Transcriptomic Profiling of Populus Roots Challenged with Fusarium Reveals Differential Responsive Patterns of Invertase and Invertase Inhibitor-Like Families within Carbohydrate Metabolism
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Fungal Inoculation
2.2. Fungal Growth, DNA Isolation, and Quantification
2.3. RNA Isolation and Library Preparation
2.4. Transcriptomic Sequencing, Data Mining, and Bioinformatics Analyses
2.5. Evolutionary Analyses and Expression Evaluation by qPCR
2.6. Invertase Extraction and Determination of Invertase Activities In Vitro
3. Results
3.1. Quantification of the Fungal Growth and Colonization in Populus Roots
3.2. Global Changes of Gene Expression in the Host Transcriptome
3.3. Differential Expression and Functional Classification of Fs-Responsive Genes
3.4. Expression Validation of Invertase and Invertase Inhibitor-Like Families and Enzyme Activities
4. Discussion
4.1. Changes in Signaling Transduction Genes
4.2. Expression of Defense Responsive and Transcriptional Regulation-Related Genes
4.3. Genes Involved in Modulation of Secondary and Sugar Metabolism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Volaire, F. A unified framework of plant adaptive strategies to drought: Crossing scales and disciplines. Glob. Chang. Biol. 2018, 24, 2929–2938. [Google Scholar] [CrossRef] [PubMed]
- Popp, J.; Lakner, Z.; Harangi-Rákos, M.; Fári, M. The effect of bioenergy expansion: Food, energy, and environment. Renew. Sustain. Energy Rev. 2014, 32, 559–578. [Google Scholar] [CrossRef]
- Anderson, J.P.; Gleason, C.A.; Foley, R.C.; Thrall, P.H.; Burdon, J.B.; Singh, K.B. Plants versus pathogens: An evolutionary arms race. Funct. Plant Biol. 2010, 37, 499. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef]
- Rodríguez-Pires, S.; Melgarejo, P.; De Cal, A.; Espeso, E.A. Proteomic Studies to Understand the Mechanisms of Peach Tissue Degradation by Monilinia laxa. Front. Plant Sci. 2020, 11, 1286. [Google Scholar] [CrossRef]
- Benedetti, M.; Pontiggia, D.; Raggi, S.; Cheng, Z.; Scaloni, F.; Ferrari, S.; Ausubel, F.M.; Cervone, F.; De Lorenzo, G. Plant immunity triggered by engineered in vivo release of oligogalacturonides, damage-associated molecular patterns. Proc. Natl. Acad. Sci. USA 2015, 112, 5533–5538. [Google Scholar] [CrossRef]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef]
- Ferrari, S.; Savatin, D.V.; Sicilia, F.; Gramegna, G.; Cervone, F.; De Lorenzo, G. Oligogalacturonides: Plant damage-associated molecular patterns and regulators of growth and development. Front. Plant Sci. 2013, 4, 49. [Google Scholar] [CrossRef]
- Medzhitov, R.; Janeway, C.A. Innate immunity: The virtues of a nonclonal system of recognition. Cell 1997, 91, 295–298. [Google Scholar] [CrossRef]
- Nicaise, V.; Roux, M.; Zipfel, C. Recent advances in PAMP-Triggered immunity against bacteria: Pattern recognition receptors watch over and raise the alarm. Plant Physiol. 2009, 150, 1638–1647. [Google Scholar] [CrossRef] [PubMed]
