Abstract
Phylogenetic analyses of combined LSU, rpb2, tub2 and ITS sequence data of representative Xylariales taxa indicated that Diabolocovidia, Didymobotryum and Vamsapriya cluster together and form a distinct clade in Xylariales. Morphological comparison also shows their distinctiveness from other families of Xylariales. Therefore, we introduce it as a novel family, Vamsapriyaceae. Based on morphological characteristics, Podosporium and Tretophragmia, which were previously classified in Ascomycota genera incertae sedis, are now included in the Vamsapriyaceae. In addition, three Vamsapriya species, V. chiangmaiensis sp. nov, V. uniseptata sp. nov, and V. indica are described and illustrated in this paper.
1. Introduction
Xylariales is a large order with both conspicuous and inconspicuous fruiting bodies, and unitunicate, perithecial ascomycetes [1,2]. Many species of Xylariales are saprobes and endophytes [3,4]. Some Xylariales species can produce secondary metabolites which are especially important for the pharmaceutical chemical industry [3,5,6].
Xylariales was established by Nannfeldt [7] to accommodate the type family Xylariaceae, along with Diatrypaceae, Hypocreaceae, Hyponectriaceae, Lasiosphaeriaceae and Polystigmataceae. The previous classification of Xylariales was mainly based on morphology [8,9,10,11,12,13]. With the development of molecular technology, the classification basis of Xylariales was gradually diversified [2,14,15,16]. Smith et al. [2] performed the first multigene analysis to find the familial relationships within Xylariales and treated the order with seven families. Lumbsch and Huhndorf [17] listed six families in Xylariales, while Senanayake et al. [18] revised Xylariales and accepted 11 families. Hyde et al. [19] redefined the families of Sordariomycetes and accepted 15 families in Xylariales based on morphology and multigene analysis, viz. Barrmaeliaceae, Cainiaceae, Clypeosphaeriaceae, Coniocessiaceae, Diatrypaceae, Graphostromataceae, Hansfordiaceae, Hypoxylaceae, Induratiaceae, Lopadostomataceae, Microdochiaceae, Polystigmataceae, Requienellaceae, Xylariaceae and Zygosporiaceae. Hyde et al. [20] introduced Fasciatisporaceae to accommodate Fasciatispora in Xylariales. However, the taxonomic position of many taxa in Xylariales are still uncertain, and they are treated as genera incertae sedis [20,21]. This may probably be due to monospecific genera with either sexual or asexual morph, with no additional collections and lack of molecular data, and sometimes due to the polyphyletic nature of some genera (such as Anthostomella and Xylaria) [22,23,24,25].
Vamsapriya was introduced by Gawas and Bhat [26] based on abundant asexual morphs of the genus, which is characterized by erect, cylindrical, dark brown, synnematous conidiophores, monotretic, clavate to cylindrical conidiogenous cells, and cylindrical or broadly fusiform or obclavate, brown to dark brown conidia [26,27,28,29,30,31,32,33]. The first sexual morph of Vamsapriya was described by Dai et al. [28], which has solitary, immersed ascomata visible as black dots, 8spored, unitunicate asci, and hyaline, fusiform apiospores. They linked the sexual morph of V. bambusicola (MFLUCC 11-0637) to the asexual morph of V. bambusicola (MFLUCC 11-0477) using ITS phylogenies [27,28]. The phylogenetic placement of Vamsapriya has been confusing. Dai et al. [27,28] and Jiang et al. [31] accepted Vamsapriya into the Xylariaceae. However, phylogenetic analyses using broader taxon sampling indicated that Vamsapriya was distant from Xylariaceae [19,34].
This study aims to resolve the phylogenetic position of Vamsapriya. Three Vamsapriya collections (V. chiangmaiensis sp. nov, V. uniseptata sp. nov, and V. indica) on bamboo from China and Thailand are described and illustrated herein. Vamsapriya, along with Diabolocovidia and Didymobotryum, formed a distinct monophyletic clade in the combined LSU, rpb2, tub2 and ITS phylogenetic analyses. A new family, Vamsapriyaceae, is thus established. Podosporium and Tretophragmia are also accepted in Vamsapriyaceae based on their morphology of hyphomycetous asexual morph.
2. Materials and Methods
2.1. Collection, Examination, Isolation and Conservation
Fresh specimens were collected from bamboo in terrestrial habitats in China and Thailand between August 2019 and September 2020. Sample collections and observations were followed by the method described in Senanayake et al. [35]. The samples were stored in envelopes and taken to the laboratory for examination. Morphological observations were done using a stereo microscope (LEICA M125 C, Wetzlar, Germany). The fungal structures were captured using a Nikon ECLIPSE Ni compound microscope (Nikon, Tokyo, Japan) fitted with a NikonDS-Ri2 digital camera (Nikon, Tokyo, Japan). The Tarosoft (R) Image Frame Work software was used to take the measurements. Adobe Photoshop CS6 software (Adobe Systems, San Jose, CA, USA) was used to do photo-plates.
Single spore isolation was carried out to obtain pure cultures following the method described in Senanayake et al. [35]. Germinated spores were transferred to pure potato dextrose agar (PDA) and cultivated under normal light at 26 °C for four weeks. Herbarium specimens were deposited in the Fungarium of Mae Fah Luang University (MFLU), Chiang Rai, Thailand, and the herbarium of the Guizhou Academy of Agriculture Sciences (GZAAS), Guiyang, China. Pure cultures were deposited in the Mae Fah Luang University Culture Collection (MFLUCC) and the Guizhou Culture Collection (GZCC). FacesofFungi (FoF) and Index Fungorum numbers were obtained as described in Jayasiri et al. [36] and Index Fungorum [37].
2.2. DNA Extraction, PCR Amplification and Sequencing
Genomic DNA was extracted from fresh fungal mycelia using the Genomic DNA Extraction Kit (GD2416 BIOMIGA, San Diego, CA, USA). Polymerase chain reactions (PCR) were carried out using a BIO-RAD T100 Thermal Cycler in a 20 μL reaction volume which contained 10 μL 2x PCR Master Mix, 7 μL ddH2O, 1 μL of each primer, and 1 μL template DNA. The PCR thermal cycle program and primers are given in Table 1. The PCR products were sent for sequencing to SinoGenoMax, Beijing, China.

Table 1.
Primers and PCR protocol used in this study.
