A New Front in Microbial Warfare—Delivery of Antifungal Effectors by the Type VI Secretion System
Abstract
1. Introduction
2. Occurrence of Type VI Secretion Systems
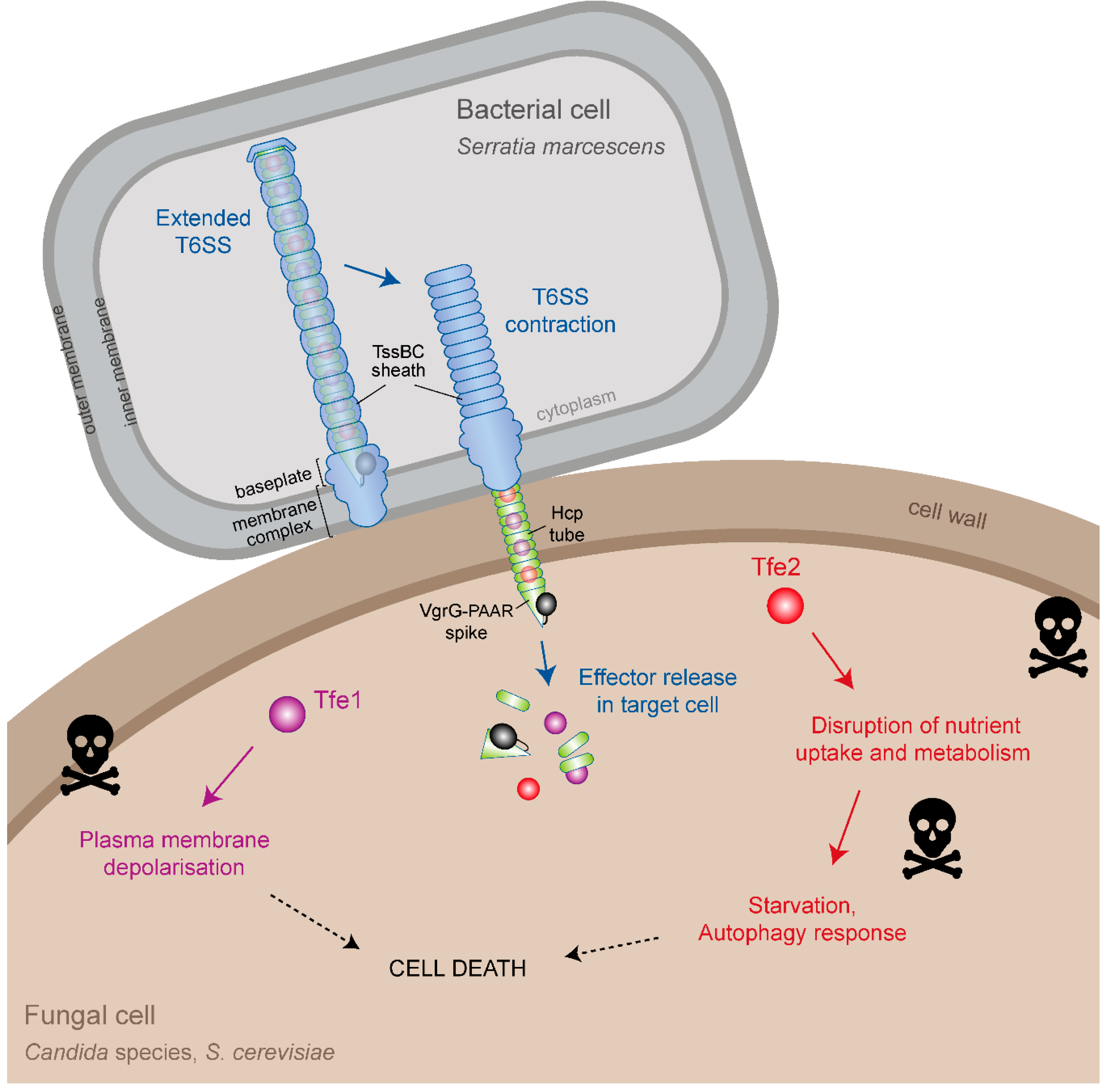
3. Effector Delivery by the Type VI Secretion System
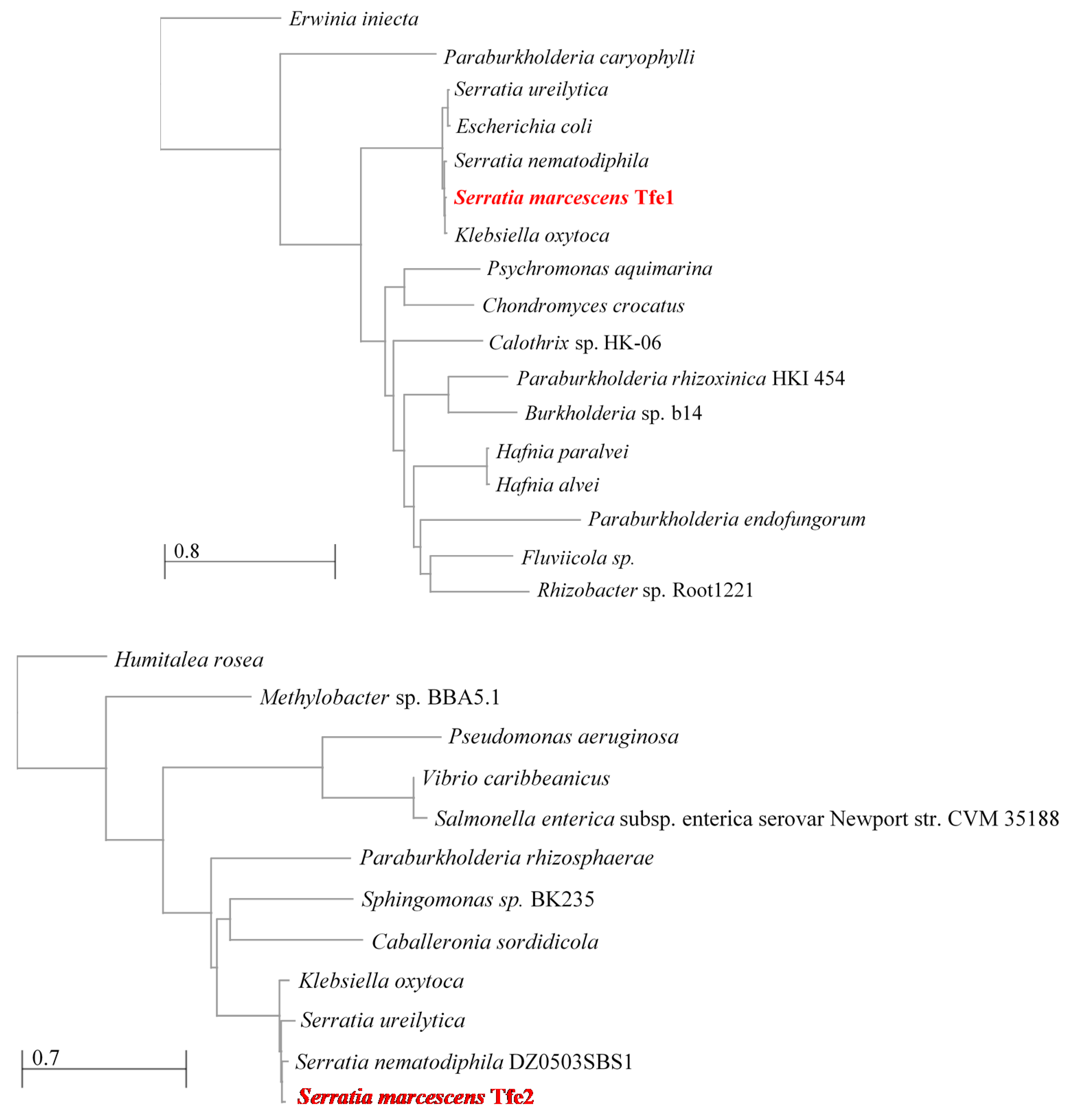
4. Identification of T6SS Antifungal Effectors
5. Mode of Action of Tfe1 and Tfe2
6. Prevalence of T6SS-Dependent Antifungal Activity
7. Roles for Antifungal T6SSs in Polymicrobial Communities?
8. Outstanding Questions
- How many bacteria with T6SSs can use these systems against fungal cells? It will also be interesting to see if T6SSs dedicated to antifungal activity exist, perhaps regulated in response to fungal cues, or if antifungal and antibacterial activity always co-occurs.