- Jwa, N.-S.; Hwang, B.K. Convergent Evolution of Pathogen Effectors toward Reactive Oxygen Species Signaling Networks in Plants. Front. Plant Sci. 2017, 8, 1687. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Naseem, M.; Kunz, M.; Dandekar, T. Plant–Pathogen Maneuvering over Apoplastic Sugars. Trends Plant Sci. 2017, 22, 740–743. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.; Guivarćh, A.; Hirsche, J.; Bauerfeind, M.A.; González, M.-C.; Hyun, T.K.; Eom, S.H.; Chriqui, D.; Engelke, T.; Großkinsky, D.K.; et al. Metabolic control of tobacco pollination by sugars and invertases. Plant Physiol. 2017, 173, 984–997. [Google Scholar] [CrossRef] [PubMed]
- Tauzin, A.S.; Giardina, T. Sucrose and invertases, a part of the plant defense response to the biotic stresses. Front. Plant Sci. 2014, 5, 293. [Google Scholar] [CrossRef]
- Proels, R.K.; Hückelhoven, R. Cell-wall invertases, key enzymes in the modulation of plant metabolism during defence responses. Mol. Plant Pathol. 2014, 15, 858–864. [Google Scholar] [CrossRef]
- Roitsch, T.; González, M.-C. Function and regulation of plant invertases: Sweet sensations. Trends Plant Sci. 2004, 9, 606–613. [Google Scholar] [CrossRef]
- Liu, S.; Lan, J.; Zhou, B.; Qin, Y.; Zhou, Y.; Xiao, X.; Yang, J.; Gou, J.; Qi, J.; Huang, Y.; et al. HbNIN2, a cytosolic alkaline/neutral-invertase, is responsible for sucrose catabolism in rubber-producing laticifers of Hevea brasiliensis (para rubber tree). New Phytol. 2015, 206, 709–725. [Google Scholar] [CrossRef]
- Barnes, W.J.; Anderson, C.T. Cytosolic invertases contribute to cellulose biosynthesis and influence carbon partitioning in seedlings of Arabidopsis thaliana. Plant J. 2018, 94, 956–974. [Google Scholar] [CrossRef]
- Maruta, T.; Miyazaki, N.; Nosaka, R.; Tanaka, H.; Padilla-Chacon, D.; Otori, K.; Kimura, A.; Tanabe, N.; Yoshimura, K.; Tamoi, M.; et al. A gain-of-function mutation of plastidic invertase alters nuclear gene expression with sucrose treatment partially via GENOMES UNCOUPLED1-mediated signaling. New Phytol. 2015, 206, 1013–1023. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Le Roy, K.; Bolouri-Moghaddam, M.-R.; Vanhaecke, M.; Lammens, W.; Rolland, F.; Van den Ende, W. Exploring the neutral invertase–oxidative stress defence connection in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 3849–3862. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yang, D.L.; Kong, Y.; Chen, Y.; Li, X.Z.; Zeng, L.J.; Li, Q.; Wang, E.T.; He, Z.H. Sugar homeostasis mediated by cell wall invertase GRAIN INCOMPLETE FILLING 1 (GIF1) plays a role in pre-existing and induced defence in rice. Mol. Plant Pathol. 2014, 15, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, S.; Choi, E.; Auh, C.K.; Park, J.B.; Kim, D.G.; Chung, Y.J.; Lee, T.K.; Lee, S. Altered invertase activities of symptomatic tissues on Beet severe curly top virus (BSCTV) infected Arabidopsis thaliana. J. Plant Res. 2013, 126, 743–752. [Google Scholar] [CrossRef]
