotic surfaces | Possible vaccine in humans | [19,20] | ||
| Secreted aspartic protease 2 Sap-2 | Nutrient acquisition, virulence factor | [21] | ||||
| Antifungal protein AFP | A. giganteus | Cannibal toxin regulating clonal growth, inhibitor of competing fungi | Prospective antifungal use in clinic or agriculture | [22,23] | ||
| Effectors | M. oryzae, Z. tritici, other plant-infecting fungi | Classical secretion route, biotrophic interphase complex, non-classical secretion route, extracellular vesicles | Virulence factors | Cognate receptor and R proteins used to elicit protective crop immunity | [24,25,26] | |
| Secondary metabolite | Triacetyl-fusarine C | A. fumigatus | Intermediates generated in mitochondria and peroxisomes plasma membrane transporter unknown | Micronutrient acquisition (iron chelator), necessary for full virulence | Biosynthetic pathway is a possible drug target, siderophores could be used to decontaminate metals from natural ecosystems | [27,28] |
| Penicillin | P. chrysogenum | Intermediates generated in peroxisome and vacuoles, possibly secreted via vesicles or unknown plasma membrane transporter | Inhibitor of competing microbes | Antimicrobial with widespread clinical use | [29] | |
| Trisporic acid | B. trispora | Unknown | Mating pheromone | Used to elevate yields of β-carotene during fermentation | [30] | |
| Enniatin | Fusarium spp., A. niger (heterologous expression host) | Unknown | Prevent predation | Antimicrobial and anti-inflammatory clinical applications | [31] | |
| Organic acid | Citric acid | A. niger | Plasma membrane transporter CexA | Micronutrient acquisition (iron chelator), inhibitor of competing microbes, overflow metabolite | Food and beverage as flavour enhancer, cleaning product, many industrial uses as a weak acid and platform chemical | [32] |
| Malic acid | Plasma membrane transporter DctA | Inhibitor of competing microbes, overflow metabolite | Industrial uses as a weak acid and platform chemical | [33] | ||
| Itaconic acid | A. terreus, U. maydis, A niger (heterologous expression host) | Plasma membrane transporter MfsA (A. terreus) or Itp1 (U. maydis) | Inhibitor of competing microbes, overflow metabolite | Industrial uses as a weak acid and platform chemical | [34] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cairns, T.C.; Zheng, X.; Zheng, P.; Sun, J.; Meyer, V. Turning Inside Out: Filamentous Fungal Secretion and Its Applications in Biotechnology, Agriculture, and the Clinic. J. Fungi 2021, 7, 535. https://doi.org/10.3390/jof7070535
Cairns TC, Zheng X, Zheng P, Sun J, Meyer V. Turning Inside Out: Filamentous Fungal Secretion and Its Applications in Biotechnology, Agriculture, and the Clinic. Journal of Fungi. 2021; 7(7):535. https://doi.org/10.3390/jof7070535
Chicago/Turabian StyleCairns, Timothy C., Xiaomei Zheng, Ping Zheng, Jibin Sun, and Vera Meyer. 2021. "Turning Inside Out: Filamentous Fungal Secretion and Its Applications in Biotechnology, Agriculture, and the Clinic" Journal of Fungi 7, no. 7: 535. https://doi.org/10.3390/jof7070535
APA StyleCairns, T. C., Zheng, X., Zheng, P., Sun, J., & Meyer, V. (2021). Turning Inside Out: Filamentous Fungal Secretion and Its Applications in Biotechnology, Agriculture, and the Clinic. Journal of Fungi, 7(7), 535. https://doi.org/10.3390/jof7070535