2.3. Phylogenetic Analyses
The sequences used in this study (Table 2) were downloaded from GenBank according to the results of blast searches and previous studies [27,28,29,30,31,32,33]. Alignments for each locus were carried out in MAFFT v7.212 [40]. AliView [41] was used for checking the alignments and changing the format. Terminal ends and ambiguous regions of the alignment were deleted manually. Four single gene alignments were combined using the Sequence Matrix [42].

Table 2.
Taxa names, strain numbers and corresponding sequences used for the molecular phylogenetic analyses.
Single gene analyses were done to compare the topologies and clade stabilities, respectively. Single and combined phylogenies were subjected to Bayesian posterior probability (BYPP), maximum likelihood (ML) and maximum parsimony (MP) analyses. The BYPP analysis was performed in MrBayes v. 3.2.6 [43]. MrModeltest v. 2.3 [44] was used to estimate the best model. GTR+I+G model was chosen for LSU and rpb2; SYM+I+G (Xylariales analysis) and GTR+G (Vamsapriya analysis) models were chosen for ITS; HKY+I+G model was chosen for tub2. Six chains were run and trees were sampled every 1000th generation, the temperature value of the heated chain was set at 0.15. The first 25% sampled trees were discarded as “burn-in”, and the remaining trees were used for calculating BYPP in the majority rule consensus tree. The ML analyses were carried out using IQ-TREE [45] on the IQ-TREE web server (http://iqtree.cibiv.univie.ac.at, 6 September 2021) under partitioned models. The best-fit substitution models were determined by W-IQ-TREE [45]: TIM3e+I+G4 for LSU; TIM3+F+I+G4 for rpb2; TIM2+F+I+G4 for tub2; SYM+I+G4 for ITS. Ultrafast bootstrap analysis was implemented with 1,000 replicates. The MP analyses were carried out with a heuristic search in PAUP v. 4.0 b10 [46]. Bootstrap analysis was used to estimate clade stability, including 1000 replicates, each with 10 replicates of random stepwise addition of taxa [47].
Phylogenetic trees were viewed using FigTree v1.4.4 [48] and modified in Adobe Illustrator CS6 software (Adobe Systems, USA). The sequences generated from our collections were deposited in GenBank.
3. Results
3.1. Phylogenetic Analyses
In Xylariales phylogenetic analyses, the final combined dataset of Xylariales consists of 84 strains representing fifteen families along with the outgroup Amphisphaeria sorbi (MFLUCC 13-0721) and A. thailandica (MFLU 18-0794) in Amphisphaeriales. The aligned sequence matrix comprises LSU (1–829), rpb2 (830–1875), tub2 (1876–3579) and ITS (3580–4305), sequence data for a total of 4305 characters, including coded alignment gaps. Among them, 1894 characters were constant, 374 variable characters were parsimony-uninformative and 2037 characters were parsimony informative. The matrix had 2693 distinct alignment patterns. The BYPP, ML, and MP analyses based on combined sequence data provided similar tree topology. For BYPP, the standard deviation of split frequencies was reached at 0.0099 after 2,980,000 generations. The most likely tree (−ln = 66,531.894) is presented (Figure 1). The MP analysis resulted in two trees with TL = 15,668, CI = 0.302, RI = 0.524, RC = 0.158, HI = 0.698.
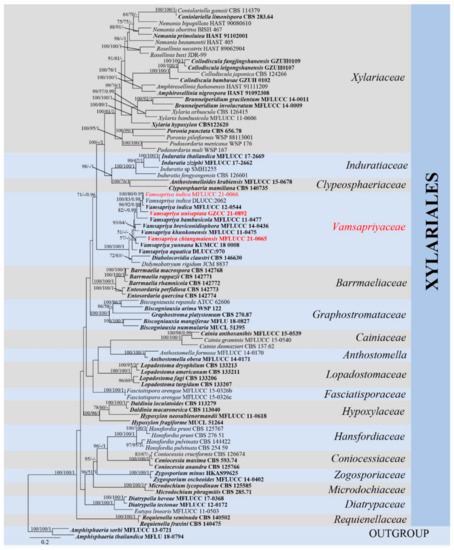
Figure 1.
Maximum likelihood (RAxML) tree, based on analysis of a combined dataset of LSU, rpb2, tub2 and ITS sequence data. The tree is rooted with Amphisphaeria sorbi (MFLUCC 13-0721) and A. thailandica (MFLU 18-0794). Bootstrap support values for ML and MP greater than 50% and Bayesian posterior probabilities greater than 0.95 are given near nodes, respectively. Ex-type strains are in bold, the new isolates are in red.
The single locus trees (Supplemental Figures S1–S4) and the multi-locus (LSU, rpb2, tub2 and ITS) tree (Figure 1) showed similar tree topology. In multigene analyses, Vamsapriya species clustered with Diabolocovidia claustri and Didymobotryum rigidum, and they formed an internal distinct clade with maximum support (ML-bs = 100%, MP-bs = 100%, BYPP = 1.00). Xylariaceae, Induratiaceae and Clypeosphaeriaceae clustered together, which is a sister to Vamsapriyaceae without significant support. Moreover, V. chiangmaiensis (MFLUCC 21-0065) formed a sister clade to V. yunnana; however, the support for this relationship in Figure 1 is extremely poor and does not exist in Figure 2, and V. uniseptata (GZCC 21-0892) is sister to V. indica. Our isolate MFLUCC 21-0066 grouped in V. indica clade with MFLUCC 12-0544 and DLUCC:2062, indicating they are phylogenetically the same species. Two Anthostomella (Xylariaceae) species, A. formosa (MFLUCC 14-0170) and A. obesa (MFLUCC 14-0171) formed a distinct clade and is sister to Cainiaceae.
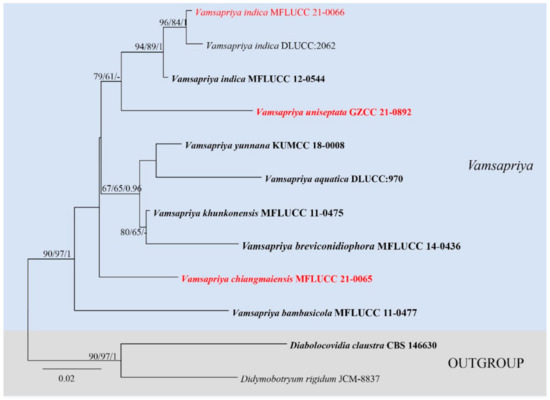
Figure 2.
