Candida auris Whole-Genome Sequence Benchmark Dataset for Phylogenomic Pipelines
Abstract
1. Introduction
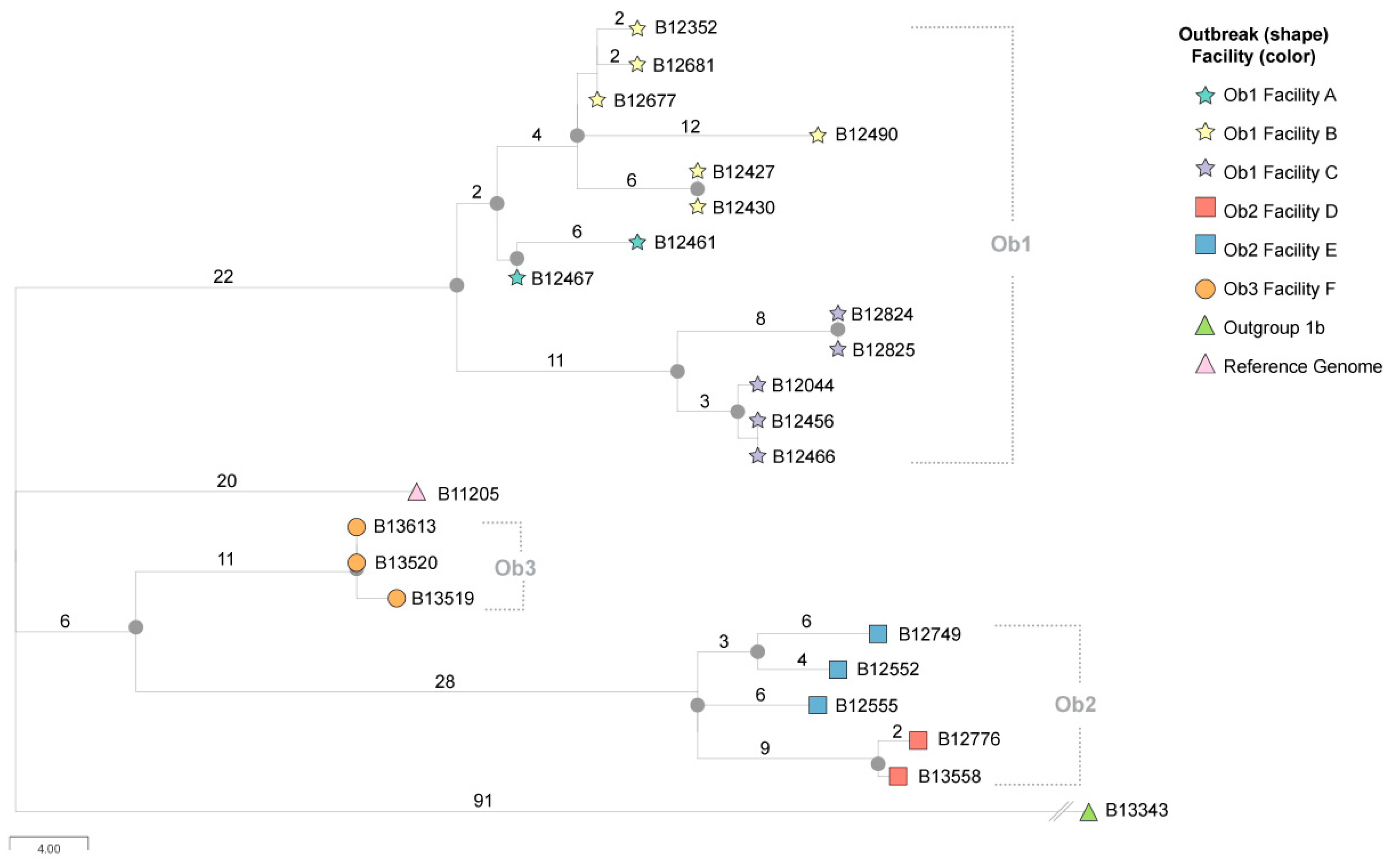
2. Materials and Methods
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Rhodes, J.; Fisher, M.C. Global epidemiology of emerging Candida auris. Curr. Opin. Microbiol. 2019, 52, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Chow, N.A.; Muñoz, J.F.; Gade, L.; Berkow, E.L.; Li, X.; Welsh, R.M.; Forsberg, K.; Lockhart, S.R.; Adam, R.; Alanio, A.; et al. Tracing the Evolutionary History and Global Expansion of Candida auris Using Population Genomic Analyses. mBio 2020, 11, e03364-19. [Google Scholar] [CrossRef] [PubMed]
- Saris, K.; Meis, J.F.; Voss, A. Candida auris. Curr. Opin. Infect. Dis. 2018, 31, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Welsh, R.M.; Bentz, M.L.; Shams, A.; Houston, H.; Lyons, A.; Rose, L.J.; Litvintseva, A.P. Survival, Persistence, and Isolation of the Emerging Multidrug-Resistant Pathogenic Yeast Candida auris on a Plastic Health Care Surface. J. Clin. Microbiol. 2017, 55, 2996–3005. [Google Scholar] [CrossRef] [PubMed]
- Van Schalkwyk, E.; Mpembe, R.S.; Thomas, J.; Shuping, L.; Ismail, H.; Lowman, W.; Karstaedt, A.S.; Chibabhai, V.; Wadula, J.; Avenant, T.; et al. Epidemiologic Shift in Candidemia Driven by Candida auris, South Africa, 2016-2017(1). Emerg. Infect. Dis. 2019, 25, 1698–1707. [Google Scholar] [CrossRef] [PubMed]
- Chow, N.A.; Gade, L.; Tsay, S.V.; Forsberg, K.; Greenko, J.A.; Southwick, K.L.; Barrett, P.M.; Kerins, J.L.; Lockhart, S.R.; Chiller, T.M.; et al. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: A molecular epidemiological survey. Lancet Infect. Dis. 2018, 18, 1377–1384. [Google Scholar] [CrossRef]
