Zinc at the Host–Fungus Interface: How to Uptake the Metal?
Abstract
1. Introduction
2. Current Knowledge on Zinc Homeostasis in Pathogenic Fungi
3. Zinc-Dependent Zinc Uptake
3.1. Saccharomyces cerevisiae
3.2. Cryptococcus neoformans and Cryptococcus gattii
4. Zinc- and pH-Dependent Zinc Uptake
4.1. Aspergillus fumigatus
4.2. Candida albicans and Candida dubliniensis
5. Zinc Uptake Not Related to Depend on pH Regulation Thus Far
Blastomyces dermatitidis, Histoplasma capsulatum, and Trichophyton mentagrophytes
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Andreini, C.; Bertini, I.; Cavallaro, G.; Holliday, G.L.; Thornton, J.M. Metal ions in biological catalysis: From enzyme databases to general principles. J. Biol. Inorg. Chem. 2008, 13, 1205–1218. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.E. Zinc proteins: Enzymes, storage proteins, transcription factors, and replication proteins. Annu. Rev. Biochem. 1992, 61, 897–946. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.; Klug, A. Zinc fingers. Sci Am. 1993, 268, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.; Learte-Aymami, S.; Mosquera, J.; Celaya, G.; Rodriguez-Larrea, D.; Vazquez, M.E.; Mascarenas, J.L. DNA-binding miniproteins based on zinc fingers. Assessment of the interaction using nanopores. Chem. Sci. 2018, 9, 4118–4123. [Google Scholar] [CrossRef]
- Oteiza, P.I. Zinc and the modulation of redox homeostasis. Free Radic. Biol. Med. 2012, 53, 1748–1759. [Google Scholar] [CrossRef]
- Staats, C.C.; Kmetzsch, L.; Schrank, A.; Vainstein, M.H. Fungal zinc metabolism and its connections to virulence. Front. Cell Infect. Microbio.l 2013, 3, 65. [Google Scholar] [CrossRef]
- Clark, H.L.; Jhingran, A.; Sun, Y.; Vareechon, C.; de Jesus Carrion, S.; Skaar, E.P.; Chazin, W.J.; Calera, J.A.; Hohl, T.M.; Pearlman, E. Zinc and Manganese Chelation by Neutrophil S100A8/A9 (Calprotectin) Limits Extracellular Aspergillus fumigatus Hyphal Growth and Corneal Infection. J. Immunol. 2016, 196, 336–344. [Google Scholar] [CrossRef]
- Subramanian Vignesh, K.; Landero Figueroa, J.A.; Porollo, A.; Caruso, J.A.; Deepe, G.S., Jr. Granulocyte macrophage-colony stimulating factor induced Zn sequestration enhances macrophage superoxide and limits intracellular pathogen survival. Immunity 2013, 39, 697–710. [Google Scholar] [CrossRef]
- Zhao, H.; Eide, D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. USA 1996, 93, 2454–2458. [Google Scholar] [CrossRef]
- Amich, J.; Leal, F.; Calera, J.A. Repression of the acid ZrfA/ZrfB zinc-uptake system of Aspergillus fumigatus mediated by PacC under neutral, zinc-limiting conditions. Int. Microbiol. 2009, 12, 39–47. [Google Scholar]
- Schneider, R.O.; Diehl, C.; dos Santos, F.M.; Piffer, A.C.; Garcia, A.W.; Kulmann, M.I.; Schrank, A.; Kmetzsch, L.; Vainstein, M.H.; Staats, C.C. Effects of zinc transporters on Cryptococcus gattii virulence. Sci. Rep. 2015, 5, 10104. [Google Scholar] [CrossRef] [PubMed]