Maximum likelihood (RAxML) tree for Vamsapriya, based on ITS sequence data. The tree is rooted with Diabolocovidia claustra (CBS 146630) and Didymobotryum rigidum (JCM-8837). Ex-type strains are in bold, the new isolates are in red.
The ITS based on Vamsapriya analyses contained 12 taxa and rooted with Diabolocovidia claustra (CBS 146630) and Didymobotryum rigidum (JCM-8837). The manually adjusted ITS alignment contained 563 characters. The best scoring RAxML tree with a final likelihood value of −1737.963458 is presented (Figure 2). Maximum parsimony analysis comprised 563 characters, of which 446 were constant, 54 were parsimony-informative, and 63 were parsimony-uninformative; the tree length is 184, CI = 0.739, RI = 0.597, RC = 0.441, HI = 0.261. The results showed our strain MFLUCC 21-0066 clustered together with V. indica (MFLUCC 12-0544 and DLUCC:2062) with good supports (ML-bs = 94%, MP-bs = 89%, BYPP = 1.00). Vamsapriya chiangmaiensis (MFLUCC 21-0065) formed a distinct clade, and Vamsapriya uniseptata (GZCC 21-0892) grouped with three V. indica (ML-bs = 79%, MP-bs = 61%).
3.2. Taxonomy
Vamsapriyaceae Y.R. Sun, Yong Wang bis & K.D. Hyde, fam. nov.
Index Fungorum number: IF558620; Facesoffungi number: FoF09926
Etymology: Name reflects the type genus
Type genus: Vamsapriya Gawas & Bhat
Saprobic on dead wood. Sexual morph: Ascomata solitary, scattered, immersed, subglobose, black, ostiolate. Peridium thin-walled, brown. Paraphyses hyaline, septate. Asci 8-spored, unitunicate, cylindrical, short pedicellate, with a J+ apical ring. Ascospores apiosporous, fusiform to broad fusiform, hyaline. Asexual morph: Hyphomycetous. Colonies on natural substrate effuse, black, velvety. Mycelium immersed, septate, branched. Synnemata present or absent; when present (Didymobotryum, Podosporium, Tretophragmia, Vamsapriya), synnemata erect, rigid, dark brown, composed of compact parallel conidiophores. Conidiophores erect, straight or curved, cylindrical, dark brown, septate. Conidiogenous cells mono- or polytretic, integrated, terminal, clavate to cylindrical, brown. Conidia catenate or solitary, acrogenous, simple, pigmented, multi-shaped, septate; when absent (Diabolocovidia, adapted from Crous et al. [49]), conidiophores micronematous, flexuous, mostly reduced to a terminal conidiogenous cell. Conidiogenous cells monoblastic, subcylindrical to clavate, pale brown, smooth. Conidia catenate, acrogenous, brown, ellipsoid to obovoid, thin-walled, aseptate.
Notes: A new family, Vamsapriyaceae, is introduced to accommodate Diabolocovidia, Didymobotryum, Podosporium, Tretophragmia, and Vamsapriya. Their phylogenetic position, which is distinct from other families, supports the establishment of the new family within Xylariales. Although the phylogeny of Podosporium and Tretophragmia could not be inferred due to the lack of molecular data, their morphological characters resemble Didymobotryum and Vamsapriya in having brown to dark, simple, straight synnemata, monotretic conidiogenous cells and solitary, obclavate, multi-septate, dark brown conidia [50,51,52,53]. We thus temporarily accept Podosporium and Tretophragmia in Vamsapriyaceae based on morphology. Sequence data are needed to resolve their phylogenetic affinities.
Vamsapriya Gawas & Bhat, Mycotaxon 94: 150 (2006) [2005]
Index Fungorum number: IF29041; Facesoffungi number: FoF00372
Type species: Vamsapriya indica Gawas & Bhat, Mycotaxon 94: 150 (2006) [2005]
Saprobic on dead wood. Sexual morph: Ascomata solitary, scattered, immersed, subglobose, black, ostiolate. Peridium thin-walled, brown. Paraphyses hyaline, septate. Asci 8-spored, unitunicate, cylindrical, straight, short pedicellate, with a J+ apical ring. Ascospores uniseriate or overlapping uniseriate, fusiform to broad fusiform, apiosporous, hyaline, pointed at both ends, surrounded by a mucilaginous sheath. Asexual morph: Hyphomycetous. Colonies on natural substrate effuse, black, velvety. Mycelium immersed, septate, branched. Conidiophores macronematous, synnematous, erect, straight or curved, dark brown, cylindrical, septate. Synnemata erect, rigid, dark brown, composed of compact parallel conidiophores. Conidiogenous cells monotretic, integrated, terminal, clavate to cylindrical. Conidia catenate or solitary, acrogenous, cylindrical, oblong, fusiform or obclavate, brown to dark brown, septate, verruculose.
Notes: Vamsapriya species are reported from tropical and subtropical regions, and most species are found in terrestrial as saprobes [26,27,28,29,30,31]. Nine species are accepted in the Vamsapriya, of which six have molecular data. Vamsapriya is the only holomorphic genus in Vamsapriyaceae, and V. bambusicola is the only species with a sexual-asexual connection in Vamsapriya. Bamboo seems to be the host preference for Vamsapriya species [26,27,28,29,30,31,32,33].
Vamsapriya indica Gawas & Bhat, Mycotaxon 94: 150 (2006) [2005]
Index Fungorum number: IF550801; Facesoffungi number: FoF00374, Figure 3

Figure 3.
Vamsapriya indica (MFLU 21-0088) (a,b) Colonies on natural substrate. (c) Conidiophore and conidia. (d–f) Conidiogenous cells and developing conidia. (g–k) Conidia. (l) Germinated conidium. Scale bars: a = 2000 µm, b = 1000 µm, c = 200 µm, d–l = 20 µm.