- Rhodes, J.; Abdolrasouli, A.; Farrer, R.A.; Cuomo, C.A.; Aanensen, D.M.; Armstrong-James, D.; Fisher, M.C.; Schelenz, S. Author Correction: Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg. Microbes Infect. 2018, 7, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, J.F.; Gade, L.; Chow, N.A.; Loparev, V.N.; Juieng, P.; Berkow, E.L.; Farrer, R.A.; Litvintseva, A.P.; Cuomo, C.A. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat. Commun. 2018, 9, 5346. [Google Scholar] [CrossRef] [PubMed]
- Chow, N.A.; De Groot, T.; Badali, H.; Abastabar, M.; Chiller, T.M.; Meis, J.F. Potential Fifth Clade of Candida auris, Iran, 2018. Emerg. Infect. Dis. 2019, 25, 1780–1781. [Google Scholar] [CrossRef] [PubMed]
- Timme, R.E.; Strain, E.; Baugher, J.D.; Davis, S.; Gonzalez-Escalona, N.; Leon, M.S.; Allard, M.W.; Brown, E.W.; Tallent, S.; Rand, H. Phylogenomic Pipeline Validation for Foodborne Pathogen Disease Surveillance. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef] [PubMed]
- Timme, R.E.; Crandall, K.; Shumway, M.; Trees, E.K.; Simmons, M.; Agarwala, R.; Davis, S.; Tillman, G.E.; Defibaugh-Chavez, S.; Carleton, H.A.; et al. Benchmark datasets for phylogenomic pipeline validation, applications for foodborne pathogen surveillance. PeerJ 2017, 5, e3893. [Google Scholar] [CrossRef] [PubMed]
- De Groot, T.; Puts, Y.; Berrio, I.; Chowdhary, A.; Meis, J.F. Development of Candida auris Short Tandem Repeat Typing and Its Application to a Global Collection of Isolates. mBio 2020, 11, e02971-19. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.J.; Shin, J.H.; Byun, S.A.; Choi, M.J.; Won, E.J.; Lee, D.; Lee, S.Y.; Chun, S.; Lee, J.H.; Choi, H.J.; et al. Candida auris Clinical Isolates from South Korea: Identification, Antifungal Susceptibility, and Genotyping. J. Clin. Microbiol. 2019, 57, 4. [Google Scholar] [CrossRef] [PubMed]
- Mohsin, J.; Hagen, F.; Al-Balushi, Z.A.M.; De Hoog, G.S.; Chowdhary, A.; Meis, J.F.; Al-Hatmi, A.M.S. The first cases of Candida auris candidaemia in Oman. Mycoses 2017, 60, 569–575. [Google Scholar] [CrossRef] [PubMed]
- NCBI Pathogen Detection System. 2017. Available online: https://www.ncbi.nlm.nih.gov/pathogens/ (accessed on 18 February 2021).