- Cabello, S.; Lorenz, C.; Crespo, S.; Cabrera, J.; Ludwig, R.; Escobar, C.; Hofmann, J. Altered sucrose synthase and invertase expression affects the local and systemic sugar metabolism of nematode-infected Arabidopsis thaliana plants. J. Exp. Bot. 2014, 65, 201–212. [Google Scholar] [CrossRef]
- Qin, G.; Zhu, Z.; Wang, W.; Cai, J.; Chen, Y.; Li, L.; Tian, S. A Tomato Vacuolar Invertase Inhibitor Mediates Sucrose Metabolism and Influences Fruit Ripening. Plant Physiol. 2016, 172, 1596–1611. [Google Scholar] [CrossRef]
- Xu, X.; Hu, Q.; Yang, W.; Jin, Y. The roles of cell wall invertase inhibitor in regulating chilling tolerance in tomato. BMC Plant Biol. 2017, 17, 195. [Google Scholar] [CrossRef]
- Jin, Y.; Ni, D.-A.; Ruan, Y.-L.; Jin, Y.; Ni, D.A.; Ruan, Y.L. Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. Plant Cell 2009, 21, 2072–2089. [Google Scholar] [CrossRef]
- Tang, X.; Su, T.; Han, M.; Wei, L.; Wang, W.; Yu, Z.; Xue, Y.; Wei, H.; Du, Y.; Greiner, S.; et al. Suppression of extracellular invertase inhibitor gene expression improves seed weight in soybean (Glycine max). J. Exp. Bot. 2017, 68, 469–482. [Google Scholar] [CrossRef]
- Lin, Y.; Liu, T.; Liu, J.; Liu, X.; Ou, Y.; Zhang, H.; Li, M.; Sonnewald, U.; Song, B.; Xie, C. Subtle Regulation of Potato Acid Invertase Activity by a Protein Complex of Invertase, Invertase Inhibitor, and SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE. Plant Physiol. 2015, 168, 1807–1819. [Google Scholar] [CrossRef]
- Veillet, F.; Gaillard, C.; Coutos-Thévenot, P.; La Camera, S. Targeting the AtCWIN1 Gene to Explore the Role of Invertases in Sucrose Transport in Roots and during Botrytis cinerea Infection. Front. Plant Sci. 2016, 7, 1899. [Google Scholar] [CrossRef] [PubMed]
- Bonfig, K.B.; Gabler, A.; Simon, U.K.; Luschin-Ebengreuth, N.; Hatz, M.; Berger, S.; Muhammad, N.; Zeier, J.; Sinha, A.K.; Roitsch, T. Post-translational derepression of invertase activity in source leaves via down-regulation of invertase inhibitor expression is part of the plant defense response. Mol. Plant 2010, 3, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
- Siemens, J.; González, M.C.; Wolf, S.; Hofmann, C.; Greiner, S.; Du, Y.; Rausch, T.; Roitsch, T.; Ludwig-Müller, J. Extracellular invertase is involved in the regulation of clubroot disease in Arabidopsis thaliana. Mol. Plant Pathol. 2011, 12, 247–262. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.-J.; van der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.-J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature. 2010, 464, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.J. The Fusarium solani species complex: Ubiquitous pathogens of agricultural importance. Mol. Plant Pathol. 2016, 17, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Hafizi, R.; Salleh, B.; Latiffah, Z. Morphological and molecular characterization of Fusarium. solani and F. oxysporum associated with crown disease of oil palm. Braz. J. Microbiol. 2013, 44, 959–968. [Google Scholar] [CrossRef]