Saprobic on dead bamboo culms. Sexual morph: Undetermined. Asexual morph: Hyphomycetous. Colonies effuse, dark brown, hairy. Conidiophores macronematous, synnematous, single, erect, cylindrical, straight or slightly flexuous, dark brown, smooth-walled. Synnemata erect, straight or slightly flexuous, dark brown, rigid, with cylindrical to clavate apical fertile part, composed of compactly arranged conidiophores, 1300–1900 um long, 80–150 μm wide at the base, 30–40 μm wide in the middle, 60–140 μm wide at the apical fertile region, with basal portion immersed. Conidiogenous cells monotretic, integrated, terminal, brown, cylindrical to clavate, apically rounded, smooth-walled, 4.5–8.5 × 3–4.5 μm ( = 6.5 × 4 μm, n = 30). Conidia catenate, acrogenous, cylindrical, rounded at the apex, taper and subtruncate at the base, olivaceous brown to brown, 2–8-septate, slightly constricted at the septa, smooth, 20–48 × 4.5–6.5 μm ( = 32 × 5.5 μm, n = 20).
Cultural characters: Conidia germinated on PDA within 12 h, germ tubes produced from both ends. Colonies reached 20 mm diam. within four weeks at 26 °C, cottony, flat, circular, edge entire, white from above, white to yellow from the below.
Material examined: Thailand, Chiang Mai Province, Mae Taeng District, Pa Pae, Mushroom Research Center, on bamboo culms, 10 September 2020, H.W. Shen, M38 (MFLU 21-0088; living culture, MFLUCC 21-0066).
Notes: Vamsapriya indica is the type species of Vamsapriya [26]. Dai et al. [27] recollected V. indica from Thailand and provided the culture characters and sequences data. Bao et al. [32] reported it from a bamboo plant in a freshwater habitat in China. Including our collection, all of these four isolates are recorded from bamboo. Morphological comparison is shown in Table 3. Our collection has longer synnemata than those of the three isolates.

Table 3.
Synopsis of characters of Vamsapriya indica collections.
Vamsapriya chiangmaiensis Y.R. Sun, Yong Wang bis & K.D. Hyde, sp. nov.
Index Fungorum number: IF558618; Facesoffungi number: FoF09927, Figure 4

Figure 4.
Vamsapriya chiangmaiensis (MFLU 21-0087, holotype) (a,b) Appearance of ascomata on host substrate. (c) Vertical section of ascoma. (d–f) Asci. (g) Paraphyses. (h) Apical ring of asci. (i–m) Ascospores. (n) Germinated ascospore. (o) Ascospore stained in Indian ink. (p,q) Colonies on PDA. Scale bars: c = 200 μm, d–g = 20 μm, f,h–o = 10 µm.
Etymology: Name reflects the collected site.
Holotype: MFLU 21-0087
Saprobic on dead bamboo culms. Sexual morph: Ascomata 650–1000 × 650–850 μm, solitary scattered, immersed within the host cortex, visible as black, circular dots, in cross section globose to subglobose. Ostiole raised, centric, periphysate ostiolar canal. Peridium composed of hyaline inner layer and dark brown to dark outer layer. Paraphyses long, hyaline, unbranched, septate, 1.5–4 μm wide ( = 2 μm, n = 15). Asci 8-spored, unitunicate, straight or slightly curved, cylindrical, short pedicellate, with apical ring, 140–190 × 6.5–12 μm ( = 160 × 9 μm, n = 15). Ascospores uniseriate, fusiform, 17–26 × 5.5–8 μm ( = 20.5 × 6.5 μm, n = 30), constricted apiosporous with a large cell 12.5–22 μm length, guttulate; basal cell 3.5–6.5 μm length, hyaline, smooth-walled, surround a gelatinous mucilaginous sheath. Asexual morph: Undetermined.
Culture characters: Ascospores germinated on PDA within 12 h, germ tubes produced from one end. Colonies reached 45 mm diam. within four weeks at 26 °C, flat, circular, cottony. White from above; brown to dark brown in the center, white to pale brown around from below.
Material examined: Thailand, Chiang Mai Province, Mae Taeng District, Mushroom Research Center, on bamboo culms, 15 July 2020, Y.R. Sun, M35 (MFLU 21-0087, holotype; ex-type living culture, MFLUCC 21-0065).
Notes: Vamsapriya chiangmaiensis is the second species that has a sexual morph in Vamsapriya. It is similar to V. bambusicola in having solitary, subglobose ascomata, 8-spored, unitunicate, cylindrical asci and fusiform hyaline ascospores. It can be distinguished by the longer asci (140–190 μm vs. 115–140 μm). In addition, polymorphic nucleotides from the ITS region showed 37 base differences, and the details are given in Table 4. Therefore, we identified V. chiangmaiensis as a new species following the suggestions for species delineation [54].

Table 4.
Nucleotide differences in the ITS regions of V. bambusicola and V. chiangmaiensis. Numbers are in reference to the nucleotide position of DNA sequences (V. bambusicola) submitted in GenBank.
Vamsapriya uniseptata N.G. Liu & K.D. Hyde , sp. nov.
Index Fungorum number: IF558619; Facesoffungi number: FoF09928, Figure 5.

Figure 5.
Vamsapriya uniseptata (GZAAS:21-0378, holotype) (a,b) Colonies on natural substrate. (c) Conidiophores and conidia. (d–f) Conidiogenous cells and developing conidia. (g–k) Conidia. (l) Germinated conidium. Scale bars: c = 100 µm, d,e,g,h = 5 µm, f = 10 µm.
Etymology: Name reflects the 1-septate conidia.
Holotype: GZAAS 21-0378
Saprobic on submerged decaying wood in terrestrial habitat. Colonies on natural substrate effuse, black, velvety. Asexual morph: Hyphomycetous. Mycelium mostly immersed, composed of septate, branched, hyaline to brown hyphae. Conidiophores macronematous, synnematous, erect, straight or broadly curved, dark brown, cylindrical, septate. Synnemata erect, rigid, dark brown, composed of compact parallel conidiophores, up to 1300 µm long, 30–50 µm wide in the middle. Conidiogenous cells monotretic, integrated, terminal, clavate, brown to dark brown. Conidia catenate, acrogenous, olivaceous brown, smooth, oblong, rounded at the apex, taper and subtruncate at the base, 1-septate at the middle, septa thickened and darkened, slightly constricted at the septa, with a large globule in each cell, 14–19 × 3.5–4.5 μm ( = 16.5 × 5 µm, n = 30). Sexual morph: Unknown.
Cultural characters: Conidia germinated on PDA within 12 h and germ tubes produced from both ends. Colonies reached 30 mm within four weeks at 26 °C, flat, circular, cottony, white from above, from below brown to dark brown in the center, white to pale brown around.
