Diversity of Culturable Fungi in Two-Phase Olive Mill Waste, a Preliminary Evaluation of Their Enzymatic Potential, and Two New Trichoderma Species
Abstract
1. Introduction
2. Materials and Methods
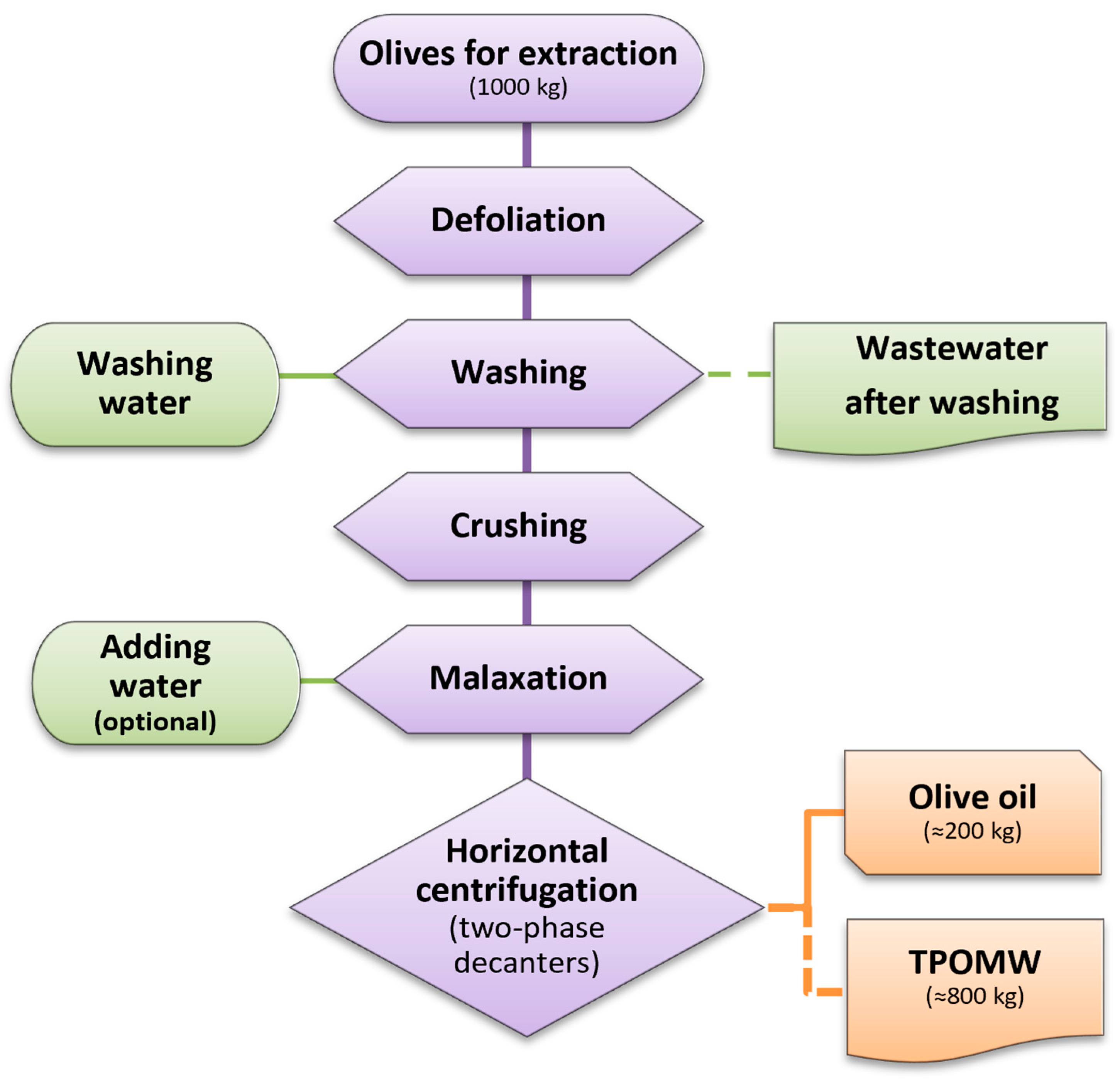
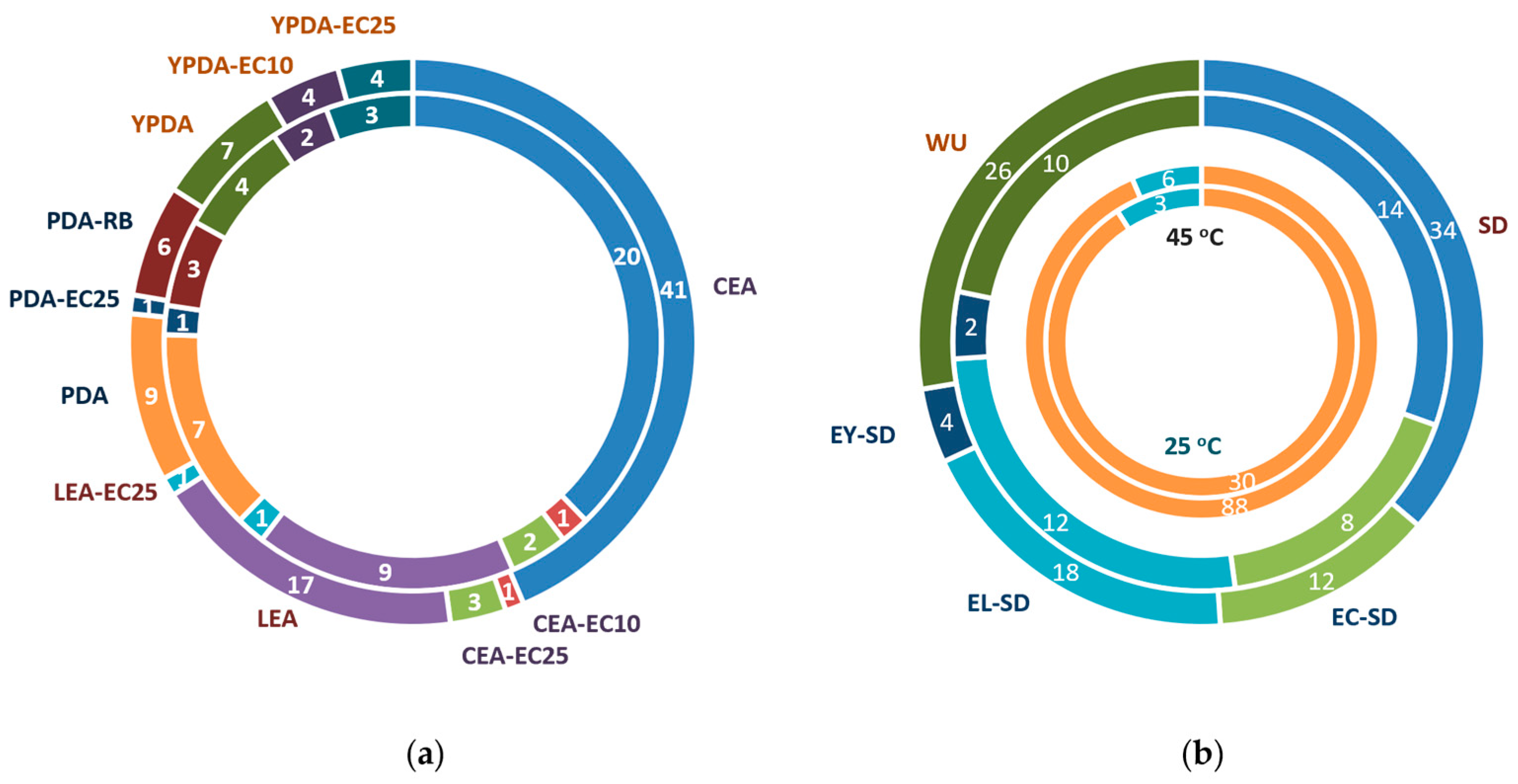
2.1. Fungal Isolation from TPOMW, Selection, and Cultivation Conditions
2.2. DNA Extraction, PCR, and Sequencing
2.3. Phylogenetic Analyses
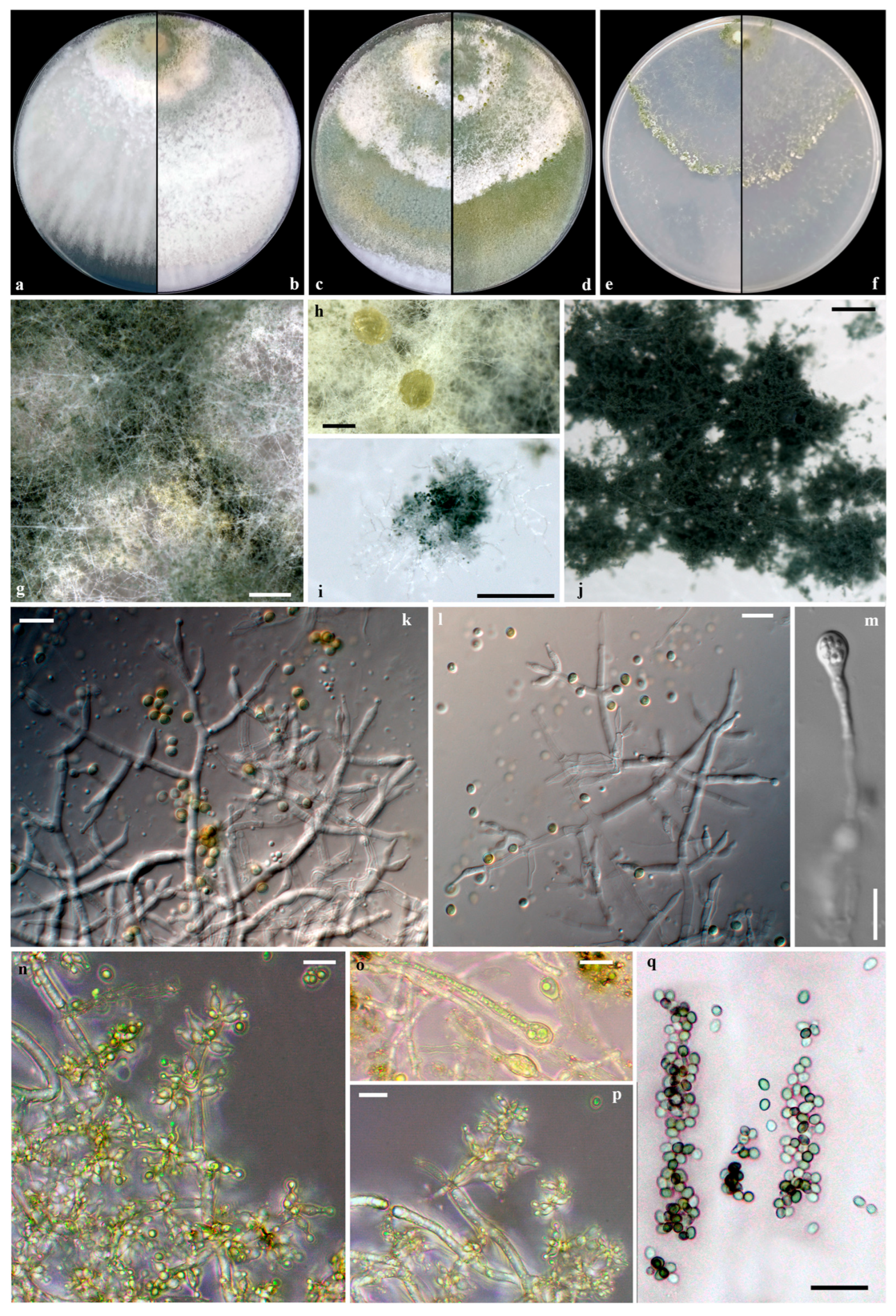
2.4. Morphological Characterization of Isolated Fungi
2.5. Enzyme Activity and Biodegradation Potential
3. Results
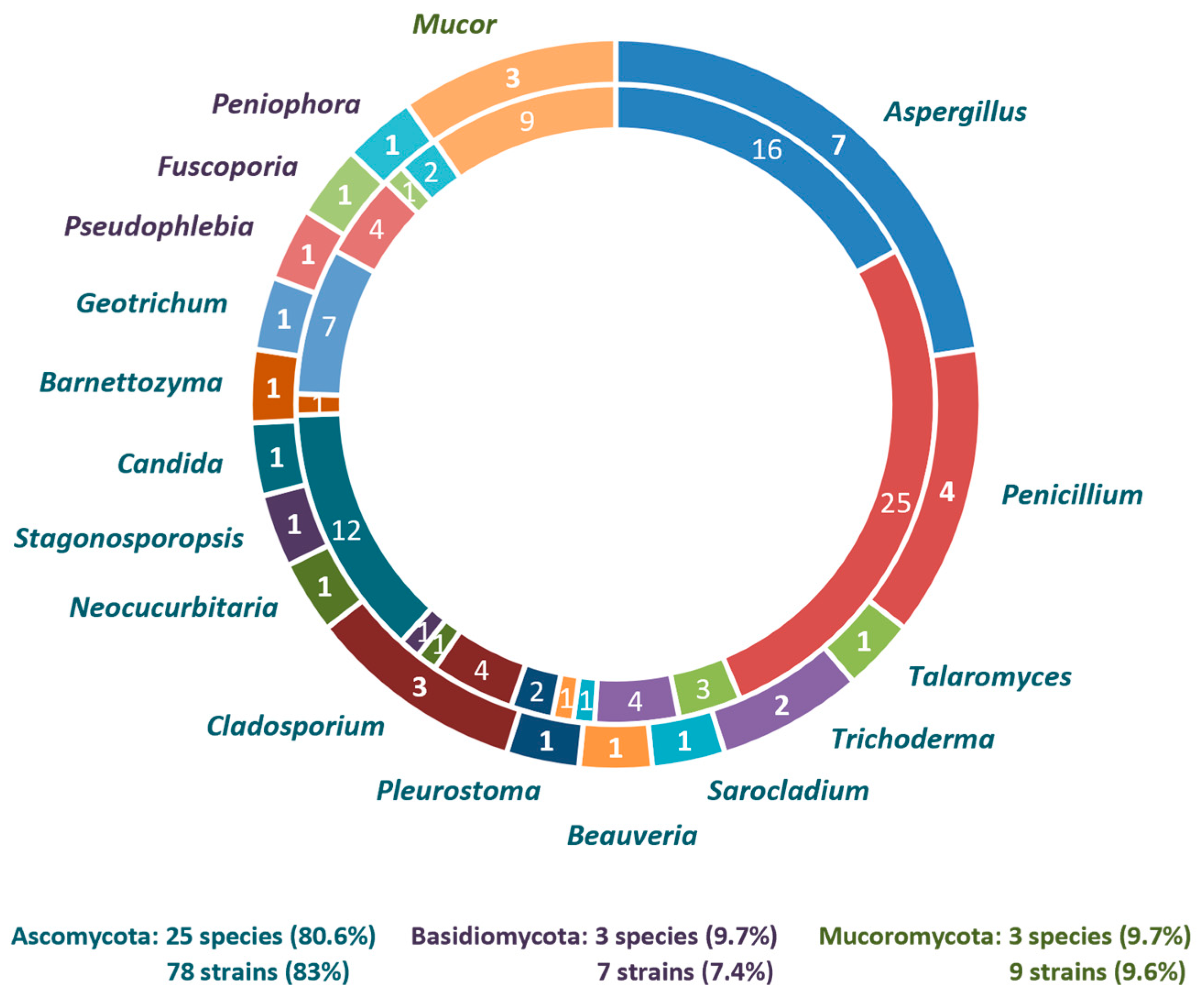
3.1. Diversity and Phylogeny of Fungi from TPOMW
Taxonomy
3.2. Preliminary Evaluation of Biodegradation Efficacies of Isolated Fungi
4. Discussion
4.1. Fungal Diversity in TPOMW
4.2. General Taxonomic and Phylogenetic Remarks on the Genus Trichoderma
4.3. Biodegradation Potential of Fungal Strains
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ntougias, S.; Bourtzis, K.; Tsiamis, G. The Microbiology of Olive Mill Wastes. BioMed Res. Int. 2013, 2013, 784591. [Google Scholar] [CrossRef]
- Souilem, S.; El-Abbassi, A.; Kiai, H.; Hafidi, A.; Sayadi, S.; Galanakis, C.M. Olive Oil Production Sector: Environmental Effects and Sustainability Challenges. In Olive Mill Waste; Galanakis, C.M., Ed.; Academic Press: London, UK, 2017; pp. 1–28. [Google Scholar] [CrossRef]
- Justino, C.I.; Pereira, R.; Freitas, A.C.; Rocha-Santos, T.A.; Panteleitchouk, T.S.; Duarte, A.C. Olive oil mill wastewaters before and after treatment: A critical review from the ecotoxicological point of view. Ecotoxicology 2012, 21, 615–629. [Google Scholar] [CrossRef]
- Martínez-Gallardo, M.R.; López, M.J.; López-González, J.A.; Jurado, M.M.; Suárez-Estrella, F.; Pérez-Murcia, M.D.; Sáez, J.A.; Moral, R.; Moreno, J. Microbial communities of the olive mill wastewater sludge stored in evaporation ponds: The resource for sustainable bioremediation. J. Environ. Manag. 2021, 279, 111810. [Google Scholar] [CrossRef] [PubMed]
- Morillo, J.A.; Antizar-Ladislao, B.; Monteoliva-Sánchez, M.; Ramos-Cormenzana, A.; Russell, N.J. Bioremediation and biovalorisation of olive-mill wastes. Appl. Microbiol. Biotechnol. 2009, 82, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Romero-García, J.M.; Niño, L.; Martínez-Patiño, C.; Álvarez, C.; Castro, E.; Negro, M.J. Biorefinery based on olive biomass: State of the art and future trends. Bioresour. Technol. 2014, 159, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Sánchez, C.; González-González, A.; Cuadros-Salcedo, F.; Cuadros-Blázquez, F. Two-phase olive mill waste: A circular economy solution to an imminent problem in Southern Europe. J. Clean. Prod. 2020, 274, 122789. [Google Scholar] [CrossRef]