- Kurakado, S.; Arai, R.; Sugita, T. Association of the Hypha-related Protein Pra1 and Zinc Transporter Zrt1 with Biofilm Formation by the Pathogenic Yeast Candida albicans. Microbiol. Immunol. 2018, 62, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Lehtovirta-Morley, L.E.; Alsarraf, M.; Wilson, D. Pan-Domain Analysis of ZIP Zinc Transporters. Int. J. Mol. Sci. 2017, 18, 2631. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Hu, Y.M.; Corkins, M.E.; Palmer, A.E.; Bird, A.J. Zinc transporters belonging to the Cation Diffusion Facilitator (CDF) family have complementary roles in transporting zinc out of the cytosol. PLoS Genet. 2018, 14, e1007262. [Google Scholar] [CrossRef]
- MacDiarmid, C.W.; Gaither, L.A.; Eide, D. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 2000, 19, 2845–2855. [Google Scholar] [CrossRef]
- Eide, D.J. Zinc transporters and the cellular trafficking of zinc. Biochim. Biophys. Acta 2006, 1763, 711–722. [Google Scholar] [CrossRef]
- Zhao, H.; Butler, E.; Rodgers, J.; Spizzo, T.; Duesterhoeft, S.; Eide, D. Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J. Biol. Chem. 1998, 273, 28713–28720. [Google Scholar] [CrossRef]
- Böttcher, B.; Palige, K.; Jacobsen, I.D.; Hube, B.; Brunke, S. Csr1/Zap1 Maintains Zinc Homeostasis and Influences Virulence in Candida dubliniensis but Is Not Coupled to Morphogenesis. Eukaryot. Cell 2015, 14, 661–670. [Google Scholar] [CrossRef]
- Vicentefranqueira, R.; Amich, J.; Marin, L.; Sanchez, C.I.; Leal, F.; Calera, J.A. The Transcription Factor ZafA Regulates the Homeostatic and Adaptive Response to Zinc Starvation in Aspergillus fumigatus. Genes (Basel) 2018, 9, 318. [Google Scholar] [CrossRef]
- Köhler, J.R.; Casadevall, A.; Perfect, J. The spectrum of fungi that infects humans. Cold Spring Harb. Perspect. Med. 2014, 5, a019273. [Google Scholar] [CrossRef]
- Taylor, L.H.; Latham, S.M.; Woolhouse, M.E. Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 983–989. [Google Scholar] [CrossRef] [PubMed]
- de Curcio, J.S.; Silva, M.G.; Silva Bailao, M.G.; Bao, S.N.; Casaletti, L.; Bailao, A.M.; de Almeida Soares, C.M. Identification of membrane proteome of Paracoccidioides lutzii and its regulation by zinc. Future Sci. OA 2017, 3, FSO232. [Google Scholar] [CrossRef] [PubMed]
- Parente, A.F.; de Rezende, T.C.; de Castro, K.P.; Bailao, A.M.; Parente, J.A.; Borges, C.L.; Silva, L.P.; Soares, C.M. A proteomic view of the response of Paracoccidioides yeast cells to zinc deprivation. Fungal Biol. 2013, 117, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Tristao, G.B.; Assuncao Ldo, P.; Dos Santos, L.P.; Borges, C.L.; Silva-Bailao, M.G.; Soares, C.M.; Cavallaro, G.; Bailao, A.M. Predicting copper-, iron-, and zinc-binding proteins in pathogenic species of the Paracoccidioides genus. Front. Microbiol. 2014, 5, 761. [Google Scholar] [CrossRef] [PubMed]
- Munoz, J.F.; Gauthier, G.M.; Desjardins, C.A.; Gallo, J.E.; Holder, J.; Sullivan, T.D.; Marty, A.J.; Carmen, J.C.; Chen, Z.; Ding, L.; et al. The Dynamic Genome and Transcriptome of the Human Fungal Pathogen Blastomyces and Close Relative Emmonsia. PLoS Genet. 2015, 11, e1005493. [Google Scholar] [CrossRef] [PubMed]
- Gerwien, F.; Skrahina, V.; Kasper, L.; Hube, B.; Brunke, S. Metals in fungal virulence. FEMS Microbiol. Rev. 2018, 42. [Google Scholar] [CrossRef]