- How broad is fungal susceptibility to T6SS action? It remains to be seen whether other species, genera and phyla can be targeted by bacterial T6SSs and whether particular T6SSs and/or effector proteins are specific for different types of fungi. For example, it is unknown if true filamentous fungi are susceptible to T6SS action. It is tempting to speculate that the composition of the fungal cell wall may play a critical role in determining the efficacy of T6SS attacks.
- What are the precise modes of action of Tfe1 and Tfe2, and yet-to-be-identified T6SS antifungal effectors, and how many toxin molecules are required to cause fungal death? It will be very interesting to discover the range of activities such effectors might have and to determine whether any effectors that act against fungal cells also act against higher eukaryotic host organisms. Indeed we speculate that some of the effectors reported to act against host cells might have originally been acquired to act against fungal competitors, as targets such as actin are conserved throughout the eukaryotic kingdom.
- Can bacteria also use T6SSs to deliver effector proteins that promote positive, mutualistic interactions between bacterial and fungal cells, rather than being solely antagonistic?
- What is the significance of T6SS-mediated bacterial-fungal interactions in “real-life” polymicrobial communities? We look forward to learning how these interactions can change the balance between health and disease, influence the gut microbiota or define the composition of environmentally-important communities.
Author Contributions
Funding
Conflicts of Interest
References
- Peleg, A.Y.; Hogan, D.A.; Mylonakis, E. Medically important bacterial-fungal interactions. Nat. Rev. Microbiol. 2010, 8, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Azoulay, E.; Timsit, J.F.; Tafflet, M.; de Lassence, A.; Darmon, M.; Zahar, J.R.; Adrie, C.; Garrouste-Orgeas, M.; Cohen, Y.; Mourvillier, B.; et al. Candida colonization of the respiratory tract and subsequent Pseudomonas ventilator-associated pneumonia. Chest 2006, 129, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Klaerner, H.G.; Uknis, M.E.; Acton, R.D.; Dahlberg, P.S.; Carlone-Jambor, C.; Dunn, D.L. Candida albicans and Escherichia coli are synergistic pathogens during experimental microbial peritonitis. J. Surg. Res. 1997, 70, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.M.; Noverr, M.C. Candida albicans-Staphylococcus aureus polymicrobial peritonitis modulates host innate immunity. Infect. Immun. 2013, 81, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.P.; Cowley, E.S.; Nobile, C.J.; Hartooni, N.; Newman, D.K.; Johnson, A.D. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr. Biol. 2014, 24, 2411–2416. [Google Scholar] [CrossRef] [PubMed]
- Harriott, M.M.; Noverr, M.C. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: Effects on antimicrobial resistance. Antimicrob. Agents Chemother. 2009, 53, 3914–3922. [Google Scholar] [CrossRef] [PubMed]
- Tampakakis, E.; Peleg, A.Y.; Mylonakis, E. Interaction of Candida albicans with an intestinal pathogen, Salmonella enterica Serovar Typhimurium. Eukaryot. Cell 2009, 8, 732–737. [Google Scholar] [CrossRef]
- Hogan, D.A.; Kolter, R. Pseudomonas-Candida interactions: An ecological role for virulence factors. Science 2002, 296, 2229–2232. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Tampakakis, E.; Fuchs, B.B.; Eliopoulos, G.M.; Moellering, R.C., Jr.; Mylonakis, E. Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2008, 105, 14585–14590. [Google Scholar] [CrossRef]