- Podgornik, M.; Bučar-Miklavčič, M.; Levart, A.; Salobir, J.; Rezar, V.; Butinar, B. Chemical characteristics of two-phase olive-mill waste and evaluation of their direct soil application in humid Mediterranean regions. Agronomy 2022, 12, 1621. [Google Scholar] [CrossRef]
- Roig, A.; Cayuela, M.L.; Sánchez-Monedero, M.A. An overview on olive mill wastes and their valorisation methods. Waste Manag. 2006, 26, 960–969. [Google Scholar] [CrossRef] [PubMed]
- Ntougias, S.; Baldrian, P.; Ehaliotis, C.; Nerud, F.; Merhautová, V.; Zervakis, G.I. Olive mill wastewater biodegradation potential of white-rot fungi–Mode of action of fungal culture extracts and effects of ligninolytic enzymes. Bioresour. Technol. 2015, 189, 121–130. [Google Scholar] [CrossRef]
- Basak, B.; Saha, S.; Chatterjee, P.K.; Ganguly, A.; Chang, S.W.; Jeon, B.H. Pretreatment of polysaccharidic wastes with cellulolytic Aspergillus fumigatus for enhanced production of biohythane in a dual-stage process. Bioresour. Technol. 2020, 299, 122592. [Google Scholar] [CrossRef]
- Öngen, G.; Güngör, G.; Kanberoglu, B. Decolourisation and dephenolisation potential of selected Aspergillus section Nigri strains–Aspergillus tubingensis in olive mill wastewater. World J. Microbiol. Biotechnol. 2007, 23, 519–524. [Google Scholar] [CrossRef]
- Rodríguez Márquez, M.; Rodríguez Gutiérrez, G.; Giménez, M.; Rizzo, P.F.; Bueno, L.; Deiana, C.; Monetta, P. Obtaining phenolic-enriched liquid fractions and compostable pomace for agriculture from alperujo using standard two-phase olive oil mill equipment. Agriculture 2024, 14, 1427. [Google Scholar] [CrossRef]
- Campaniello, D.; Speranza, B.; Altieri, C.; Sinigaglia, M.; Bevilacqua, A.; Corbo, M.R. Removal of phenols in table olive processing wastewater by using a mixed inoculum of Candida boidinii and Bacillus pumilus: Effects of inoculation dynamics, temperature, pH, and effluent age on the abatement efficiency. Microorganisms 2021, 9, 1783. [Google Scholar] [CrossRef]
- Baffi, M.A.; Romo-Sánchez, S.; Úbeda-Iranzo, J.; Briones-Pérez, A.I. Fungi isolated from olive ecosystems and screening of their potential biotechnological use. New Biotechnol. 2012, 29, 451–456. [Google Scholar] [CrossRef]
- Bavaro, S.L.; Susca, A.; Frisvad, J.C.; Tufariello, M.; Chytiri, A.; Perrone, G.; Mita, G.; Logrieco, A.F.; Bleve, G. Isolation, characterization, and selection of molds associated to fermented black table olives. Front. Microbiol. 2017, 8, 1356. [Google Scholar] [CrossRef]
- Slama, H.B.; Chenari Bouket, A.; Alenezi, F.N.; Khardani, A.; Luptakova, L.; Vallat, A.; Oszako, T.; Rateb, M.E.; Belbahri, L. Olive mill and olive pomace evaporation pond’s by-products: Toxic level determination and role of indigenous microbiota in toxicity alleviation. Appl. Sci. 2021, 11, 5131. [Google Scholar] [CrossRef]