Material examined: China, Guizhou Province, Xingyi City, Qingshuihe Town, 8 August 2019, N.G. Liu, Q1 (GZAAS 21-0378, holotype; ex-type living culture, GZCC 21-0892).
Notes: Vamsapriya uniseptata is distinguishable by having smaller, 1-septate conidia, while other Vamsapriya species have elongated phragmoconidia. In the BLASTn search, the closest match of the ITS sequence of V. uniseptata is V. khunkonensis (MFLUCC 13-0497, MFLUCC 11-0475 (93.4%)), followed by V. indica (MFLUCC 12-0544 (91.7%)). The LSU sequence of V. uniseptata is V. indica (DLUCC:2062 (99.8%)) and V. khunkonensis (MFLUCC 11-0475 (99.7%)). Vamsapriya uniseptata can be distinguished from V. khunkonensis in the multigene phylogenetic analyses. The ITS region of V. indica (MFLUCC 13-0497) differs by 23 base pairs (527 bp without gaps). Based on distinct morphology and phylogeny, V. uniseptata is introduced as a novel taxon.
3.3. Other Accepted Genera in Vamsapriyaceae
Diabolocovidia Crous, Persoonia 44: 331 (2020)
Index Fungorum number: IF835401; Facesoffungi number: FoF09929.
Parasitic on leaves in terrestrial habitats. Mycelium composed of septate, branched, hyaline to pale brown hyphae. Asexual morph: Hyphomycetous. Conidiophores solitary, erect, flexuous, mostly reduced to a terminal conidiogenous cell. Conidiogenous cells monoblastic, terminal, subcylindrical to clavate, pale brown, smooth. Conidia catenate, acrogenous, brown, ellipsoid to obovoid, thin-walled, un-septate, verruculose [49]. Sexual morph: Unknown.
Type species: Diabolocovidia claustri Crous
Notes: Diabolocovidia is a monotypic genus introduced by Crous et al. [49] to accommodate Diabolocovidia claustri, which was isolated from leaves of Serenoa repens in the U.S.A. Diabolocovidia claustri is characterized by mononematous, micronematous conidiophores in Xylariaceae. In their phylogenetic analyses, Diabolocovidia is basal to Vamsapriya [49]. Diabolocovidia is the only genus without synnemata in Vamsapriyaceae.
Didymobotryum Sacc., Syll. fung. (Abellini) 4: 626 (1886)
Index Fungorum number: IF8009; Facesoffungi number: FoF09930.
Saprobic on decaying plants materials in terrestrial habitats. Colonies on natural substrate effuse, olivaceous to dark brown, velvety. Mycelium mostly immersed, composed of septate, branched, thick-walled, subhyaline hyphae. Asexual morph: Hyphomycetous. Conidiophores macronematous, synnematous, erect, straight or broadly curved, dark brown, cylindrical, septate. Synnemata erect, rigid, dark brown, composed of compact parallel conidiophores. Conidiogenous cells monotretic, integrated, integrated or discrete, cylindrical to clavate, olivaceous brown to dark brown. Conidia catenate, dry, acrogenous, cylindrical, verrucose, 1-septate, slightly constricted at the septa, olivaceous brown to brown. Sexual morph: Unknown.
Type species: Didymobotryum rigidum (Berk. & Broome) Sacc.
Notes: Didymobotryum was introduced by Saccardo [55] typified by D. rigidum. Didymobotryum species have a worldwide distribution [56,57,58,59]. Six species are accepted in the Index Fungorum [37] but only D. rigidum has molecular data (ITS: LC228650, LSU: LC228707).
Podosporium Schwein., Trans. Am. phil. Soc., New Series 4(2): 278 (1832) [1834]
Index Fungorum number: IF9487; Facesoffungi number: FoF09931.
Saprobic on decaying plants materials in terrestrial habitats. Colonies on natural substrate effuse, brown, velvety. Mycelium mostly immersed, composed of septate, flexuous branched hyphae. Asexual morph: Hyphomycetous. Conidiophores arranged in synnemata, brown, septate, sometimes branched at the apex. Synnemata erect, rigid, brown to dark. Conidiogenous cells mono- or polytretic, integrated or discrete, subulate or cylindrical, darkly pigmented. Conidia solitary, obclavate or bacilliform, multi-septate, brown to dark brown. Sexual morph: Unknown.
Type species: Podosporium rigidum Schwein.
Notes: Podosporium was introduced by Schweinitz [60] with P. rigidum as the type species. Since then, many Podosporium species have been discovered worldwide [60,61,62,63]. Most of them are saprobes in terrestrial habitats [60,61,62,63]. There are 67 species listed in the Index Fungorum [37] but no sequence data are available.
Tretophragmia Subram. & Natarajan, Proc. Natl. Inst. Sci. India, B, Biol. Sci. 39: 550 (1974) [1973]
Index Fungorum number: IF10265; Facesoffungi number: FoF09932.
Saprobic on plants materials in terrestrial habitats. Colonies on natural substrate effuse, dark, velvety. Asexual morph: Hyphomycetous. Conidiophores macronematous, synnematous, brown, septate, erect, straight or broadly curved. Synnemata rigid, brown to dark, simple, erect, straight, consisting of a stalk and a capitate, broadened, fertile head. Conidiogenous cells monotretic, subulate or cylindrical, darkly pigmented. Conidia solitary, obclavate to fusiform or irregular in shape, straight, curved or bent, multi-septate, dark brown. Sexual morph: Unknown.
Type species: Tretophragmia nilgirensis (Subram.) Subram. & Natarajan
Notes: Tretophragmia was introduced in 1974. The asexual morph of Tretophragmia is similar to Didymobotryum, Podosporium and Vamsapriya, while no sexual morph is reported. Seifert et al. [53] treated Tretophragmia as a synonym of Podosporium. However, Tretophragmia is accepted in the Index Fungorum [37] and the MycoBank [64] as a separate genus. So far, only two species of Tretophragmia have been described [37] and no sequence data are available. Thus, based on morphology and until DNA sequences data are available, we regard this as a separate genus.