- Chen, Y.; Yin, H.; Gao, M.; Zhu, H.; Zhang, Q.; Wang, Y. Comparative Transcriptomics Atlases Reveals Different Gene Expression Pattern Related to Fusarium Wilt Disease Resistance and Susceptibility in Two Vernicia Species. Front. Plant Sci. 2016, 7, 1974. [Google Scholar] [CrossRef]
- Ma, L.-J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O’Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium Pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef]
- Su, T.; Han, M.; Min, J.; Zhou, H.; Zhang, Q.; Zhao, J.; Fang, Y. Functional Characterization of Invertase Inhibitors PtC/VIF1 and 2 Revealed Their Involvements in the Defense Response to Fungal Pathogen in Populus trichocarpa. Front. Plant Sci. 2020, 10, 1654. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Bocock, P.N.; Morse, A.M.; Dervinis, C.; Davis, J.M. Evolution and diversity of invertase genes in Populus trichocarpa. Planta 2008, 227, 565–576. [Google Scholar] [CrossRef]
- Mengiste, T. Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 2012, 50, 267–294. [Google Scholar] [CrossRef]
- Bueno, C.J.; Fischer, I.H.; Rosa, D.D.; Firmino, A.C.; Harakava, R.; Oliveira, C.M.G.; Furtado, E.L. Fusarium solani f. sp. passiflorae: A new forma specialis causing collar rot in yellow passion fruit. Plant Pathol. 2014, 63, 382–389. [Google Scholar] [CrossRef]
- Alwahshi, K.J.; Saeed, E.E.; Sham, A.; Alblooshi, A.A.; Alblooshi, M.M.; El-Tarabily, K.A.; Abuqamar, S.F. Molecular identification and disease management of date palm sudden decline syndrome in the united arab emirates. Int. J. Mol. Sci. 2019, 20, 923. [Google Scholar] [CrossRef] [PubMed]
- Cock, J.M.; Vanoosthuyse, V.; Gaude, T. Receptor kinase signalling in plants and animals: Distinct molecular systems with mechanistic similarities. Curr. Opin. Cell Biol. 2002, 14, 230–236. [Google Scholar] [CrossRef]
- Morillo, S.A.; Tax, F.E. Functional analysis of receptor-like kinases in monocots and dicots. Curr. Opin. Plant Biol. 2006, 9, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef]
- Jose, J.; Ghantasala, S.; Roy Choudhury, S. Arabidopsis Transmembrane Receptor-Like Kinases (RLKs): A Bridge between Extracellular Signal and Intracellular Regulatory Machinery. Int. J. Mol. Sci. 2020, 21, 4000. [Google Scholar] [CrossRef] [PubMed]
- Zan, Y.; Ji, Y.; Zhang, Y.; Yang, S.; Song, Y.; Wang, J. Genome-wide identification, characterization and expression analysis of populus leucine-rich repeat receptor-like protein kinase genes. BMC Genom. 2013, 14, 318. [Google Scholar] [CrossRef]
- Yang, Y.; Labbé, J.; Muchero, W.; Yang, X.; Jawdy, S.S.; Kennedy, M.; Johnson, J.; Sreedasyam, A.; Schmutz, J.; Tuskan, G.A.; et al. Genome-wide analysis of lectin receptor-like kinases in Populus. BMC Genom. 2016, 17, 699. [Google Scholar] [CrossRef]
- Liao, H.-L.; Bonito, G.; Rojas, J.A.; Hameed, K.; Wu, S.; Schadt, C.W.; Labbé, J.; Tuskan, G.A.; Martin, F.; Grigoriev, I.V.; et al. Fungal Endophytes of Populus trichocarpa Alter Host Phenotype, Gene Expression, and Rhizobiome Composition. Mol. Plant Microbe Interact. 2019, 32, 853–864. [Google Scholar] [CrossRef]