- Lamrani, K.; Lakhtar, H.; Ismaili-Alaoui, M.; Ettalibi, M.; Boiron, P.; Augur, C.; Gaime-Perraud, I.; Roussos, S. Production of fumagillin by Aspergillus fumigatus isolated from traditional trituration units, “Maasra”, in Morocco. Micol. Apl. Int. 2008, 20, 35–41. [Google Scholar]
- Bouhia, Y.; Hafidi, M.; Ouhdouch, Y.; El Boukhari, M.E.M.; El Fels, L.; Zeroual, Y.; Lyamlouli, K. Microbial community succession and organic pollutants removal during olive mill waste sludge and green waste co-composting. Front. Microbiol. 2022, 12, 814553. [Google Scholar] [CrossRef]
- Giannoutsou, E.P.; Meintanis, C.; Karagouni, A.D. Identification of yeast strains isolated from a two-phase decanter system olive oil waste and investigation of their ability for its fermentation. Bioresour. Technol. 2004, 93, 301–306. [Google Scholar] [CrossRef]
- Morillo, J.A.; Aguilera, M.; Antízar-Ladislao, B.; Fuentes, S.; Ramos-Cormenzana, A.; Russell, N.J.; Monteoliva-Sánchez, M. Molecular microbial and chemical investigation of the bioremediation of two-phase olive mill waste using laboratory-scale bioreactors. Appl. Microbiol. Biotechnol. 2008, 79, 309–317. [Google Scholar] [CrossRef]
- Tortosa, G.; Torralbo, F.; Maza-Márquez, P.; Aranda, E.; Calvo, C.; González-Murua, C.; Bedmar, E.J. Assessment of the diversity and abundance of the total and active fungal population and its correlation with humification during two-phase olive mill waste (“alperujo”) composting. Bioresour. Technol. 2020, 295, 122267. [Google Scholar] [CrossRef]
- Kee, S.H.; Chiongson, J.B.V.; Saludes, J.P.; Vigneswari, S.; Ramakrishna, S.; Bhubalan, K. Bioconversion of agro-industry sourced biowaste into biomaterials via microbial factories–A viable domain of circular economy. Environ. Pollut. 2021, 271, 116311. [Google Scholar] [CrossRef]
- Mann, J.; Markham, J.L.; Peiris, P.; Nair, N.; Spooner-Hart, R.N.; Holford, P. Screening and selection of fungi for bioremediation of olive mill wastewater. World J. Microbiol. Biotechnol. 2010, 26, 567–571. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Minh, B.Q.; Lanfear, R.; Ly-Trong, N.; Trifinopoulos, J.; Schrempf, D.; Schmidt, H.A. IQ-TREE version 2.2.0: Tutorials and manual phylogenomic software by maximum likelihood. Nucleic Acids Res. 2022, 44, W232–W235. [Google Scholar] [CrossRef]
- Miller, M.A.; Schwartz, T.; Pickett, B.E.; He, S.; Klem, E.B.; Scheuermann, R.H.; Passarotti, M.; Kaufman, S.; O’Leary, M.A. A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evol. Bioinform. 2015, 11, EBO-S21501. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Saroj, P.; Narasimhulu, K. Characterization of thermophilic fungi producing extracellular lignocellulolytic enzymes for lignocellulosic hydrolysis under solid-state fermentation. Bioresour. Bioprocess. 2018, 5, 31. [Google Scholar] [CrossRef]