- Kamizono, A.; Nishizawa, M.; Teranishi, Y.; Murata, K.; Kimura, A. Identification of a gene conferring resistance to zinc and cadmium ions in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 1989, 219, 161–167. [Google Scholar] [CrossRef]
- Teblick, A.; Jansens, H.; Dams, K.; Somville, F.J.; Jorens, P.G. Boerhaave’s syndrome complicated by a Saccharomyces cerevisiae pleural empyema. Case report and review of the literature. Acta Clin. Belg. 2017, 73, 377–381. [Google Scholar] [CrossRef]
- Ipson, M.A.; Blanco, C.L. Saccharomyces cerevisiae sepsis in a 35-week-old premature infant. A case report. J. Perinatol. 2001, 21, 459–460. [Google Scholar] [CrossRef][Green Version]
- Henry, S.; D’Hondt, L.; Andre, M.; Holemans, X.; Canon, J.L. Saccharomyces cerevisiae fungemia in a head and neck cancer patient: A case report and review of the literature. Acta Clin. Belg. 2004, 59, 220–222. [Google Scholar] [CrossRef]
- Miyabe, S.; Izawa, S.; Inoue, Y. The Zrc1 is involved in zinc transport system between vacuole and cytosol in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2001, 282, 79–83. [Google Scholar] [CrossRef] [PubMed]
- MacDiarmid, C.W.; Milanick, M.A.; Eide, D.J. Biochemical properties of vacuolar zinc transport systems of Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 39187–39194. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Eide, D.J. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997, 17, 5044–5052. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Bird, A.J.; Chung, L.M.; Newton, M.A.; Winge, D.R.; Eide, D.J. Differential control of Zap1-regulated genes in response to zinc deficiency in Saccharomyces cerevisiae. BMC Genomics 2008, 9, 370. [Google Scholar] [CrossRef]
- Evans-Galea, M.V.; Blankman, E.; Myszka, D.G.; Bird, A.J.; Eide, D.J.; Winge, D.R. Two of the five zinc fingers in the Zap1 transcription factor DNA binding domain dominate site-specific DNA binding. Biochemistry 2003, 42, 1053–1061. [Google Scholar] [CrossRef]
- Bird, A.; Evans-Galea, M.V.; Blankman, E.; Zhao, H.; Luo, H.; Winge, D.R.; Eide, D.J. Mapping the DNA binding domain of the Zap1 zinc-responsive transcriptional activator. J. Biol. Chem. 2000, 275, 16160–16166. [Google Scholar] [CrossRef]
- Bird, A.J.; McCall, K.; Kramer, M.; Blankman, E.; Winge, D.R.; Eide, D.J. Zinc fingers can act as Zn2+ sensors to regulate transcriptional activation domain function. EMBO J. 2003, 22, 5137–5146. [Google Scholar] [CrossRef]
- Herbig, A.; Bird, A.J.; Swierczek, S.; McCall, K.; Mooney, M.; Wu, C.Y.; Winge, D.R.; Eide, D.J. Zap1 activation domain 1 and its role in controlling gene expression in response to cellular zinc status. Mol. Microbiol. 2005, 57, 834–846. [Google Scholar] [CrossRef]
- Frey, A.G.; Eide, D.J. Roles of two activation domains in Zap1 in the response to zinc deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 2011, 286, 6844–6854. [Google Scholar] [CrossRef]
- Frey, A.G.; Eide, D.J. Zinc-responsive coactivator recruitment by the yeast Zap1 transcription factor. Microbiologyopen 2012, 1, 105–114. [Google Scholar] [CrossRef]