- Muchero, W.; Sondreli, K.L.; Chen, J.-G.; Urbanowicz, B.R.; Zhang, J.; Singan, V.; Yang, Y.; Brueggeman, R.S.; Franco-Coronado, J.; Abraham, N.; et al. Association mapping, transcriptomics, and transient expression identify candidate genes mediating plant–pathogen interactions in a tree. Proc. Natl. Acad. Sci. USA 2018, 115, 11573–11578. [Google Scholar] [CrossRef]
- Cristina, M.; Petersen, M.; Mundy, J. Mitogen-Activated Protein Kinase Signaling in Plants. Annu. Rev. Plant Biol. 2010, 61, 621–649. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Mewalal, R.; Gunter, L.E.; Palla, K.J.; Carter, K.; Jacobson, D.A.; Jones, P.C.; Garcia, B.J.; Weighill, D.A.; Hyatt, P.D.; et al. Defining the genetic components of callus formation: A GWAS approach. PLoS ONE 2018, 13, e0202519. [Google Scholar] [CrossRef] [PubMed]
- Halford, N.; Hey, S. Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem. J. 2009, 419, 247–259. [Google Scholar] [CrossRef]
- Bedini, A.; Mercy, L.; Schneider, C.; Franken, P.; Lucic-Mercy, E. Unraveling the Initial Plant Hormone Signaling, Metabolic Mechanisms and Plant Defense Triggering the Endomycorrhizal Symbiosis Behavior. Front. Plant Sci. 2018, 9, 1800. [Google Scholar] [CrossRef] [PubMed]
- Di, X.; Takken, F.L.W.; Tintor, N. How Phytohormones Shape Interactions between Plants and the Soil-Borne Fungus Fusarium oxysporum. Front. Plant Sci. 2016, 7, 170. [Google Scholar] [CrossRef] [PubMed]
- Kidd, B.N.; Kadoo, N.Y.; Dombrecht, B.; Tekeoglu, M.; Gardiner, D.M.; Thatcher, L.F.; Aitken, E.A.B.; Schenk, P.M.; Manners, J.M.; Kazan, K. Auxin Signaling and Transport Promote Susceptibility to the Root-Infecting Fungal Pathogen Fusarium oxysporum in Arabidopsis. Mol. Plant Microbe Interact. 2011, 24, 733–748. [Google Scholar] [CrossRef]
- Li, G.; Lin, R.; Egekwu, C.; Blakeslee, J.; Lin, J.; Pettengill, E.; Murphy, A.S.; Peer, W.A.; Islam, N.; Babst, B.A.; et al. Seasonal nitrogen remobilization and the role of auxin transport in poplar trees. J. Exp. Bot. 2020, 71, 4512–4530. [Google Scholar] [CrossRef]
- Berrocal-Lobo, M.; Molina, A.; Solano, R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 2002, 29, 23–32. [Google Scholar] [CrossRef]
- Xia, W.; Yu, H.; Cao, P.; Luo, J.; Wang, N. Identification of TIFY family genes and analysis of their expression profiles in response to phytohormone treatments and melampsora larici-populina infection in poplar. Front. Plant Sci. 2017, 8, 493. [Google Scholar] [CrossRef]
- Sels, J.; Mathys, J.; De Coninck, B.M.A.; Cammue, B.P.A.; De Bolle, M.F.C. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef]
- Su, T.; Han, M.; Cao, D.; Xu, M. Molecular and Biological Properties of Snakins: The Foremost Cysteine-Rich Plant Host Defense Peptides. J. Fungi. 2020, 6, 220. [Google Scholar] [CrossRef]