- Eichlerová, I.; Baldrian, P. Ligninolytic Enzyme Production and Decolorization Capacity of Synthetic Dyes by Saprotrophic White Rot, Brown Rot, and Litter Decomposing Basidiomycetes. J. Fungi 2020, 6, 301. [Google Scholar] [CrossRef]
- Walther, G.; Pawłowska, J.; Alastruey-Izquierdo, A.; Wrzosek, M.; Rodriguez-Tudela, J.L.; Dolatabadi, S.; Chakrabarti, A.; De Hoog, G.S. DNA barcoding in Mucorales: An inventory of biodiversity. Persoonia 2013, 30, 11–47. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Moreno, G.; Blanco, M.N.; Checa, J.; Platas, G.; Peláez, F. Taxonomic and phylogenetic revision of three rare irpicoid species within the Meruliaceae. Mycol. Prog. 2011, 10, 481–491. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J. Relationships among genera of the Saccharomycotina (Ascomycota) from multigene phylogenetic analysis of type species. FEMS Yeast Res. 2013, 13, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.Y.; Shang, Y.J.; Wei, X.Y.; Groenewald, M.; Robert, V.; Zhang, R.P.; Li, A.H.; Han, P.J.; Ji, F.; Li, J.N.; et al. Taxonomic revision of Geotrichum and Magnusiomyces, with the descriptions of five new Geotrichum species from China. Mycology 2024, 15, 400–423. [Google Scholar] [CrossRef]
- Sun, X.R.; Xu, M.Y.; Kong, W.L.; Wu, F.; Zhang, Y.; Xie, X.L.; Li, D.W.; Wu, X.Q. Fine identification and classification of a novel beneficial Talaromyces fungal species from Masson pine rhizosphere soil. J. Fungi 2022, 8, 155. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Qiao, M.; Lv, Y.; Du, X.; Zhang, K.Q.; Yu, Z. New species of Trichoderma isolated as endophytes and saprobes from Southwest China. J. Fungi 2021, 7, 467. [Google Scholar] [CrossRef]
- Li, Q.R.; Tan, P.; Jiang, Y.L.; Hyde, K.D.; Mckenzie, E.H.; Bahkali, A.H.; Kang, J.C.; Wang, Y. A novel Trichoderma species isolated from soil in Guizhou, T. guizhouense. Mycol. Prog. 2013, 12, 167–172. [Google Scholar] [CrossRef]
- Cao, Z.J.; Qin, W.T.; Zhao, J.; Liu, Y.; Wang, S.X.; Zheng, S.Y. Three new Trichoderma species in Harzianum clade associated with the contaminated substrates of edible fungi. J. Fungi 2022, 8, 1154. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Voglmayr, H. Biodiversity of Trichoderma (Hypocreaceae) in southern Europe and Macaronesia. Stud. Mycol. 2015, 80, 1–87. [Google Scholar] [CrossRef]
- Robbertse, B.; Strope, P.K.; Chaverri, P.; Gazis, R.; Ciufo, S.; Domrachev, M.; Schoch, C.L. Improving taxonomic accuracy for fungi in public sequence databases: Applying ‘one name one species’ in well-defined genera with Trichoderma/Hypocrea as a test case. Database 2017, 2017, bax072. [Google Scholar] [CrossRef]
- Barrera, V.A.; Iannone, L.; Romero, A.I.; Chaverri, P. Expanding the Trichoderma harzianum species complex: Three new species from Argentine natural and cultivated ecosystems. Mycologia 2021, 113, 1136–1155. [Google Scholar] [CrossRef]