- Hogan, D.A.; Vik, A.; Kolter, R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 2004, 54, 1212–1223. [Google Scholar] [CrossRef]
- Trunk, K.; Peltier, J.; Liu, Y.C.; Dill, B.D.; Walker, L.; Gow, N.A.R.; Stark, M.J.R.; Quinn, J.; Strahl, H.; Trost, M.; et al. The type VI secretion system deploys antifungal effectors against microbial competitors. Nat. Microbiol. 2018, 3, 920–931. [Google Scholar] [CrossRef] [PubMed]
- Coulthurst, S. The type VI secretion system: A versatile bacterial weapon. Microbiology 2019, 165, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Bingle, L.E.; Bailey, C.M.; Pallen, M.J. Type VI secretion: A beginner’s guide. Curr. Opin. Microbiol. 2008, 11, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Boyer, F.; Fichant, G.; Berthod, J.; Vandenbrouck, Y.; Attree, I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: What can be learned from available microbial genomic resources? BMC Genom. 2009, 10, 104. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.B.; Wexler, A.G.; Harding, B.N.; Whitney, J.C.; Bohn, A.J.; Goo, Y.A.; Tran, B.Q.; Barry, N.A.; Zheng, H.; Peterson, S.B.; et al. A type VI secretion-related pathway in bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 2014, 16, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Verster, A.J.; Ross, B.D.; Radey, M.C.; Bao, Y.; Goodman, A.L.; Mougous, J.D.; Borenstein, E. The landscape of type VI secretion across human gut microbiomes reveals its role in community composition. Cell Host Microbe 2017, 22, 411–419. [Google Scholar] [CrossRef] [PubMed]
- MacIntyre, D.L.; Miyata, S.T.; Kitaoka, M.; Pukatzki, S. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. USA 2010, 107, 19520–19524. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; West, T.E.; Boyer, F.; Chiang, W.C.; Carl, M.A.; Hood, R.D.; Rohmer, L.; Tolker-Nielsen, T.; Skerrett, S.J.; Mougous, J.D. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 2010, 6, e1001068. [Google Scholar] [CrossRef] [PubMed]
- Si, M.; Zhao, C.; Burkinshaw, B.; Zhang, B.; Wei, D.; Wang, Y.; Dong, T.G.; Shen, X. Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc. Natl. Acad. Sci. USA 2017, 114, E2233–E2242. [Google Scholar] [CrossRef]
- Flaugnatti, N.; Le, T.T.; Canaan, S.; Aschtgen, M.S.; Nguyen, V.S.; Blangy, S.; Kellenberger, C.; Roussel, A.; Cambillau, C.; Cascales, E.; et al. A phospholipase A1 antibacterial type VI secretion effector interacts directly with the C-terminal domain of the VGRG spike protein for delivery. Mol. Microbiol. 2016, 99, 1099–1118. [Google Scholar] [CrossRef]
- Sana, T.G.; Berni, B.; Bleves, S. The T6SSS of Pseudomonas aeruginosa strain PAO1 and their effectors: Beyond bacterial-cell targeting. Front. Cell. Infect. Microbiol. 2016, 6, 61. [Google Scholar] [CrossRef] [PubMed]
- Sana, T.G.; Flaugnatti, N.; Lugo, K.A.; Lam, L.H.; Jacobson, A.; Baylot, V.; Durand, E.; Journet, L.; Cascales, E.; Monack, D.M. Salmonella typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc. Natl. Acad. Sci. USA 2016, 113, E5044–E5051. [Google Scholar] [CrossRef] [PubMed]
- Spiewak, H.L.; Shastri, S.; Zhang, L.; Schwager, S.; Eberl, L.; Vergunst, A.C.; Thomas, M.S. Burkholderia cenocepacia utilizes a type VI secretion system for bacterial competition. MicrobiologyOpen 2019, e774. [Google Scholar] [CrossRef] [PubMed]