- Vailleau, F.; Daniel, X.; Tronchet, M.; Montillet, J.-L.; Triantaphylides, C.; Roby, D. A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc. Natl. Acad. Sci. USA 2002, 99, 10179–10184. [Google Scholar] [CrossRef] [PubMed]
- Mehrtens, F.; Kranz, H.; Bednarek, P.; Weisshaar, B. The Arabidopsis Transcription Factor MYB12 Is a Flavonol-Specific Regulator of Phenylpropanoid Biosynthesis. Plant Physiol. 2005, 138, 1083–1096. [Google Scholar] [CrossRef]
- Meng, F.; Yang, C.; Cao, J.; Chen, H.; Pang, J.; Zhao, Q.; Wang, Z.; Qing Fu, Z.; Liu, J. A bHLH transcription activator regulates defense signaling by nucleo-cytosolic trafficking in rice. J. Integr. Plant Biol. 2020, 62, 1552–1573. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Tsolakidou, M.-D.; Pantelides, L.S.; Tzima, A.K.; Kang, S.; Paplomatas, E.J.; Tsaltas, D. Disruption and Overexpression of the Gene Encoding ACC (1-Aminocyclopropane-1-Carboxylic Acid) Deaminase in Soil-Borne Fungal Pathogen Verticillium dahliae Revealed the Role of ACC as a Potential Regulator of Virulence and Plant Defense. Mol. Plant Microbe Interact. 2019, 32, 639–653. [Google Scholar] [CrossRef]
- Dixon, R.A.; Achnine, L.; Kota, P.; Liu, C.-J.; Reddy, M.S.S.; Wang, L. The phenylpropanoid pathway and plant defence-a genomics perspective. Mol. Plant Pathol. 2002, 3, 371–390. [Google Scholar] [CrossRef] [PubMed]
- Baldacci-Cresp, F.; Behr, M.; Kohler, A.; Badalato, N.; Morreel, K.; Goeminne, G.; Mol, A.; de Almeida Engler, J.; Boerjan, W.; El Jaziri, M.; et al. Molecular Changes Concomitant with Vascular System Development in Mature Galls Induced by Root-Knot Nematodes in the Model Tree Host Populus tremula × P. alba. Int. J. Mol. Sci. 2020, 21, 406. [Google Scholar] [CrossRef]
- Pichersky, E.; Raguso, R.A. Why do plants produce so many terpenoid compounds? New Phytol. 2018, 220, 692–702. [Google Scholar] [CrossRef]
- Hou, J.; Wu, Q.; Zuo, T.; Guo, L.; Chang, J.; Chen, J.; Wang, Y.; He, W. Genome-wide transcriptomic profiles reveal multiple regulatory responses of poplar to Lonsdalea quercina infection. Trees 2016, 30, 1389–1402. [Google Scholar] [CrossRef]
- Bezrutczyk, M.; Yang, J.; Eom, J.-S.; Prior, M.; Sosso, D.; Hartwig, T.; Szurek, B.; Oliva, R.; Vera-Cruz, C.; White, F.F.; et al. Sugar flux and signaling in plant-microbe interactions. Plant J. 2018, 93, 675–685. [Google Scholar] [CrossRef]
- Doidy, J.; Grace, E.; Kühn, C.; Simon-Plas, F.; Casieri, L.; Wipf, D. Sugar transporters in plants and in their interactions with fungi. Trends Plant Sci. 2012, 17, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wu, Q.; He, W.; He, T.; Wu, Q.; Miao, Y. Combined De Novo Transcriptome and Metabolome Analysis of Common Bean Response to Fusarium oxysporum f. sp. phaseoli Infection. Int. J. Mol. Sci. 2019, 20, 6278. [Google Scholar] [CrossRef] [PubMed]