- Chaverri, P.; Branco-Rocha, F.; Jaklitsch, W.; Gazis, R.; Degenkolb, T.; Samuels, G.J. Systematics of the Trichoderma harzianum species complex and the re-identification of commercial biocontrol strains. Mycologia 2015, 107, 558–590. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.J.; Zhao, J.; Liu, Y.; Wang, S.X.; Zheng, S.Y.; Qin, W.T. Diversity of Trichoderma species associated with green mold contaminating substrates of Lentinula edodes and their interaction. Front. Microbiol. 2024, 14, 1288585. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhuang, W.Y. Trichoderma brevicrassum strain TC967 with capacities of diminishing cucumber disease caused by Rhizoctonia solani and promoting plant growth. Biol. Control 2020, 142, 104151. [Google Scholar] [CrossRef]
- Valenzuela-Lopez, N.; Cano-Lira, J.F.; Stchigel, A.M.; Rivero-Menendez, O.; Alastruey-Izquierdo, A.; Guarro, J. Neocucurbitaria keratinophila: An emerging opportunistic fungus causing superficial mycosis in Spain. Med. Mycol. 2019, 57, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Carlucci, A.; Raimondo, M.L.; Cibelli, F.; Phillips, A.J.; Lops, F. Pleurostomophora richardsiae, Neofusicoccum parvum and Phaeoacremonium aleophilum associated with a decline of olives in southern Italy. Phytopathol. Mediterr. 2013, 52, 517–527. [Google Scholar] [CrossRef]
- Garibaldi, A.; Tabone, G.; Luongo, I.; Gullino, M.L. First report of Stagonosporopsis ailanthicola causing leaf spot on Delphinium consolida in Italy. J. Plant Pathol. 2022, 104, 1553. [Google Scholar] [CrossRef]
- Chen, K.; Zhuang, W.Y. Discovery from a large-scaled survey of Trichoderma in soil of China. Sci. Rep. 2017, 7, 9090. [Google Scholar] [CrossRef]
- Cai, F.; Druzhinina, I.S. In honor of John Bissett: Authoritative guidelines on molecular identification of Trichoderma. Fungal Divers. 2021, 107, 1–69. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Mohamed, S.G.A.; Alsaadi, B.H.; Althubyani, M.M.; Awari, Z.I.; Hussein, H.G.A.; Aljohani, A.A.; Albasri, J.F.; Faraj, S.A.; Mohamed, G.A. Secondary metabolites, biological activities, and industrial and biotechnological importance of Aspergillus sydowii. Mar. Drugs 2023, 21, 441. [Google Scholar] [CrossRef]
- Yang, J.K.; Xiong, W.; Chen, F.Y.; Xu, L.; Han, Z.G. Aromatic amino acids in the cellulose binding domain of Penicillium crustosum endoglucanase EGL1 differentially contribute to the cellulose affinity of the enzyme. PLoS ONE 2017, 12, e0176444. [Google Scholar] [CrossRef]
- Marques, G.L.; dos Santos Reis, N.; Silva, T.P.; Ferreira, M.L.O.; Aguiar-Oliveira, E.; de Oliveira, J.R.; Franco, M. Production and characterisation of xylanase and endoglucanases produced by Penicillium roqueforti ATCC 10110 through the solid-state fermentation of rice husk residue. Waste Biomass Valorization 2018, 9, 2061–2069. [Google Scholar] [CrossRef]