- Bird, A.J.; Blankman, E.; Stillman, D.J.; Eide, D.J.; Winge, D.R. The Zap1 transcriptional activator also acts as a repressor by binding downstream of the TATA box in ZRT2. EMBO J. 2004, 23, 1123–1132. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Eide, D. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 23203–23210. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.H.; Frey, A.G.; Eide, D.J. Transcriptional regulation of the Zrg17 zinc transporter of the yeast secretory pathway. Biochem J. 2011, 435, 259–266. [Google Scholar] [CrossRef] [PubMed]
- MacDiarmid, C.W.; Milanick, M.A.; Eide, D.J. Induction of the ZRC1 metal tolerance gene in zinc-limited yeast confers resistance to zinc shock. J. Biol. Chem. 2003, 278, 15065–15072. [Google Scholar] [CrossRef] [PubMed]
- Kumanovics, A.; Poruk, K.E.; Osborn, K.A.; Ward, D.M.; Kaplan, J. YKE4 (YIL023C) encodes a bidirectional zinc transporter in the endoplasmic reticulum of Saccharomyces cerevisiae. J. Biol. Chem. 2006, 281, 22566–22574. [Google Scholar] [CrossRef]
- Miyabe, S.; Izawa, S.; Inoue, Y. Expression of ZRC1 coding for suppressor of zinc toxicity is induced by zinc-starvation stress in Zap1-dependent fashion in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2000, 276, 879–884. [Google Scholar] [CrossRef]
- Ellis, C.D.; Wang, F.; MacDiarmid, C.W.; Clark, S.; Lyons, T.; Eide, D.J. Zinc and the Msc2 zinc transporter protein are required for endoplasmic reticulum function. J. Cell. Biol. 2004, 166, 325–335. [Google Scholar] [CrossRef]
- Lyons, T.J.; Gasch, A.P.; Gaither, L.A.; Botstein, D.; Brown, P.O.; Eide, D.J. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl. Acad. Sci. USA 2000, 97, 7957–7962. [Google Scholar] [CrossRef]
- Wu, Y.H.; Taggart, J.; Song, P.X.; MacDiarmid, C.; Eide, D.J. An MSC2 Promoter-lacZ Fusion Gene Reveals Zinc-Responsive Changes in Sites of Transcription Initiation That Occur across the Yeast Genome. PLoS ONE 2016, 11, e0163256. [Google Scholar] [CrossRef]
- Frey, A.G.; Bird, A.J.; Evans-Galea, M.V.; Blankman, E.; Winge, D.R.; Eide, D.J. Zinc-regulated DNA binding of the yeast Zap1 zinc-responsive activator. PLoS ONE 2011, 6, e22535. [Google Scholar] [CrossRef]
- Gitan, R.S.; Shababi, M.; Kramer, M.; Eide, D.J. A cytosolic domain of the yeast Zrt1 zinc transporter is required for its post-translational inactivation in response to zinc and cadmium. J. Biol. Chem. 2003, 278, 39558–39564. [Google Scholar] [CrossRef] [PubMed]
- MacDougall, L.; Kidd, S.E.; Galanis, E.; Mak, S.; Leslie, M.J.; Cieslak, P.R.; Kronstad, J.W.; Morshed, M.G.; Bartlett, K.H. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, USA. Emerg. Infect. Dis. 2007, 13, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Bielska, E.; May, R.C. What makes Cryptococcus gattii a pathogen? FEMS Yeast Res. 2016, 16, fov106. [Google Scholar] [CrossRef] [PubMed]
- Gullo, F.P.; Rossi, S.A.; Sardi Jde, C.; Teodoro, V.L.; Mendes-Giannini, M.J.; Fusco-Almeida, A.M. Cryptococcosis: Epidemiology, fungal resistance, and new alternatives for treatment. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1377–1391. [Google Scholar] [CrossRef]