- Weber, B.S.; Hennon, S.W.; Wright, M.S.; Scott, N.E.; de Berardinis, V.; Foster, L.J.; Ayala, J.A.; Adams, M.D.; Feldman, M.F. Genetic dissection of the type VI secretion system in acinetobacter and identification of a novel peptidoglycan hydrolase, TagX, required for its biogenesis. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Bernal, P.; Llamas, M.A.; Filloux, A. Type VI secretion systems in plant-associated bacteria. Environ. Microbiol. 2018, 20, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Brackmann, M.; Nazarov, S.; Wang, J.; Basler, M. Using force to punch holes: Mechanics of contractile nanomachines. Trends Cell Biol. 2017, 27, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Cianfanelli, F.R.; Monlezun, L.; Coulthurst, S.J. Aim, load, fire: The type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 2016, 24, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Clemens, D.L.; Lee, B.Y.; Horwitz, M.A. The francisella type VI secretion system. Front. Cell. Infect. Microbiol. 2018, 8, 121. [Google Scholar] [CrossRef]
- Nguyen, V.S.; Douzi, B.; Durand, E.; Roussel, A.; Cascales, E.; Cambillau, C. Towards a complete structural deciphering of type VI secretion system. Curr. Opin. Struct. Biol. 2018, 49, 77–84. [Google Scholar] [CrossRef]
- Hachani, A.; Wood, T.E.; Filloux, A. Type VI secretion and anti-host effectors. Curr. Opin. Microbiol. 2016, 29, 81–93. [Google Scholar] [CrossRef]
- Russell, A.B.; Peterson, S.B.; Mougous, J.D. Type VI secretion system effectors: Poisons with a purpose. Nat. Rev. Microbiol. 2014, 12, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Boer, W.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef] [PubMed]
- Barret, M.; Frey-Klett, P.; Guillerm-Erckelboudt, A.Y.; Boutin, M.; Guernec, G.; Sarniguet, A. Effect of wheat roots infected with the pathogenic fungus Gaeumannomyces graminis var. Tritici on gene expression of the biocontrol bacterium Pseudomonas fluorescens Pf29Arp. Mol. Plant Microbe Interact. 2009, 22, 1611–1623. [Google Scholar] [CrossRef] [PubMed]
- Marchi, M.; Boutin, M.; Gazengel, K.; Rispe, C.; Gauthier, J.P.; Guillerm-Erckelboudt, A.Y.; Lebreton, L.; Barret, M.; Daval, S.; Sarniguet, A. Genomic analysis of the biocontrol strain Pseudomonas fluorescens Pf29Arp with evidence of T3SS and T6SS gene expression on plant roots. Environ. Microbiol. Rep. 2013, 5, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Haapalainen, M.; Mosorin, H.; Dorati, F.; Wu, R.F.; Roine, E.; Taira, S.; Nissinen, R.; Mattinen, L.; Jackson, R.; Pirhonen, M.; et al. Hcp2, a secreted protein of the phytopathogen Pseudomonas syringae pv. Tomato DC3000, is required for fitness for competition against bacteria and yeasts. J. Bacteriol. 2012, 194, 4810–4822. [Google Scholar] [CrossRef] [PubMed]
- Arnoldo, A.; Curak, J.; Kittanakom, S.; Chevelev, I.; Lee, V.T.; Sahebol-Amri, M.; Koscik, B.; Ljuma, L.; Roy, P.J.; Bedalov, A.; et al. Identification of small molecule inhibitors of Pseudomonas aeruginosa exoenzyme s using a yeast phenotypic screen. PLoS Genet. 2008, 4, e1000005. [Google Scholar] [CrossRef]
- Hood, R.D.; Singh, P.; Hsu, F.; Guvener, T.; Carl, M.A.; Trinidad, R.R.; Silverman, J.M.; Ohlson, B.B.; Hicks, K.G.; Plemel, R.L.; et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 2010, 7, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Robb, C.S.; Robb, M.; Nano, F.E.; Boraston, A.B. The structure of the toxin and type six secretion system substrate Tse2 in complex with its immunity protein. Structure 2016, 24, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Murdoch, S.L.; Trunk, K.; English, G.; Fritsch, M.J.; Pourkarimi, E.; Coulthurst, S.J. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J. Bacteriol. 2011, 193, 6057–6069. [Google Scholar] [CrossRef]
- Fritsch, M.J.; Trunk, K.; Diniz, J.A.; Guo, M.; Trost, M.; Coulthurst, S.J. Proteomic identification of novel secreted antibacterial toxins of the Serratia marcescens type VI secretion system. Mol. Cell. Proteom. 2013, 12, 2735–2749. [Google Scholar] [CrossRef]