- He, R.; Bai, X.; Cai, P.; Sun, C.; Zhang, D.; Chen, S. Genome sequence of Talaromyces piceus 9-3 provides insights into lignocellulose degradation. 3 Biotech 2017, 7, 368. [Google Scholar] [CrossRef]
- Zhang, X.; Kong, D.; Liu, X.; Xie, H.; Lou, X.; Zeng, C. Combined microbial degradation of crude oil under alkaline conditions by Acinetobacter baumannii and Talaromyces sp. Chemosphere 2021, 273, 129666. Chemosphere 2021, 273, 129666. [Google Scholar] [CrossRef] [PubMed]
- Tarayre, C.; Bauwens, J.; Brasseur, C.; Mattéotti, C.; Millet, C.; Guiot, P.A.; Destain, J.; Vandenbol, M.; Portetelle, D.; De Pauw, E.; et al. Isolation and cultivation of xylanolytic and cellulolytic Sarocladium kiliense and Trichoderma virens from the gut of the termite Reticulitermes santonensis. Environ. Sci. Pollut. Res. Int. 2015, 22, 4369–4382. [Google Scholar] [CrossRef] [PubMed]
- Isaie, M.; Padmavathi, T. Optimization of process parameters for biosynthesis of cellulase by Cladosporium cladosporioides using agro wastes. Int. J. Pharma Bio Sci. 2013, 4, B1129–B1138. [Google Scholar]
- Grujić, M.; Dojnov, B.; Potočnik, I.; Atanasova, L.; Duduk, B.; Srebotnik, E.; Druzhinina, I.S.; Kubicek, C.P.; Vujčić, Z. Superior cellulolytic activity of Trichoderma guizhouense on raw wheat straw. World J. Microbiol. Biotechnol. 2019, 35, 194. [Google Scholar] [CrossRef]
- Xia, Y.; Wang, J.; Guo, C.; Xu, H.; Wang, W.; Yang, M.; Shen, Q.; Zhang, R.; Miao, Y. Exploring the multi-level regulation of lignocellulases in the filamentous fungus Trichoderma guizhouense NJAU4742 from an omics perspective. Microb. Cell Factories 2022, 21, 144. [Google Scholar] [CrossRef]
- Liu, D.; Li, J.; Zhao, S.; Zhang, R.; Wang, M.; Miao, Y.; Shen, Y.; Shen, Q. Secretome diversity and quantitative analysis of cellulolytic Aspergillus fumigatus Z5 in the presence of different carbon sources. Biotechnol. Biofuels 2013, 6, 149. [Google Scholar] [CrossRef]
- Dias, L.M.; Dos Santos, B.V.; Albuquerque, C.J.B.; Baeta, B.E.L.; Pasquini, D.; Baffi, M.A. Biomass sorghum as a novel substrate in solid-state fermentation for the production of hemicellulases and cellulases by Aspergillus niger and A. fumigatus. J. Appl. Microbiol. 2018, 124, 708–718. [Google Scholar] [CrossRef]
- Vivekanand, V.; Dwivedi, P.; Sharma, A.; Sabharwal, N.; Singh, R.P. Enhanced delignification of mixed wood pulp by Aspergillus fumigatus laccase mediator system. World J. Microbiol. Biotechnol. 2008, 24, 2799. [Google Scholar] [CrossRef]