- Lunn, D.; Phan, T.D.; Tucker, G.A.; Lycett, G.W. Cell wall composition of tomato fruit changes during development and inhibition of vesicle trafficking is associated with reduced pectin levels and reduced softening. Plant Physiol. Biochem. 2013, 66, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Fatima, U.; Senthil-Kumar, M. Plant and pathogen nutrient acquisition strategies. Front. Plant Sci. 2015, 6, 750. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Q.; Hou, B.-H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Fotopoulos, V.; Gilbert, M.J.; Pittman, J.K.; Marvier, A.C.; Buchanan, A.J.; Sauer, N.; Hall, J.L.; Williams, L.E. The monosaccharide transporter gene, AtSTP4, and the cell-wall invertase, Atbetafruct1, are induced in Arabidopsis during infection with the fungal biotroph Erysiphe cichoracearum. Plant Physiol. 2003, 132, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Santi, S.; De Marco, F.; Polizzotto, R.; Grisan, S.; Musetti, R. Recovery from stolbur disease in grapevine involves changes in sugar transport and metabolism. Front. Plant Sci. 2013, 4, 171. [Google Scholar] [CrossRef] [PubMed]
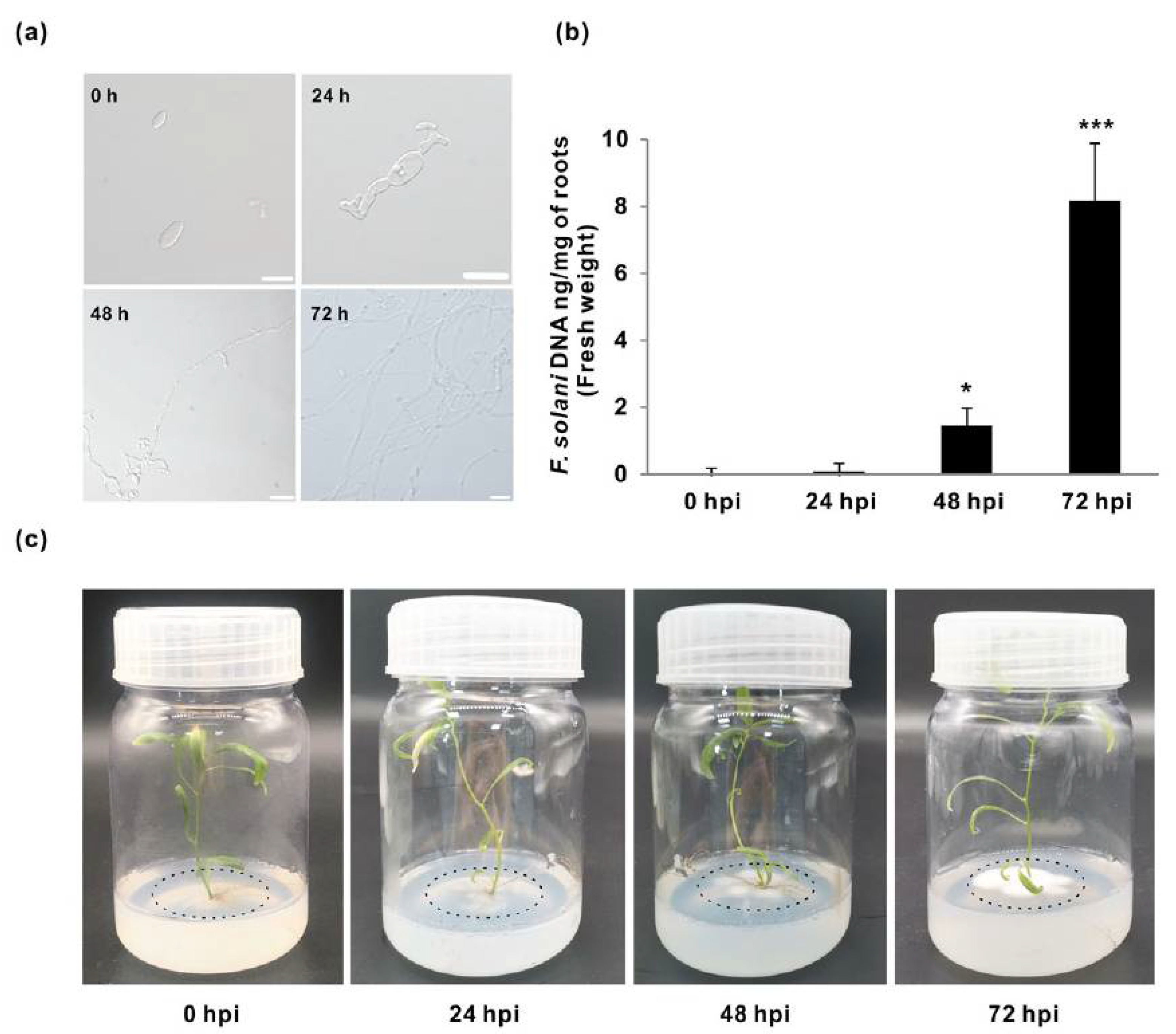

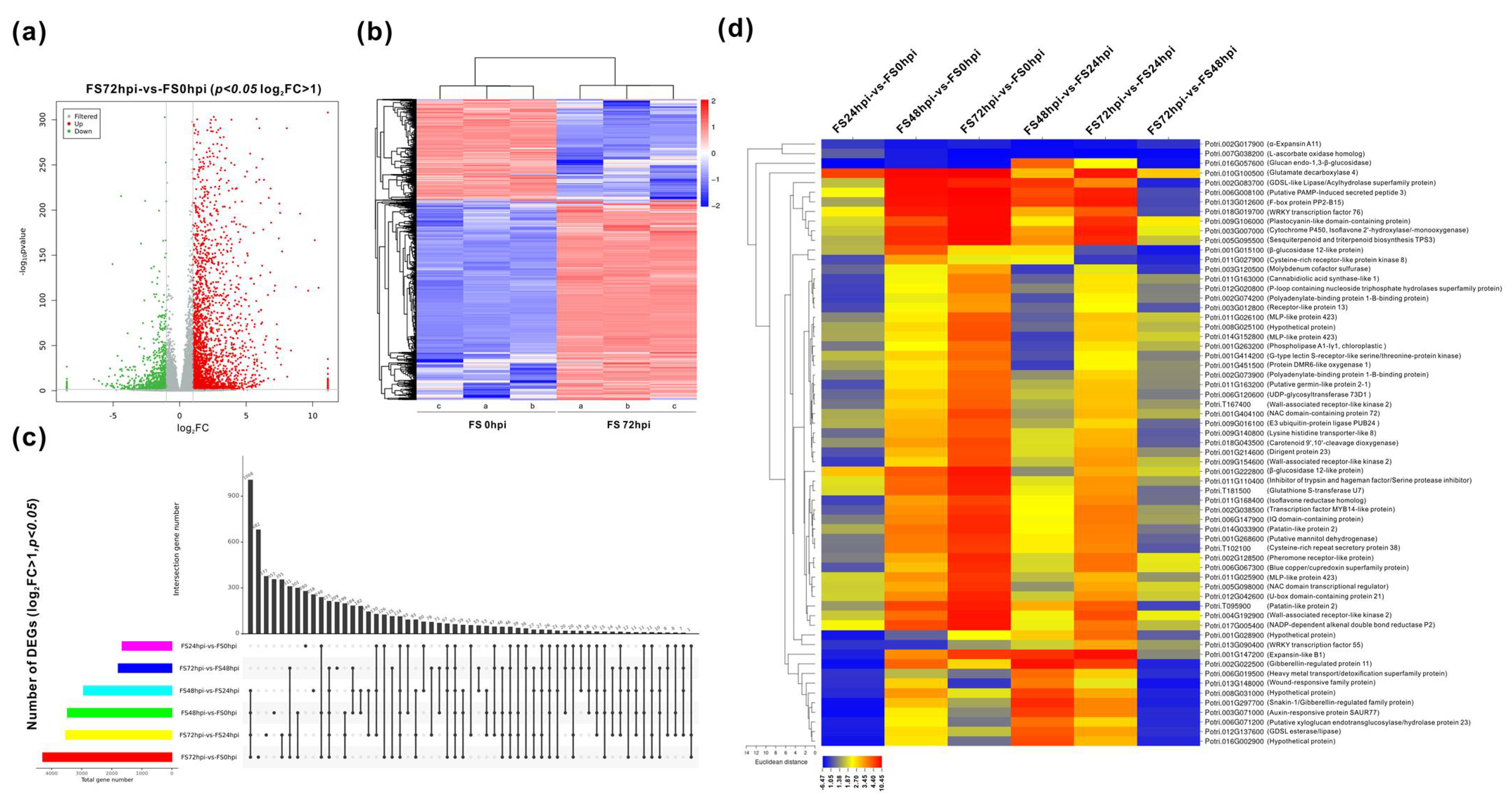
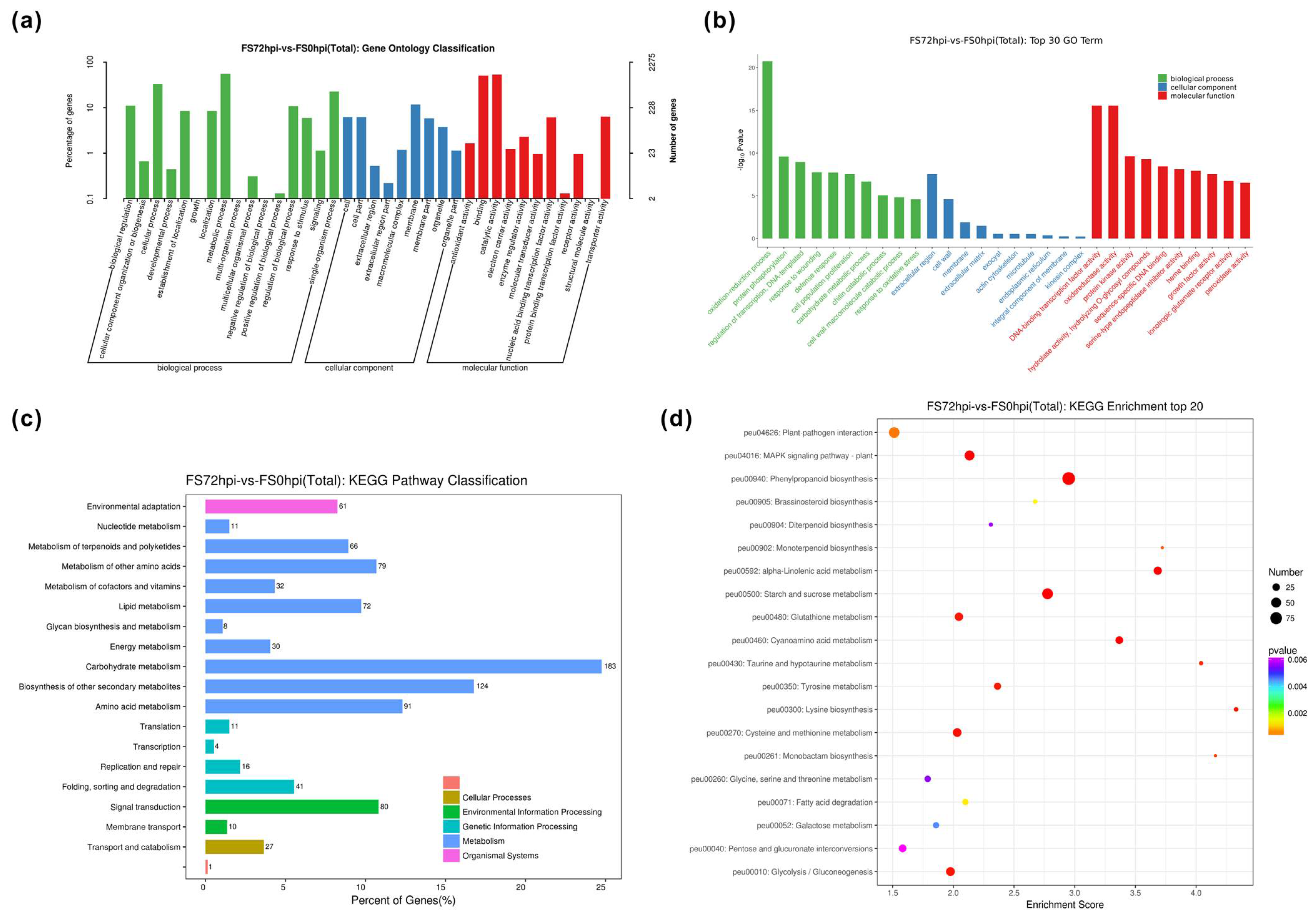

| Sample | Raw Reads | Raw Bases | Clean Reads | Clean Bases | Valid Bases | Q30 1 | GC |
|---|---|---|---|---|---|---|---|
| FS0a | 43.13M | 6.47G | 42.39M | 6.00G | 92.66% | 93.13% | 44.35% |
| FS0b | 45.83M | 6.88G | 45.03M | 6.38G | 92.79% | 92.98% | 44.29% |
| FS0c | 42.16M | 6.32G | 41.42M | 5.86G | 92.58% | 93.10% | 44.64% |
| FS24a | 43.55M | 6.53G | 42.73M | 5.99G | 91.77% | 92.73% | 44.62% |
| FS24b | 49.27M | 7.39G | 48.46M | 6.86G | 92.81% | 93.10% | 44.48% |
| FS24c | 58.47M | 8.77G | 57.49M | 8.15G | 92.91% | 93.19% | 44.46% |
| FS48a | 54.86M | 8.23G | 53.96M | 7.67G | 93.14% | 93.11% | 47.30% |
| FS48b | 49.66M | 7.45G | 48.87M | 6.94G | 93.12% | 93.37% | 47.36% |
| FS48c | 47.25M | 7.09G | 46.51M | 6.59G | 93.00% | 93.45% | 47.66% |
| FS72a | 48.22M | 7.23G | 47.45M | 6.71G | 92.74% | 93.33% | 48.69% |
| FS72b | 53.16M | 7.97G | 52.29M | 7.40G | 92.75% | 93.35% | 48.71% |
| FS72c | 48.26M | 7.24G | 47.45M | 6.71G | 92.72% | 93.20% | 48.62% |
| Groups | Up | Down | Total | Total (%) |
|---|---|---|---|---|
| FS24/FS0 | 979 | 680 | 1659 | 4.01 |
| FS48/FS0 | 2669 | 810 | 3479 | 8.42 |
| FS72/FS0 | 2870 | 1426 | 4296 | 10.39 |
| FS48/FS24 | 2172 | 768 | 2940 | 7.11 |
| FS72/FS24 | 2347 | 1193 | 3540 | 8.56 |
| FS72/FS48 | 739 | 1048 | 1787 | 4.32 |
| Groups | Total | Up | Down | Total | Up | Down |
|---|---|---|---|---|---|---|
| Number of GO Term | Number of DEG | |||||
| FS24/FS0 | 61 | 59 | 47 | 888 | 565 | 323 |
| FS48/FS0 | 79 | 76 | 62 | 1855 | 1447 | 408 |
| FS72/FS0 | 90 | 76 | 65 | 2275 | 1501 | 774 |
| FS48/FS24 | 101 | 98 | 79 | 1551 | 1105 | 446 |
| FS72/FS24 | 91 | 86 | 75 | 1876 | 1184 | 692 |
| FS72/FS48 | 57 | 50 | 51 | 923 | 362 | 561 |
| KEEG Term | ID | FS24/FS0 (All/Up/Down) | FS48/FS0 (All/Up/Down) | FS72/FS0 (All/Up/Down) |
|---|---|---|---|---|
| Phenylpropanoid biosynthesis | path:peu00940 | 27/16/11 | 69/59/10 | 92/75/17 |
| Glycolysis/Gluconeogenesis | path:peu00010 | 4/3/1 | 34/30/4 | 36/33/3 |
| Sugar metabolism 2 | path:peu00520 | 8/4/4 | 36/34/2 | 29/24/5 |