- Schneider, R.O.; Fogaca Nde, S.; Kmetzsch, L.; Schrank, A.; Vainstein, M.H.; Staats, C.C. Zap1 regulates zinc homeostasis and modulates virulence in Cryptococcus gattii. PLoS ONE 2012, 7, e43773. [Google Scholar] [CrossRef]
- Do, E.; Hu, G.; Caza, M.; Kronstad, J.W.; Jung, W.H. The ZIP family zinc transporters support the virulence of Cryptococcus neoformans. Med. Mycol. 2016, 54, 605–615. [Google Scholar] [CrossRef]
- Cho, M.; Hu, G.; Caza, M.; Horianopoulos, L.C.; Kronstad, J.W.; Jung, W.H. Vacuolar zinc transporter Zrc1 is required for detoxification of excess intracellular zinc in the human fungal pathogen Cryptococcus neoformans. J. Microbiol. 2018, 56, 65–71. [Google Scholar] [CrossRef]
- Garcia, A.W.A.; Kinskovski, U.P.; Diehl, C.; Reuwsaat, J.C.V.; Souza, H.M.; Pinto, H.B.; Trentin, D.S.; Oliveira, H.C.; Rodrigues, M.L.; Becker, E.M.; et al. Participation of Zip3, a ZIP domain-containing protein, in stress response and virulence in Cryptococcus gattii. Fungal Genet. Biol. 2020, 144. [Google Scholar] [CrossRef]
- Dos Santos, F.M.; Piffer, A.C.; Schneider, R.O.; Ribeiro, N.S.; Garcia, A.W.A.; Schrank, A.; Kmetzsch, L.; Vainstein, M.H.; Staats, C.C. Alterations of zinc homeostasis in response to Cryptococcus neoformans in a murine macrophage cell line. Future Microbiol. 2017, 12, 491–504. [Google Scholar] [CrossRef]
- Vuong, M.F.; Waymack, J.R. Aspergillosis. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Martínez, C.E.; Motto, H.L. Solubility of lead, zinc and copper added to mineral soils. Environ. Pollut. 2000, 107, 153–158. [Google Scholar] [CrossRef]
- Tapiero, H.; Tew, K.D. Trace elements in human physiology and pathology: Zinc and metallothioneins. Biomed. Pharmacother. 2003, 57, 399–411. [Google Scholar] [CrossRef]
- Moreno, M.A.; Ibrahim-Granet, O.; Vicentefranqueira, R.; Amich, J.; Ave, P.; Leal, F.; Latge, J.P.; Calera, J.A. The regulation of zinc homeostasis by the ZafA transcriptional activator is essential for Aspergillus fumigatus virulence. Mol. Microbiol. 2007, 64, 1182–1197. [Google Scholar] [CrossRef] [PubMed]
- Vicentefranqueira, R.; Moreno, M.A.; Leal, F.; Calera, J.A. The zrfA and zrfB genes of Aspergillus fumigatus encode the zinc transporter proteins of a zinc uptake system induced in an acid, zinc-depleted environment. Eukaryot. Cell. 2005, 4, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Amich, J.; Vicentefranqueira, R.; Mellado, E.; Ruiz-Carmuega, A.; Leal, F.; Calera, J.A. The ZrfC alkaline zinc transporter is required for Aspergillus fumigatus virulence and its growth in the presence of the Zn/Mn-chelating protein calprotectin. Cell. Microbiol. 2014, 16, 548–564. [Google Scholar] [CrossRef]
- Amich, J.; Vicentefranqueira, R.; Leal, F.; Calera, J.A. Aspergillus fumigatus survival in alkaline and extreme zinc-limiting environments relies on the induction of a zinc homeostasis system encoded by the zrfC and aspf2 genes. Eukaryot. Cell. 2010, 9, 424–437. [Google Scholar] [CrossRef]
- Amich, J.; Calera, J.A. Zinc acquisition: A key aspect in Aspergillus fumigatus virulence. Mycopathologia 2014, 178, 379–385. [Google Scholar] [CrossRef]