- Ldquo; FastQC. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 18 February 2021).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Argimón, S.; AbuDahab, K.; Goater, R.J.E.; Fedosejev, A.; Bhai, J.; Glasner, C.; Feil, E.J.; Holden, M.T.G.; Yeats, C.A.; Grundmann, H.; et al. Microreact: Visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genom. 2016, 2, e000093. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, G.; Bloch, D.; Murray, K.; Kratz, M.; Parton, H.; Ackelsberg, J.; Antwi, M.; Del Rosso, P.; Dorsinville, M.; Kubinson, H.; et al. Candida auris Colonization After Discharge to a Community Setting: New York City, 2017–2019. Open Forum Infect. Dis. 2021, 8, ofaa620. [Google Scholar] [CrossRef] [PubMed]
- Taylor, T.L.; Volkening, J.D.; DeJesus, E.; Simmons, M.; Dimitrov, K.M.; Tillman, G.E.; Suarez, D.L.; Afonso, C.L. Rapid, multiplexed, whole genome and plasmid sequencing of foodborne pathogens using long-read nanopore technology. Sci. Rep. 2019, 9, 16350. [Google Scholar] [CrossRef] [PubMed]
- McTavish, E.J.; Pettengill, J.; Davis, S.; Rand, H.; Strain, E.; Allard, M.; Timme, R.E. TreeToReads—A pipeline for simulating raw reads from phylogenies. BMC Bioinform. 2017, 18, 178. [Google Scholar] [CrossRef] [PubMed]
| BioSample | Isolate ID | SRA Run | Outbreak ID | Facility ID | Subclade | Date Collected | Study * |
|---|---|---|---|---|---|---|---|
| SAMN10139509 | B12352 | SRR7909282 | 1 | B | Ic | 2016 | 30293877 |
| SAMN10139317 | B12681 | SRR7909214 | 1 | B | Ic | 2017 | 30293877 |
| SAMN10139320 | B12677 | SRR7909135 | 1 | B | Ic | 2017 | 30293877 |
| SAMN10139376 | B12490 | SRR7909210 | 1 | B | Ic | 2016 | 30293877 |
| SAMN10139493 | B12427 | SRR7909184 | 1 | B | Ic | 2016 | 30293877 |
| SAMN10139490 | B12430 | SRR7909233 | 1 | B | Ic | 2016 | 30293877 |
| SAMN10139465 | B12461 | SRR7909234 | 1 | A | Ic | 2016 | 30293877 |
| SAMN10139461 | B12467 | SRR7909297 | 1 | A | Ic | 2016 | 30293877 |
| SAMN10139268 | B12824 | SRR7909313 | 1 | C | Ic | 2017 | 30293877 |
| SAMN10139267 | B12825 | SRR7909385 | 1 | C | Ic | 2017 | 30293877 |
| SAMN10139529 | B12044 | SRR7909147 | 1 | C | Ic | 2016 | 30293877 |
| SAMN10139470 | B12456 | SRR7909156 | 1 | C | Ic | 2016 | 30293877 |
| SAMN10139462 | B12466 | SRR7909413 | 1 | C | Ic | 2016 | 30293877 |
| SAMN10139189 | B13613 | SRR7909284 | 3 | F | Ic | 2017 | 30293877 |
| SAMN10139194 | B13520 | SRR7909394 | 3 | F | Ic | 2017 | 30293877 |
| SAMN10139195 | B13519 | SRR7909408 | 3 | F | Ic | 2017 | 30293877 |
| SAMN10139295 | B12749 | SRR7909166 | 2 | E | Ic | 2017 | 30293877 |
| SAMN10139364 | B12552 | SRR7909192 | 2 | E | Ic | 2017 | 30293877 |
| SAMN10139361 | B12555 | SRR7909183 | 2 | E | Ic | 2017 | 30293877 |
| SAMN10139289 | B12776 | SRR7909308 | 2 | D | Ic | 2017 | 30293877 |
| SAMN10139190 | B13558 | SRR7909246 | 2 | D | Ic | 2017 | 30293877 |