4. Discussion
In this study, three Vamsapriya species, V. chiangmaiensis, V. indica and V. uniseptata were collected from bamboo in terrestrial habitats. In our phylogenetic analyses of combined LSU, rpb2, tub2 and ITS sequence data, Diabolocovidia, Didymobotryum and Vamsapriya formed a distinct clade in Xylariales. Morphological comparison also shows their distinctiveness from other families in Xylariales. Therefore, we propose Vamsapriyaceae as a new family in Xylariales. The sexual morph of Vamsapriya differs from those of Xylariaceae in having hyaline apiospores [28,30]. It is noteworthy that the sexual morph of Vamsapriya is similar to Induratiaceae in having 8-spored asci with J+ apical ring and hyaline, apiospores, but Induratia (Induratiaceae) differs in having geniculosporium asexual morphs [34]. Apioclypea is morphologically similar to the sexual morph of Vamsapriya in having 8-spored, pedunculate, cylindrical asci and biseriate, fusiform, hyaline ascospores with a mucilaginous sheath, but its asexual morph is unknown [19,21].
Clypeosphaeriaceae and Induratiaceae are two other families that are phylogenetically related to Vamsapriyaceae, but they are distinct in morphology. Apioclypea, Aquasphaeria, Brunneiapiospora, Clypeosphaeria, Crassoascus, and Palmaria (Clypeosphaeriaceae) lack asexual morph descriptions and Diabolocovidia, Didymobotryum, Podosporium and Tretophragmia (Vamsapriyaceae) do not have sexual morph descriptions for the comparisons in Table 5 and Table 6.

Table 5.
Asexual morph comparison of the genera in Clypeosphaeriaceae, Induratiaceae and Vamsapriyaceae.

Table 6.
Sexual morph comparison of the genera in Clypeosphaeriaceae, Induratiaceae and Vamsapriyaceae.
Diabolocovidia claustri was isolated on leaves of Serenoa repens by Crous et al. [49]. Although it has a close phylogenetic relationship with Vamsapriya, they are quite different in morphology. Diabolocovidia has micronematous rather than synnematous conidiophores, blastic rather than tretic conidiogenous cells, and ellipsoid to obovoid, aseptate conidia [49]. The phenomenon that Diabolocovidia mixes with synnematous and tretic genera like Didymobotryum and Vamsapriya reminds us of an example that Vanakripa with blastic conidiogenous resides in the phialidic genus Conioscypha [65]. These probably indicate the polyphyletic nature of some hyphomycetous groups. However, since D. claustri is the only species represented by one isolate in Diabolocovidia, we suggest using more collections to confirm its phylogenetic placement in the future.
When introducing Vamsapriya, Gawas and Bhat [26] pointed out Vamsapriya (conidia catenate, cylindrical to vermiform, phragmosporous) exhibits a combination of morphological characters of Didymobotryum (conidia catenate, ellipsoidal-cylindrical, didymosporous) [51,53,54] and Podosporium (conidia solitary, obclavate, phragmosporous) [56,61,63]. However, as more species are added to these three genera, such generic concepts based on conidial morphology have been dispelled. For example, V. uniseptata resembles species of Didymobotryum in having catenate, oblong, and 1-septate conidia, but it clusters with the type species of Vamsapriya, V. indica. Vamsapriya breviconidiophora and V. yunnana resemble Podosporium species in having obclavate, solitary, and multi-septate conidia, but they are grouped with V. aquatica, which has catenate, cylindrical to obclavate, multi-septate conidia in the phylogenetic tree. Either the authors did not follow the generic concepts strictly when introducing species, or these three genera are probably congeneric. We tend to infer the latter; however, the conclusion requires a detailed re-examination of herbarium specimens and molecular data.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof7110891/s1, Figure S1: Maximum likelihood (RAxML) tree based on ITS sequence data. The tree is rooted with Amphisphaeria sorbi (MFLUCC 13-0721) and A. thailandica (MFLU 18-0794). Figure S2: Maximum likelihood (RAxML) tree based on LSU sequence data. The tree is rooted with Amphisphaeria sorbi (MFLUCC 13-0721) and A. thailandica (MFLU 18-0794). Figure S3: Maximum likelihood (RAxML) tree based on tub2 sequence data. The tree is rooted with Amphisphaeria thailandica (MFLU 18-0794). Figure S4: Maximum likelihood (RAxML) tree based on rpb2 sequence data. The tree is rooted with Amphisphaeria thailandica (MFLU 18-0794).
Author Contributions
Methodology, Y.-R.S.; Software, Y.-R.S.; Supervision, R.S.J., K.D.H. and Y.W.; Writing—original draft, Y.-R.S.; Writing—review & editing, N.-G.L., M.C.S., R.S.J., K.D.H. and Y.W. All authors have read and agreed to the published version of the manuscript.