- Tilburn, J.; Sarkar, S.; Widdick, D.A.; Espeso, E.A.; Orejas, M.; Mungroo, J.; Penalva, M.A.; Arst, H.N., Jr. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995, 14, 779–790. [Google Scholar] [CrossRef]
- Wheeler, K.A.; Hurdman, B.F.; Pitt, J.I. Influence of pH on the growth of some toxigenic species of Aspergillus, Penicillium and Fusarium. Int. J. Food Microbiol. 1991, 12, 141–149. [Google Scholar] [CrossRef]
- Narendranath, N.V.; Power, R. Relationship between pH and medium dissolved solids in terms of growth and metabolism of Lactobacilli and Saccharomyces cerevisiae during ethanol production. Appl. Environ. Microbiol. 2005, 71, 2239–2243. [Google Scholar] [CrossRef]
- Lamb, T.M.; Mitchell, A.P. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2003, 23, 677–686. [Google Scholar] [CrossRef]
- Lamb, T.M.; Xu, W.; Diamond, A.; Mitchell, A.P. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 2001, 276, 1850–1856. [Google Scholar] [CrossRef] [PubMed]
- Serrano, R.; Ruiz, A.; Bernal, D.; Chambers, J.R.; Ariño, J. The transcriptional response to alkaline pH in Saccharomyces cerevisiae: Evidence for calcium-mediated signalling. Mol. Microbiol. 2002, 46, 1319–1333. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, B.; Greenberger, P.A.; Fink, J.N.; Kurup, V.P. Immunological characterization of Aspf2, a major allergen from Aspergillus fumigatus associated with allergic bronchopulmonary aspergillosis. Infect. Immun. 1998, 66, 5175–5182. [Google Scholar] [CrossRef] [PubMed]
- Segurado, M.; López-Aragón, R.; Calera, J.A.; Fernández-Abalos, J.M.; Leal, F. Zinc-regulated biosynthesis of immunodominant antigens from Aspergillus spp. Infect. Immun. 1999, 67, 2377–2382. [Google Scholar] [CrossRef]
- Vicentefranqueira, R.; Leal, F.; Marin, L.; Sanchez, C.I.; Calera, J.A. The interplay between zinc and iron homeostasis in Aspergillus fumigatus under zinc-replete conditions relies on the iron-mediated regulation of alternative transcription units of zafA and the basal amount of the ZafA zinc-responsiveness transcription factor. Environ. Microbiol. 2019, 21, 2787–2808. [Google Scholar] [CrossRef]
- Cai, Z.; Du, W.; Zhang, Z.; Guan, L.; Zeng, Q.; Chai, Y.; Dai, C.; Lu, L. The Aspergillus fumigatus transcription factor AceA is involved not only in Cu but also in Zn detoxification through regulating transporters CrpA and ZrcA. Cell Microbiol 2018, 20, e12864. [Google Scholar] [CrossRef]
- Guinea, J. Global trends in the distribution of Candida species causing candidemia. Clin. Microbiol. Infect. 2014, 20, 5–10. [Google Scholar] [CrossRef]
- Vila, T.; Sultan, A.S.; Montelongo-Jauregui, D.; Jabra-Rizk, M.A. Oral Candidiasis: A Disease of Opportunity. J. Fungi (Basel) 2020, 6, 15. [Google Scholar] [CrossRef]
- Moran, G.P.; Coleman, D.C.; Sullivan, D.J. Candida albicans versus Candida dubliniensis: Why Is C. albicans More Pathogenic? Int J. Microbiol. 2012, 2012, 205921. [Google Scholar] [CrossRef]
- Sullivan, D.J.; Moran, G.P.; Pinjon, E.; Al-Mosaid, A.; Stokes, C.; Vaughan, C.; Coleman, D.C. Comparison of the epidemiology, drug resistance mechanisms, and virulence of Candida dubliniensis and Candida albicans. FEMS Yeast Res. 2004, 4, 369–376. [Google Scholar] [CrossRef]