- Ye, J.S.; Yin, H.; Qiang, J.; Peng, H.; Qin, H.M.; Zhang, N.; He, B.Y. Biodegradation of anthracene by Aspergillus fumigatus. J. Hazard. Mater. 2011, 185, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Thakor, R.; Mistry, H.; Tapodhan, K.; Bariya, H. Efficient biodegradation of Congo red dye using fungal consortium incorporated with Penicillium oxalicum and Aspergillus tubingensis. Folia Microbiol. 2022, 67, 33–43. [Google Scholar] [CrossRef]
- El-Naggar, N.E.A.; Haroun, S.A.; El-Weshy, E.M.; Metwally, E.A.; Sherief, A.A. Mathematical modeling for bioprocess optimization of a protein drug, uricase, production by Aspergillus welwitschiae strain 1–4. Sci. Rep. 2019, 9, 12971. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Shah, M.; Hamayun, M.; Qadir, M.; Iqbal, A. Heavy metal tolerant endophytic fungi Aspergillus welwitschiae improves growth, ceasing metal uptake and strengthening antioxidant system in Glycine max L. Environ. Sci. Pollut. Res. 2022, 29, 15501–15515. [Google Scholar] [CrossRef]
- Lawal, O.T.; Sanni, D.M.; Olajuyigbe, F.M. Characterization of thermotolerant and thermostable rhodanese from high cyanide-degrading and agricultural wastes-utilizing Aspergillus welwitschiae LOT1 isolated from battery-effluent contaminated soil. Biocatal. Agric. Biotechnol. 2023, 52, 102807. [Google Scholar] [CrossRef]
- Patil, S.M.; Chandanshive, V.V.; Rane, N.R.; Khandare, R.V.; Watharkar, A.D.; Govindwar, S.P. Bioreactor with Ipomoea hederifolia adventitious roots and its endophyte Cladosporium cladosporioides for textile dye degradation. Environ. Res. 2016, 146, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Santana, N.B.; Dias, J.C.T.; Rezende, R.P.; Franco, M.; Oliveira, L.K.S.; Souza, L.O. Production of xylitol and bio-detoxification of cocoa pod husk hemicellulose hydrolysate by Candida boidinii XM02G. PLoS ONE 2018, 13, e0195206. [Google Scholar] [CrossRef] [PubMed]
- Demiray, E.; Açıkel, E.; Ertuğrul Karatay, S.; Dönmez, G. Saccharomyces cerevisiae and newly isolated Candida boidinii co-fermentation of industrial tea waste for improved bioethanol production. Energy Sources Part A 2022, 44, 1160–1172. [Google Scholar] [CrossRef]
- Nouri, H.; Azin, M.; Mousavi, S.L. Enhanced ethanol production from sugarcane bagasse hydrolysate with high content of inhibitors by an adapted Barnettozyma californica. Environ. Prog. Sustain. Energy 2018, 37, 1169–1175. [Google Scholar] [CrossRef]
- Ali, S.S.; Sun, J.; Koutra, E.; El-Zawawy, N.; Elsamahy, T.; El-Shetehy, M. Construction of a novel cold-adapted oleaginous yeast consortium valued for textile azo dye wastewater processing and biorefinery. Fuel 2021, 285, 119050. [Google Scholar] [CrossRef]
- Asses, N.; Ayed, L.; Bouallagui, H.; Sayadi, S.; Hamdi, M. Biodegradation of different molecular-mass polyphenols derived from olive mill wastewaters by Geotrichum candidum. Int. Biodeterior. Biodegrad. 2009, 63, 407–413. [Google Scholar] [CrossRef]