| SAMN10142006 | B13343 | SRR7909249 | NA | G | Ib | 2017 | 30293877 |
| BioSample | Isolate ID | SRA Run | Total Reads (Millions) | Avg. GC Content (%) | Assembly Length | Contigs | N50 |
|---|---|---|---|---|---|---|---|
| SAMN10139509 | B12352 | SRR7909282 | 3.4 | 45% | 12,219,463 | 997 | 20,257 |
| SAMN10139317 | B12681 | SRR7909214 | 6.2 | 47% | 11,914,250 | 1030 | 19,891 |
| SAMN10139320 | B12677 | SRR7909135 | 6.2 | 46% | 12,125,777 | 1006 | 20,610 |
| SAMN10139376 | B12490 | SRR7909210 | 3.6 | 43% | 12,215,651 | 989 | 21,267 |
| SAMN10139493 | B12427 | SRR7909184 | 2.8 | 42% | 12,209,708 | 1030 | 20,335 |
| SAMN10139490 | B12430 | SRR7909233 | 3.2 | 43% | 12,212,626 | 990 | 20,510 |
| SAMN10139465 | B12461 | SRR7909234 | 5.2 | 43% | 12,223,488 | 1033 | 20,401 |
| SAMN10139461 | B12467 | SRR7909297 | 5.4 | 44% | 12,227,902 | 1049 | 20,090 |
| SAMN10139268 | B12824 | SRR7909313 | 5 | 43% | 12,217,316 | 1073 | 19,453 |
| SAMN10139267 | B12825 | SRR7909385 | 6.4 | 44% | 12,235,136 | 1011 | 21,091 |
| SAMN10139529 | B12044 | SRR7909147 | 3.2 | 43% | 12,219,487 | 979 | 21,004 |
| SAMN10139470 | B12456 | SRR7909156 | 4.4 | 43% | 12,218,763 | 1049 | 19,233 |
| SAMN10139462 | B12466 | SRR7909413 | 5 | 43% | 12,223,757 | 1038 | 19,885 |
| SAMN10139189 | B13613 | SRR7909284 | 4.8 | 43% | 12,225,913 | 976 | 21,224 |
| SAMN10139194 | B13520 | SRR7909394 | 4.2 | 43% | 12,215,973 | 1016 | 20,348 |
| SAMN10139195 | B13519 | SRR7909408 | 2.8 | 44% | 12,161,014 | 1039 | 20,040 |
| SAMN10139295 | B12749 | SRR7909166 | 7.6 | 44% | 12,234,239 | 993 | 21,085 |
| SAMN10139364 | B12552 | SRR7909192 | 4.6 | 43% | 12,217,679 | 983 | 21,519 |
| SAMN10139361 | B12555 | SRR7909183 | 3.6 | 43% | 12,215,609 | 982 | 21,427 |
| SAMN10139289 | B12776 | SRR7909308 | 6.8 | 42% | 12,228,293 | 989 | 21,510 |
| SAMN10139190 | B13558 | SRR7909246 | 5.2 | 44% | 12,222,520 | 982 | 21,174 |
| SAMN10142006 | B13343 | SRR7909249 | 4.2 | 41% | 12,213,890 | 997 | 21,547 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Welsh, R.M.; Misas, E.; Forsberg, K.; Lyman, M.; Chow, N.A. Candida auris Whole-Genome Sequence Benchmark Dataset for Phylogenomic Pipelines. J. Fungi 2021, 7, 214. https://doi.org/10.3390/jof7030214
Welsh RM, Misas E, Forsberg K, Lyman M, Chow NA. Candida auris Whole-Genome Sequence Benchmark Dataset for Phylogenomic Pipelines. Journal of Fungi. 2021; 7(3):214. https://doi.org/10.3390/jof7030214
Chicago/Turabian StyleWelsh, Rory M., Elizabeth Misas, Kaitlin Forsberg, Meghan Lyman, and Nancy A. Chow. 2021. "Candida auris Whole-Genome Sequence Benchmark Dataset for Phylogenomic Pipelines" Journal of Fungi 7, no. 3: 214. https://doi.org/10.3390/jof7030214
APA StyleWelsh, R. M., Misas, E., Forsberg, K., Lyman, M., & Chow, N. A. (2021). Candida auris Whole-Genome Sequence Benchmark Dataset for Phylogenomic Pipelines. Journal of Fungi, 7(3), 214. https://doi.org/10.3390/jof7030214