- Kim, M.J.; Kil, M.; Jung, J.H.; Kim, J. Roles of zinc-responsive transcription factor Csr1 in filamentous growth of the pathogenic yeast Candida albicans. J. Microbiol. Biotechnol. 2008, 18, 242–247. [Google Scholar] [PubMed]
- Ness, F.; Bourot, S.; Regnacq, M.; Spagnoli, R.; Berges, T.; Karst, F. SUT1 is a putative Zn[II]2Cys6-transcription factor whose upregulation enhances both sterol uptake and synthesis in aerobically growing Saccharomyces cerevisiae cells. Eur J. Biochem. 2001, 268, 1585–1595. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Solis, N.V.; Ehrlich, R.L.; Woolford, C.A.; Filler, S.G.; Mitchell, A.P. Activation and alliance of regulatory pathways in Candida albicans during mammalian infection. PLoS Biol. 2015, 13, e1002076. [Google Scholar] [CrossRef] [PubMed]
- Loboda, D.; Rowinska-Zyrek, M. Zinc binding sites in Pra1, a zincophore from Candida albicans. Dalton Trans. 2017, 46, 13695–13703. [Google Scholar] [CrossRef]
- Citiulo, F.; Jacobsen, I.D.; Miramón, P.; Schild, L.; Brunke, S.; Zipfel, P.; Brock, M.; Hube, B.; Wilson, D. Candida albicans Scavenges Host Zinc via Pra1 during Endothelial Invasion. PLOS Pathogens 2012, 8, e1002777. [Google Scholar] [CrossRef]
- Nobile, C.J.; Nett, J.E.; Hernday, A.D.; Homann, O.R.; Deneault, J.S.; Nantel, A.; Andes, D.R.; Johnson, A.D.; Mitchell, A.P. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 2009, 7, e1000133. [Google Scholar] [CrossRef]
- Bensen, E.S.; Martin, S.J.; Li, M.; Berman, J.; Davis, D.A. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol Microbiol 2004, 54, 1335–1351. [Google Scholar] [CrossRef]
- Ramon, A.M.; Fonzi, W.A. Diverged binding specificity of Rim101p, the Candida albicans ortholog of PacC. Eukaryot. Cell. 2003, 2, 718–728. [Google Scholar] [CrossRef]
- Davis, D.; Edwards, J.E., Jr.; Mitchell, A.P.; Ibrahim, A.S. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 2000, 68, 5953–5959. [Google Scholar] [CrossRef]
- Ramon, A.M.; Porta, A.; Fonzi, W.A. Effect of environmental pH on morphological development of Candida albicans is mediated via the PacC-related transcription factor encoded by PRR2. J. Bacteriol. 1999, 181, 7524–7530. [Google Scholar] [CrossRef]
- Crawford, C.; Lehtovirta-Morley, L.E.; Alamir, O.; Niemiec, M.J.; Alawfi, B.; Alsarraf, M.; Skrahina, V.; Costa, A.C.B.P.; Anderson, A.; Yellagunda, S.; et al. Biphasic zinc compartmentalisation in a human fungal pathogen. PLoS Pathog. 2018, 14, e1007013. [Google Scholar] [CrossRef]
- Garnaud, C.; Garcia-Oliver, E.; Wang, Y.; Maubon, D.; Bailly, S.; Despinasse, Q.; Champleboux, M.; Govin, J.; Cornet, M. The Rim Pathway Mediates Antifungal Tolerance in Candida albicans through Newly Identified Rim101 Transcriptional Targets, Including Hsp90 and Ipt1. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef] [PubMed]
- Finkel, J.S.; Xu, W.; Huang, D.; Hill, E.M.; Desai, J.V.; Woolford, C.A.; Nett, J.E.; Taff, H.; Norice, C.T.; Andes, D.R.; et al. Portrait of Candida albicans adherence regulators. PLoS Pathog. 2012, 8, e1002525. [Google Scholar] [CrossRef] [PubMed]