| α-Linolenic acid metabolism | path:peu00592 | 2/0/2 | 27/24/3 | 31/28/3 |
| Cyanoamino acid metabolism | path:peu00460 | 11/6/5 | 23/17/6 | 26/21/5 |
| Starch/sucrose metabolism | path:peu00500 | 20/13/7 | 48/35/13 | 61/40/21 |
| Plant–pathogen interaction | path:peu04626 | 20/13/7 | 53/49/4 | 59/50/9 |
| MAPK signaling pathway | path:peu04016 | 15/12/3 | 36/34/2 | 49/48/1 |
| Hormone signal transduction | path:peu04075 | 19/12/7 | 50/0/50 | 51/40/11 |
| Glutathione metabolism | path:peu00480 | 11/7/4 | 15/15/0 | 32/26/6 |
| Pentose and glucuronate | path:peu00040 | 16/10/6 | 27/24/3 | 26/9/17 |
| Galactose metabolism | path:peu00052 | 7/7/0 | 13/13/0 | 16/11/5 |
| Cys and Met metabolism | path:peu00270 | 8/4/4 | 27/24/3 | 35/24/11 |
| Gly, Ser, and Thr metabolism | path:peu00260 | 3/1/1 | 16/11/5 | 17/10/7 |
| Tyr metabolism | path:peu00350 | 3/0/3 | 21/19/2 | 22/16/6 |
| Carotenoid biosynthesis | path:peu00906 | 6/4/2 | 12/0/12 | 12/8/4 |
| Zeatin biosynthesis | path:peu00908 | 11/5/6 | 11/7/4 | 11/5/6 |
| Carbon fixation | path:peu00710 | 9/0/9 | 14/0/14 | 10/9/1 |
| Brassinosteroid biosynthesis | path:peu00905 | 6/0/6 | 10/7/3 | 9/2/7 |
| Monoterpenoid biosynthesis | path:peu00902 | 3/3/0 | 5/5/0 | 7/7/0 |
| Lys biosynthesis | path:peu00300 | 1/1/0 | 6/6/0 | 9/6/3 |
| Fatty acid degradation | path:peu00071 | n.a | 16/15/1 | 16/16/0 |
| Diterpenoid biosynthesis | path:peu00904 | 5/3/2 | 9/0/9 | 8/5/3 |
| Steroid biosynthesis | path:peu00100 | 4/3/1 | 5/2/3 | 4/0/4 |
| Taurine and hypotaurine | path:peu00430 | 1/1/0 | 6/6/0 | 8/8/0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, T.; Zhou, B.; Cao, D.; Pan, Y.; Hu, M.; Zhang, M.; Wei, H.; Han, M. Transcriptomic Profiling of Populus Roots Challenged with Fusarium Reveals Differential Responsive Patterns of Invertase and Invertase Inhibitor-Like Families within Carbohydrate Metabolism. J. Fungi 2021, 7, 89. https://doi.org/10.3390/jof7020089
Su T, Zhou B, Cao D, Pan Y, Hu M, Zhang M, Wei H, Han M. Transcriptomic Profiling of Populus Roots Challenged with Fusarium Reveals Differential Responsive Patterns of Invertase and Invertase Inhibitor-Like Families within Carbohydrate Metabolism. Journal of Fungi. 2021; 7(2):89. https://doi.org/10.3390/jof7020089
Chicago/Turabian StyleSu, Tao, Biyao Zhou, Dan Cao, Yuting Pan, Mei Hu, Mengru Zhang, Haikun Wei, and Mei Han. 2021. "Transcriptomic Profiling of Populus Roots Challenged with Fusarium Reveals Differential Responsive Patterns of Invertase and Invertase Inhibitor-Like Families within Carbohydrate Metabolism" Journal of Fungi 7, no. 2: 89. https://doi.org/10.3390/jof7020089
APA StyleSu, T., Zhou, B., Cao, D., Pan, Y., Hu, M., Zhang, M., Wei, H., & Han, M. (2021). Transcriptomic Profiling of Populus Roots Challenged with Fusarium Reveals Differential Responsive Patterns of Invertase and Invertase Inhibitor-Like Families within Carbohydrate Metabolism. Journal of Fungi, 7(2), 89. https://doi.org/10.3390/jof7020089