- Durand, E.; Nguyen, V.S.; Zoued, A.; Logger, L.; Pehau-Arnaudet, G.; Aschtgen, M.S.; Spinelli, S.; Desmyter, A.; Bardiaux, B.; Dujeancourt, A.; et al. Biogenesis and structure of a type VI secretion membrane core complex. Nature 2015, 523, 555–560. [Google Scholar] [CrossRef]
- Wang, J.; Brackmann, M.; Castano-Diez, D.; Kudryashev, M.; Goldie, K.N.; Maier, T.; Stahlberg, H.; Basler, M. Cryo-EM structure of the extended type VI secretion system sheath-tube complex. Nat. Microbiol. 2017, 2, 1507–1512. [Google Scholar] [CrossRef]
- Cherrak, Y.; Rapisarda, C.; Pellarin, R.; Bouvier, G.; Bardiaux, B.; Allain, F.; Malosse, C.; Rey, M.; Chamot-Rooke, J.; Cascales, E.; et al. Biogenesis and structure of a type VI secretion baseplate. Nat. Microbiol. 2018, 3, 1404–1416. [Google Scholar] [CrossRef]
- Santin, Y.G.; Doan, T.; Lebrun, R.; Espinosa, L.; Journet, L.; Cascales, E. In vivo TSSA proximity labelling during type VI secretion biogenesis reveals TagA as a protein that stops and holds the sheath. Nat. Microbiol. 2018, 3, 1304–1313. [Google Scholar] [CrossRef]
- Dong, T.G.; Ho, B.T.; Yoder-Himes, D.R.; Mekalanos, J.J. Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2013, 110, 2623–2628. [Google Scholar] [CrossRef]
- Russell, A.B.; Singh, P.; Brittnacher, M.; Bui, N.K.; Hood, R.D.; Carl, M.A.; Agnello, D.M.; Schwarz, S.; Goodlett, D.R.; Vollmer, W.; et al. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 2012, 11, 538–549. [Google Scholar] [CrossRef]
- Russell, A.B.; LeRoux, M.; Hathazi, K.; Agnello, D.M.; Ishikawa, T.; Wiggins, P.A.; Wai, S.N.; Mougous, J.D. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 2013, 496, 508–512. [Google Scholar] [CrossRef]
- Salomon, D.; Kinch, L.N.; Trudgian, D.C.; Guo, X.; Klimko, J.A.; Grishin, N.V.; Mirzaei, H.; Orth, K. Marker for type VI secretion system effectors. Proc. Natl. Acad. Sci. USA 2014, 111, 9271–9276. [Google Scholar] [CrossRef]
- Mougous, J.D.; Cuff, M.E.; Raunser, S.; Shen, A.; Zhou, M.; Gifford, C.A.; Goodman, A.L.; Joachimiak, G.; Ordonez, C.L.; Lory, S.; et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 2006, 312, 1526–1530. [Google Scholar] [CrossRef]
- Salomon, D.; Gonzalez, H.; Updegraff, B.L.; Orth, K. Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS ONE 2013, 8, e61086. [Google Scholar] [CrossRef]
- Mougous, J.D.; Gifford, C.A.; Ramsdell, T.L.; Mekalanos, J.J. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell Biol. 2007, 9, 797–803. [Google Scholar] [CrossRef]
- Bachmann, V.; Kostiuk, B.; Unterweger, D.; Diaz-Satizabal, L.; Ogg, S.; Pukatzki, S. Bile salts modulate the mucin-activated type VI secretion system of pandemic Vibrio cholerae. PLoS Negl. Trop. Dis. 2015, 9, e0004031. [Google Scholar] [CrossRef]
- Serrano, R.; Kielland-Brandt, M.C.; Fink, G.R. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-atpases. Nature 1986, 319, 689–693. [Google Scholar] [CrossRef]
- Kjellerup, L.; Gordon, S.; Cohrt, K.O.; Brown, W.D.; Fuglsang, A.T.; Winther, A.L. Identification of antifungal H+-ATPase inhibitors with effect on plasma membrane potential. Antimicrob. Agents Chemother. 2017, 61, e00032-17. [Google Scholar] [CrossRef]
- Maresova, L.; Urbankova, E.; Gaskova, D.; Sychrova, H. Measurements of plasma membrane potential changes in Saccharomyces cerevisiae cells reveal the importance of the TOK1 channel in membrane potential maintenance. FEMS Yeast Res. 2006, 6, 1039–1046. [Google Scholar] [CrossRef]
- Hinnebusch, A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005, 59, 407–450. [Google Scholar] [CrossRef]
- Tripathi, G.; Wiltshire, C.; Macaskill, S.; Tournu, H.; Budge, S.; Brown, A.J. Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 2002, 21, 5448–5456. [Google Scholar] [CrossRef]