- Liuzzi, J.P.; Lichten, L.A.; Rivera, S.; Blanchard, R.K.; Aydemir, T.B.; Knutson, M.D.; Ganz, T.; Cousins, R.J. Interleukin-6 regulates the zinc transporter Zip14 in liver and contributes to the hypozincemia of the acute-phase response. Proc. Natl. Acad. Sci. USA 2005, 102, 6843–6848. [Google Scholar] [CrossRef] [PubMed]
- Fanning, S.; Xu, W.; Solis, N.; Woolford, C.A.; Filler, S.G.; Mitchell, A.P. Divergent targets of Candida albicans biofilm regulator Bcr1 in vitro and in vivo. Eukaryot Cell 2012, 11, 896–904. [Google Scholar] [CrossRef]
- Kumar, R.; Breindel, C.; Saraswat, D.; Cullen, P.J.; Edgerton, M. Candida albicans Sap6 amyloid regions function in cellular aggregation and zinc binding, and contribute to zinc acquisition. Sci. Rep. 2017, 7, 2908. [Google Scholar] [CrossRef]
- Dade, J.; DuBois, J.C.; Pasula, R.; Donnell, A.M.; Caruso, J.A.; Smulian, A.G.; Deepe, G.S., Jr. HcZrt2, a zinc responsive gene, is indispensable for the survival of Histoplasma capsulatum in vivo. Med. Mycol. 2016, 54, 865–875. [Google Scholar] [CrossRef]
- Kujoth, G.C.; Sullivan, T.D.; Merkhofer, R.; Lee, T.J.; Wang, H.; Brandhorst, T.; Wuthrich, M.; Klein, B.S. CRISPR/Cas9-Mediated Gene Disruption Reveals the Importance of Zinc Metabolism for Fitness of the Dimorphic Fungal Pathogen Blastomyces dermatitidis. MBio 2018, 9. [Google Scholar] [CrossRef]
- Zhang, X.; Dai, P.; Gao, Y.; Gong, X.; Cui, H.; Jin, Y.; Zhang, Y. Transcriptome sequencing and analysis of zinc-uptake-related genes in Trichophyton mentagrophytes. BMC Genomics 2017, 18, 888. [Google Scholar] [CrossRef]
- Wilson, D. An evolutionary perspective on zinc uptake by human fungal pathogens. Metallomics 2015, 7, 979–985. [Google Scholar] [CrossRef]
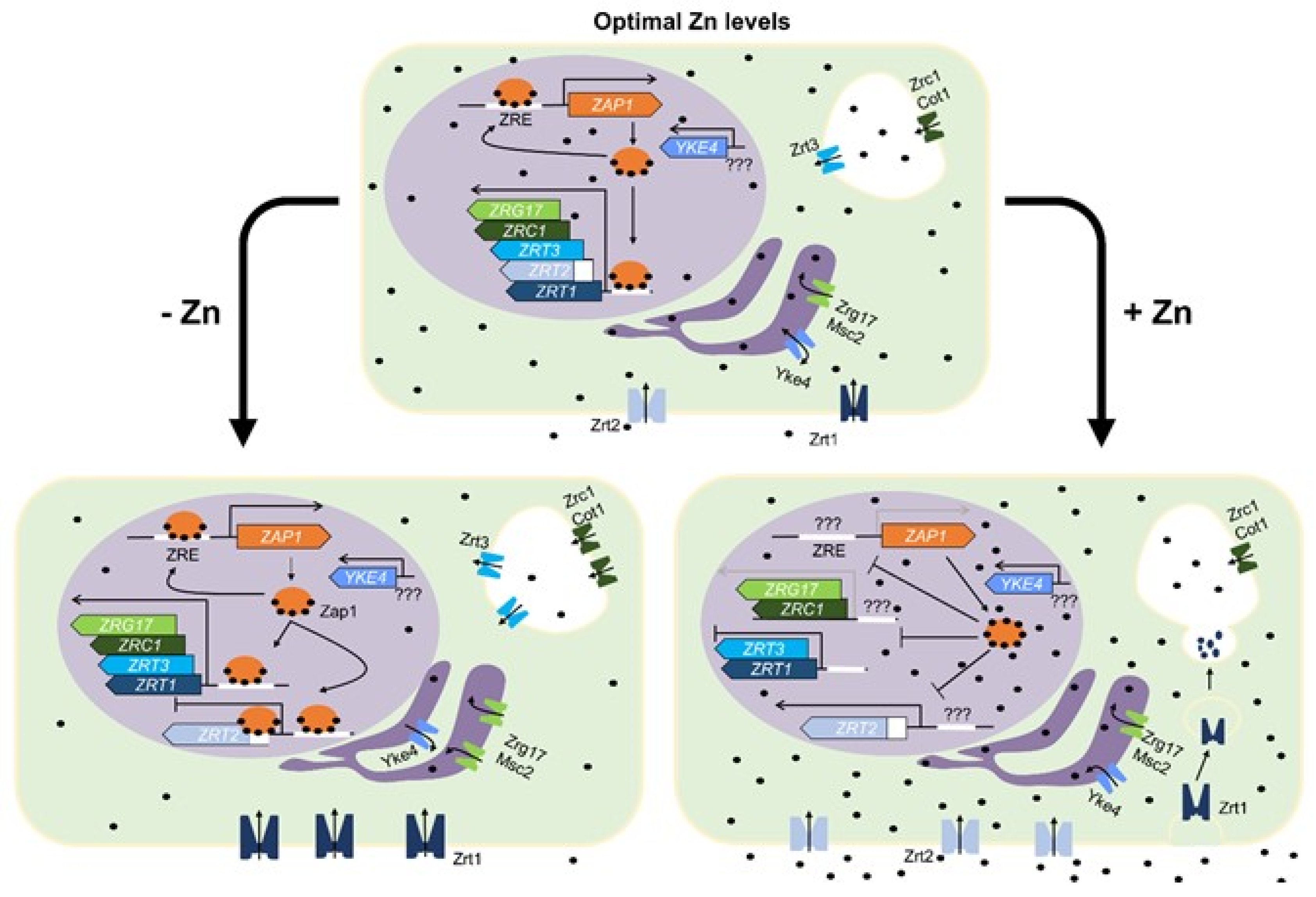

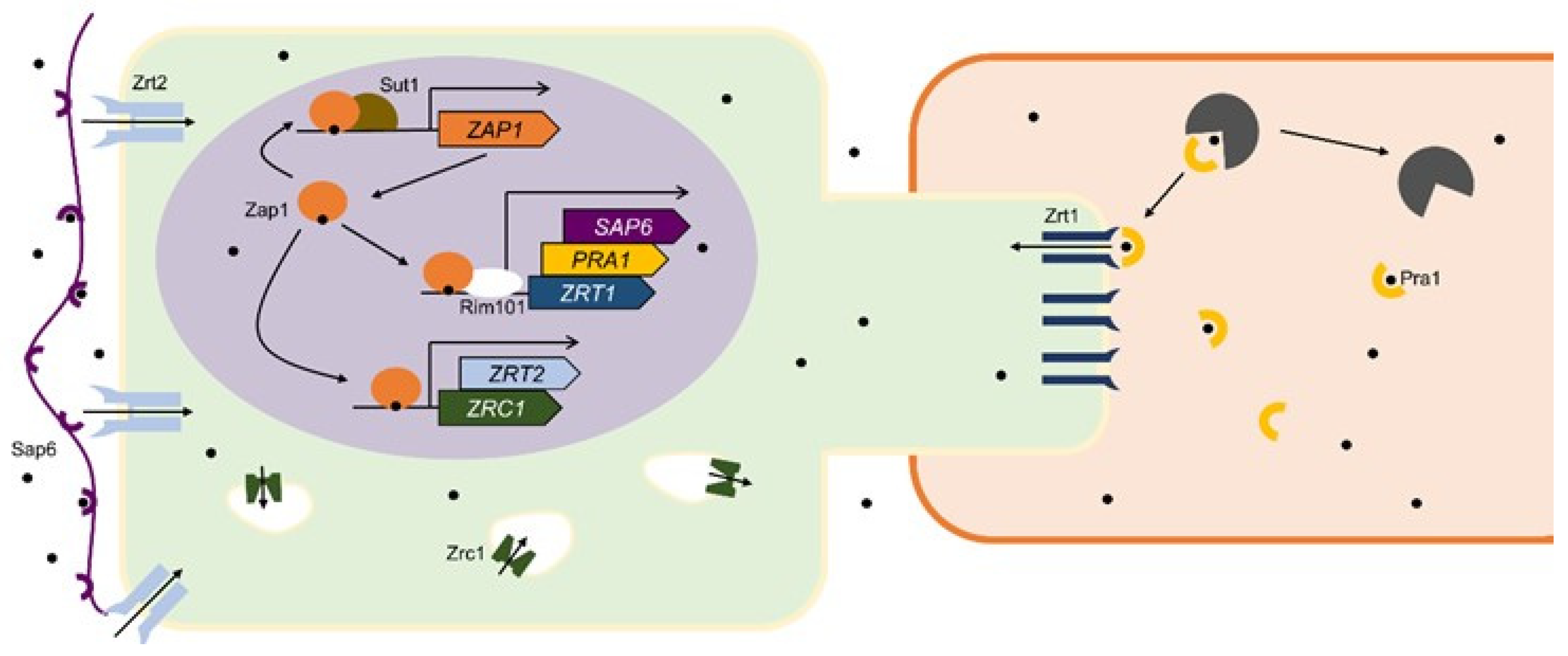
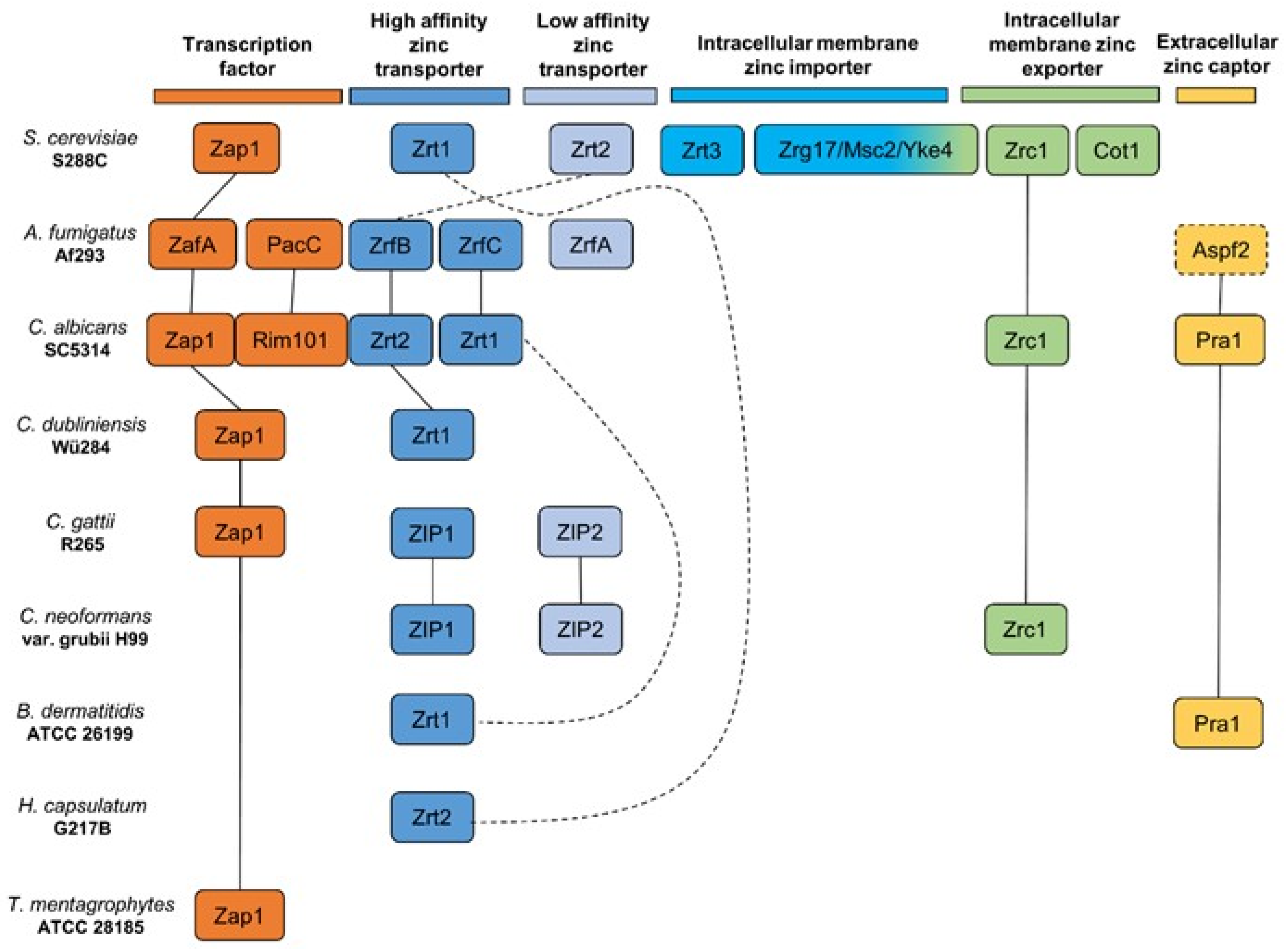
| Zinc Dependent | Zinc and pH Dependent | Not Enough Data 1 |
|---|---|---|
| Saccharomyces cerevisiae | Aspergillus fumigatus | Paracoccidioides brasiliensis |
| Cryptococcus gattii | Candida albicans | Paracoccidioides lutzii |
| Cryptococcus neoformans | Candida dubliniensis | Emmonsia parva |
| Emmonsia crescens | ||
| Trichophyton mentagrophytes | ||
| Histoplasma capsulatum | ||
| Blastomyces dermatitidis |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soares, L.W.; Bailão, A.M.; Soares, C.M.d.A.; Bailão, M.G.S. Zinc at the Host–Fungus Interface: How to Uptake the Metal? J. Fungi 2020, 6, 305. https://doi.org/10.3390/jof6040305
Soares LW, Bailão AM, Soares CMdA, Bailão MGS. Zinc at the Host–Fungus Interface: How to Uptake the Metal? Journal of Fungi. 2020; 6(4):305. https://doi.org/10.3390/jof6040305
Chicago/Turabian StyleSoares, Lucas Weba, Alexandre Melo Bailão, Célia Maria de Almeida Soares, and Mirelle Garcia Silva Bailão. 2020. "Zinc at the Host–Fungus Interface: How to Uptake the Metal?" Journal of Fungi 6, no. 4: 305. https://doi.org/10.3390/jof6040305
APA StyleSoares, L. W., Bailão, A. M., Soares, C. M. d. A., & Bailão, M. G. S. (2020). Zinc at the Host–Fungus Interface: How to Uptake the Metal? Journal of Fungi, 6(4), 305. https://doi.org/10.3390/jof6040305