Funding
The study was funded by Guizhou Science Technology Department International Cooperation Basic project ([2018]5806), National Natural Science Foundation of China (No. 31972222, 31560489), Program of Introducing Talents of Discipline to Universities of China (111 Program, D20023), and Talent project of Guizhou Science and Technology Cooperation Platform [2017]5788-5, [2019]5641 and [2020]5001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Shaun Pennycook for checking the nomenclature. We also thank the support of the Thailand Research Foundation. Ya-Ru Sun would like to thank Hong-Wei Shen for collecting the samples. Samantha C. Karunarathna is thanked for giving suggestions in this study. Kevin D. Hyde would like to thank the Thailand Research grant entitled “Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion” (grant no: RDG6130001).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Eriksson, O.E.; Winka, K. Supraordinal taxa of Ascomycota. Myconet 1997, 1, 1–16. [Google Scholar]
- Smith, G.J.D.; Liew, E.C.; Hyde, K.D. The Xylariales: A monophyletic order containing 7 families. Fungal Diversity 2003, 13, 185–218. [Google Scholar]
- Stadler, M. Importance of secondary metabolites in the Xylariaceae as parameters for assessment of their taxonomy, phylogeny, and functional biodiversity. Curr. Res. Environ. Appl. Mycol. 2011, 1, 75–133. [Google Scholar] [CrossRef]
- U’Ren, J.M.; Miadlikowska, J.; Zimmerman, N.B.; Lutzoni, F.; Stajich, J.E.; Arnold, A.E. Contributions of North American endophytes to the phylogeny, ecology, and taxonomy of Xylariaceae (Sordariomycetes, Ascomycota). Mol. Phylogenet. Evol. 2016, 98, 210–232. [Google Scholar] [CrossRef] [PubMed]
- Helaly, S.E.; Thongbai, B.; Stadler, M. Diversity of biologically active secondary metabolites from endophytic and saprotrophic fungi of the ascomycete order Xylariales. Nat. Product Rep. 2018, 35, 992–1014. [Google Scholar] [CrossRef]
- Becker, K.; Stadler, M. Recent progress in biodiversity research on the Xylariales and their secondary metabolism. J. Antibiot. 2021, 74, 1–23. [Google Scholar] [CrossRef]
- Nannfeldt, J.A. Studien uber die Morphologie und Systematik der nicht-lichenisierten inoperculaten Discomyceten. Nova Acta Regiae Societatis Scientiarum Upsaliensis Ser. IV 1932, 8, 1–368. [Google Scholar]
- Munk, A. The system of the Pyrenomycetes. Dansk Botanisk Arkin 1953, 15, 1–163. [Google Scholar]
- Müller, E.; von Arx, J.A. Die Gattungen der didymosporen Pyrenomyceten. Beiträge Kryptogamenflora Schweiz 1962, 11, 1–922. [Google Scholar]
- Muller, E. Pyrenomycetes: Meliolales, coronophorales, sphaeriales. In The Fungi, an Advanced Treatise; Academic Press: London, UK, 1973. [Google Scholar]
- Wehmeyer, L.E. The Pyrenomycetous fungi. Mycol. Mem. 1975, 6, 1–250. [Google Scholar]
- Barr, M.E. Prodromus to nonlichenized, pyrenomycetous members of class Hymenoascomycetes. Mycotaxon 1990, 39, 43–184. [Google Scholar]
- Hawksworth, D.L.; Kirk, P.M.; Sutton, B.C.; Pegler, D.N. Ainsworth and Bisby’s Dictionary of the Fungi, 8th ed.; CAB International: Oxon, UK, 1995. [Google Scholar]
- Kang, J.C.; Kong, R.Y.C.; Hyde, K.D. Studies on the Amphisphaeriales 1. Amhisphaeriaceae (sensu stricto) and its phylogenetic relationships inferred from 5.8S rDNA and ITS2 sequences. Fungal Divers. 1998, 1, 147–157. [Google Scholar]
- Kang, J.C.; Kong, R.Y.C.; Hyde, K.D. Phylogeny of Amhisphaeriaceae (sensu stricto) and related taxa revisited based on nrDNA sequences. Mycotaxon 2002, 81, 321–330. [Google Scholar]
- Eriksson, O.E.; Baral, H.O.; Currah, R.S.; Hansen, K.; Kurtzman, C.P.; Rambold, G.; Laessoe, T. Outline of Ascomycota—2003. Myconet 2003, 9, 1–89. [Google Scholar]
- Lumbsch, H.T.; Huhndorf, S.M. Myconet volume 14 part one. Outline of ascomycota—2009. Fieldiana Life Earth Sci. 2010, 1, 1–922. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Bhat, J.D.; Jones, E.B.G.; McKenzie, E.H.C.; Dai, D.Q.; Daranagama, D.A.; Dayarathne, M.C.; Goonasekara, I.D.; et al. Towards unraveling relationships in Xylariomycetidae (Sordariomycetes). Fungal Divers. 2015, 73, 73–144. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Jones, E.B.G.; Bundhun, D.; Chen, Y.J.; Bao, D.F.; Boonmee, S.; Calabon, M.S.; et al. Refined families of Sordariomycetes. Mycosphere 2020, 11, 305–1059. [Google Scholar] [CrossRef]
- Hyde, K.D.; Dong, Y.; Phookamsak, R.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Liu, N.G.; Abeywickrama, P.D.; Mapook, A.; Wei, D.P.; et al. Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020, 100, 5–277. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Bhat, J.D.; Dayarathne, M.C.; Huang, S.K.; Norphanphoun, C.; Senanayake, I.C.; Perera, R.H.; et al. Families of Sordariomycetes. Fungal Divers. 2016, 79, 1–317. [Google Scholar] [CrossRef]
- Fröhlich, J.; Hyde, K.D. Palm Microfungi. Fungal Divers. Res. Ser. 2000, 3, 1–393. [Google Scholar]
- Daranagama, D.A.; Camporesi, E.; Liu, X.Z.; Bhat, D.J.; Chamyuang, S.; Bahkali, A.H.; Stadler, M.; Hyde, K.D. Tristratiperidium microsporum gen. et sp. nov. (Xylariales) on dead leaves of Arundo plinii. Mycol. Prog. 2015, 15, 1–8. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Hardy, G.E.S.J.; Crane, C.; Barrett, S.; Cano-Lira, J.F.; le Roux, J.J.; Thangavel, R.; Guarro, J.; et al. Fungal Planet description sheetS: 469–557. Persoonia 2016, 37, 218–403. [Google Scholar] [CrossRef]
- Fiuza, P.O.; Silva, C.; Santos, T.A.B.; Raja, H.; Castañeda-Ruiz, R.F.; Gusmão, L.F.P. Roselymyces, a new asexual genus of the Xylariales (Ascomycota) from Brazil. Sydowia 2018, 70, 59–65. [Google Scholar]
- Gawas, P.; Bhat, D.J. Vamsapriya indica gen. et sp. nov., a bambusicolous, synnematous fungus from India. Mycotaxon 2005, 94, 149–154. [Google Scholar]