- Reggiori, F.; Klionsky, D.J. Autophagic processes in yeast: Mechanism, machinery and regulation. Genetics 2013, 194, 341–361. [Google Scholar] [CrossRef]
- Deutscher, D.; Meilijson, I.; Kupiec, M.; Ruppin, E. Multiple knockout analysis of genetic robustness in the yeast metabolic network. Nat. Genet. 2006, 38, 993–998. [Google Scholar] [CrossRef]
- Sopko, R.; Huang, D.; Preston, N.; Chua, G.; Papp, B.; Kafadar, K.; Snyder, M.; Oliver, S.G.; Cyert, M.; Hughes, T.R.; et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell 2006, 21, 319–330. [Google Scholar] [CrossRef]
- Heisler, D.B.; Kudryashova, E.; Grinevich, D.O.; Suarez, C.; Winkelman, J.D.; Birukov, K.G.; Kotha, S.R.; Parinandi, N.L.; Vavylonis, D.; Kovar, D.R.; et al. Actin-directed toxin. ACD toxin-produced actin oligomers poison formin-controlled actin polymerization. Science 2015, 349, 535–539. [Google Scholar] [CrossRef]
- Suarez, G.; Sierra, J.C.; Erova, T.E.; Sha, J.; Horneman, A.J.; Chopra, A.K. A type VI secretion system effector protein, VGRG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 2010, 192, 155–168. [Google Scholar] [CrossRef]
- Aubert, D.F.; Xu, H.; Yang, J.; Shi, X.; Gao, W.; Li, L.; Bisaro, F.; Chen, S.; Valvano, M.A.; Shao, F. A burkholderia type VI effector deamidates Rho GTPases to activate the pyrin inflammasome and trigger inflammation. Cell Host Microbe 2016, 19, 664–674. [Google Scholar] [CrossRef]
- Coyne, M.J.; Roelofs, K.G.; Comstock, L.E. Type VI secretion systems of human gut bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genom. 2016, 17, 58. [Google Scholar] [CrossRef]
- Anderson, M.C.; Vonaesch, P.; Saffarian, A.; Marteyn, B.S.; Sansonetti, P.J. Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host Microbe 2017, 21, 769–776. [Google Scholar] [CrossRef]
- Zhao, W.; Caro, F.; Robins, W.; Mekalanos, J.J. Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science 2018, 359, 210–213. [Google Scholar] [CrossRef]
- Chatzidaki-Livanis, M.; Geva-Zatorsky, N.; Comstock, L.E. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc. Natl. Acad. Sci. USA 2016, 113, 3627–3632. [Google Scholar] [CrossRef]
- Mahlen, S.D. Serratia infections: From military experiments to current practice. Clin. Microbiol. Rev. 2011, 24, 755–791. [Google Scholar] [CrossRef]
- Hoarau, G.; Mukherjee, P.K.; Gower-Rousseau, C.; Hager, C.; Chandra, J.; Retuerto, M.A.; Neut, C.; Vermeire, S.; Clemente, J.; Colombel, J.F.; et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s Disease. mBio 2016, 7. [Google Scholar] [CrossRef]
- Limon, J.J.; Skalski, J.H.; Underhill, D.M. Commensal fungi in health and disease. Cell Host Microbe 2017, 22, 156–165. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trunk, K.; Coulthurst, S.J.; Quinn, J. A New Front in Microbial Warfare—Delivery of Antifungal Effectors by the Type VI Secretion System. J. Fungi 2019, 5, 50. https://doi.org/10.3390/jof5020050
Trunk K, Coulthurst SJ, Quinn J. A New Front in Microbial Warfare—Delivery of Antifungal Effectors by the Type VI Secretion System. Journal of Fungi. 2019; 5(2):50. https://doi.org/10.3390/jof5020050
Chicago/Turabian StyleTrunk, Katharina, Sarah J. Coulthurst, and Janet Quinn. 2019. "A New Front in Microbial Warfare—Delivery of Antifungal Effectors by the Type VI Secretion System" Journal of Fungi 5, no. 2: 50. https://doi.org/10.3390/jof5020050
APA StyleTrunk, K., Coulthurst, S. J., & Quinn, J. (2019). A New Front in Microbial Warfare—Delivery of Antifungal Effectors by the Type VI Secretion System. Journal of Fungi, 5(2), 50. https://doi.org/10.3390/jof5020050