- Dai, D.Q.; Bahkali, A.H.; Li, Q.R.; Bhat, D.J.; Wijayawardene, N.N.; Li, W.J.; Chukeatirote, E.; Zhao, R.L.; Xu, J.C.; Hyde, K.D. Vamsapriya (Xylariaceae) redescribed, with two new species and molecular sequence data. Cryptogamie Mycologie 2014, 35, 339–357. [Google Scholar] [CrossRef]
- Dai, D.Q.; Phookamsak, R.; Wijayawardene, N.N.; Li, W.J.; Bhat, D.J.; Xu, J.C.; Taylor, J.E.; Hyde, K.D.; Chukeatirote, E. Bambusicolous fungi. Fungal Divers. 2017, 82, 1–105. [Google Scholar] [CrossRef]
- Castañeda-Ruiz, R.F.; Zhang, X.G.; Li, D.W.; Gusmão, L.F.P.; Pérez-Martínez, S.; Sosa, D. Notes on Vamsapriya and V. camagueyensis comb. nov. Mycotaxon 2017, 132, 553–557. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Abreu, V.P.; Bazzicalupo, A.; Thilini, C.K.W.; Clericuzio, M.; Dayarathne, M.C.; Dissanayake, A.J.; Ekanayaka, A.H.; He, M.Q.; et al. Fungal diversity notes 603–708: Taxonomic and phylogenetic notes on genera and species. Fungal Divers. 2017, 87, 1–235. [Google Scholar] [CrossRef]
- Jiang, H.B.; Phookamsak, R.; Bhat, D.J.; Khan, S.; Bahkali, A.; Elgorban, A.M.; Hyde, K.D. Vamsapriya yunnana, a new species of Vamsapriya (Xylariaceae; Xylariales) associated with bamboo from Yunnan; China. Phytotaxa 2018, 356, 61–70. [Google Scholar] [CrossRef]
- Bao, D.F.; Hyde, K.D.; McKenzie, E.H.; Jeewon, R.; Su, H.Y.; Nalumpang, S.; Luo, Z.L. Biodiversity of Lignicolous Freshwater Hyphomycetes from China and Thailand and Description of Sixteen Species. J. Fungi 2021, 7, 669. [Google Scholar] [CrossRef]
- Ling, Y.; Li, H.H.; Xia, J.W.; Zhang, X.G.; Li, Z. Vamsapriya jinniuensis sp. nov.; and a first record of Garnaudia elegans from southern China. Mycotaxon 2018, 133, 367–372. [Google Scholar] [CrossRef]
- Samarakoon, M.C.; Thongbai, B.; Hyde, K.D.; Brönstrup, M.; Beutling, U.; Lambert, C.; Miller, A.N.; Liu, J.K.; Promputtha, I.; Stadler, M. Elucidation of the life cycle of the endophytic genus Muscodor and its transfer to Induratia in Induratiaceae fam. nov.; based on a polyphasic taxonomic approach. Fungal Divers. 2020, 101, 177–210. [Google Scholar] [CrossRef]
- Senanayake, I.; Rathnayake, A.; Marasinghe, D.; Calabon, M.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection; Examination; Isolation; Sporulation and Preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.A.; Bha, T.J.; Buyck, B.; Cai, L.; Dai, Y.C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The Faces of Fungi database: Fungal names linked with morphology; Phylogeny and human impacts. Fungal Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- Index Fungorum. Available online: http://www.indexfungorum.org/Names/Names.asp (accessed on 6 September 2021).
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nylander, J. MrModeltest (Version 2.2); Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods); Version 4.0 b10; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree. Version 1.4.2; University of Edinburgh: Edinburgh, UK, 2014. [Google Scholar]
- Crous, P.W.; Wingfield, M.J.; Chooi, Y.H.; Gilchrist, C.L.; Lacey, E.; Pitt, J.I.; Roets, F.; Swart, W.J.; Cano-Lira, J.F.; Valenzuela-Lopez, N. Fungal Planet description sheets:1042–1111. Pers. Mol. Phylogeny Evol. Fungi 2020, 44, 301. [Google Scholar] [CrossRef]
- Seifert, K. Contributions toward a mycobiota of Indonesia: Synnematous hyphomycetes. Mem. N. Y. Bot. Gard 1990, 59, 109–154. [Google Scholar]
- Zhang, Y.D.; Ma, J.; Ma, L.G.; Zhang, X.G. A new species of Podosporium and a new record from southern China. Mycotaxon 2010, 114, 401–405. [Google Scholar] [CrossRef]
- Wang, J.Y.; Zhang, K.; Yang, C.L.; Xia, J.W.; Ma, Y.R.; Gao, J.M.; Li, X.Y.; Zhang, X.G.; Cai, Y.M. Podosporium bacilliforme sp. nov. and a first record of Phaeoblastophora peckii from southern China. Mycotaxon 2016, 131, 841–846. [Google Scholar] [CrossRef]
- Seifert, K.; Morgan-Jones, G.; Gams, W.; Kendrick, B. The Genera of Hyphomycetes; CBS-KNA W Fungal Biodiversity Centre: Utrecht, The Netherlands, 2011. [Google Scholar]
- Jeewon, R.; Hyde, K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 2016, 7, 1669–1677. [Google Scholar] [CrossRef]
- Saccardo, P.A. Sylloge Hyphomycetum. Sylloge Fungorum 1886, 4, 807. [Google Scholar]
- Berkeley, M.J.; Broome, C.E. Enumeration of the fungi of Ceylon. Part II; containing the remainder of the hymenomycetes; with the remaining established tribes of fungi. J. Linn. Soc. Bot. 1873, 14, 29–140. [Google Scholar] [CrossRef]
- Ellis, M.B. Dematiaceous Hyphomycetes; Commonwealth Mycological Institute: Kew, UK, 1971. [Google Scholar]
- Zhuang, W.Y. Higher Fungi of Tropical China; Mycotaxon Ltd.: Ithaca, NY, USA, 2001. [Google Scholar]
- Wang, X.M.; Liu, G.N.; Chen, S.S.; Du, X.F. Records of Beltrania rhombica and Didymobotryum rigidum from China. Mycotaxon 2017, 132, 525–529. [Google Scholar] [CrossRef]
- Von Schweinitz, L.D. Synopsis fungorum in America boreali media degentium. Trans. Am. Philos. Soc. 1832, 4, 141–316. [Google Scholar] [CrossRef]
- Ciferri, R. Observations on meliolicolous hyphales from Santo Domingo. Sydowia 1955, 9, 302. [Google Scholar]
- Mercado, S. Nuevos e interesantes hifomicetes enterblásticos de Cuba. Acta Bot. Cuba 1983, 16, 1–8. [Google Scholar]
- Crous, P.W.; Wingfield, M.J.; Kendrick, W.B. Foliicolous dematiaceous hyphomycetes from Syzygium cordatum. Can. J. Bot. 1995, 73, 224–234. [Google Scholar] [CrossRef][Green Version]
- Mycobank. Available online: https://www.mycobank.org/ (accessed on 6 September 2021).
- Yang, H.; Dong, W.; Yu, X.D.; Bhat, D.J.; Boonmee, S.; Zhang, H. Four freshwater dematiaceous hyphomycetes in Sordariomycetes with two new species of Parafuscosporella. Phytotaxa 2020, 44, 19–34. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).