- Maldonado, R.R.; Lopes, D.B.; Aguiar-Oliveira, E.; Kamimura, E.S.; Macedo, G.A. A review on Geotrichum lipases: Production, purification, immobilization and applications. Chem. Biochem. Eng. Q. 2016, 30, 439–454. [Google Scholar] [CrossRef]
- Bourret, T.B.; Kramer, E.K.; Rogers, J.D.; Glawe, D.A. Isolation of Geotrichum candidum pathogenic to tomato (Solanum lycopersicum) in Washington State. North Am. Fungi 2013, 8, 1–7. [Google Scholar] [CrossRef][Green Version]
- Xiao, P.; Mori, T.; Kamei, I.; Kondo, R. Metabolism of organochlorine pesticide heptachlor and its metabolite heptachlor epoxide by white rot fungi, belonging to genus Phlebia. FEMS Microbiol. Lett. 2011, 314, 140–146. [Google Scholar] [CrossRef]
- Tri, C.L.; Khuong, L.D.; Kamei, I. The improvement of sodium hydroxide pretreatment in bioethanol production from Japanese bamboo Phyllostachys edulis using the white rot fungus Phlebia sp. MG-60. Int. Biodeterior. Biodegrad. 2018, 133, 86–92. [Google Scholar] [CrossRef]
- Catto, A.L.; Montagna, L.S.; Almeida, S.H.; Silveira, R.M.; Santana, R.M. Wood plastic composites weathering: Effects of compatibilization on biodegradation in soil and fungal decay. Int. Biodeterior. Biodegrad. 2016, 109, 11–22. [Google Scholar] [CrossRef]
- Brebu, M. Environmental degradation of plastic composites with natural fillers—A review. Polymers 2020, 12, 166. [Google Scholar] [CrossRef]
- Wei, H.; Wang, W.; Yarbrough, J.M.; Baker, J.O.; Laurens, L.; Van Wychen, S.; Chen, X.; Taylor, L.E., II; Xu, Q.; Himmel, M.E.; et al. Genomic, proteomic, and biochemical analyses of oleaginous Mucor circinelloides: Evaluating its capability in utilizing cellulolytic substrates for lipid production. PLoS ONE 2013, 8, e71068. [Google Scholar] [CrossRef]
- Yamazaki, K.I.; Takagi, K.; Kataoka, R.; Kotake, M.; Yamada, T.; Kiyota, H. Novel phosphorylation of aldrin-trans-diol by dieldrin-degrading fungus Mucor racemosus strain DDF. Int. Biodeterior. Biodegrad. 2014, 92, 36–40. [Google Scholar] [CrossRef]
- Rodrigues Reis, C.E.; Bento, H.B.; Carvalho, A.K.; Rajendran, A.; Hu, B.; De Castro, H.F. Critical applications of Mucor circinelloides within a biorefinery context. Crit. Rev. Biotechnol. 2019, 39, 555–570. [Google Scholar] [CrossRef]
- Ule, O.; Ogbonna, D.N.; Okparama, R.N.; Nrior, R.R. Myco-enhanced bioremediation in open field crude oil contaminated soil using Mucor racemosus and Aspergillus niger. Curr. J. Appl. Sci. Technol. 2021, 40, 119–141. [Google Scholar] [CrossRef]
- Almeida, R.R.; Pinto, N.A.R.; Soares, I.C.; Ferreira, L.B.C.; Lima, L.L.; Leitão, A.A.; de Lima Guimarães, L.G. Production and physicochemical properties of fungal chitosans with efficacy to inhibit mycelial growth activity of pathogenic fungi. Carbohydr. Res. 2023, 525, 108762. [Google Scholar] [CrossRef]
- Amobonye, A.; Bhagwat, P.; Pandey, A.; Singh, S.; Pillai, S. Biotechnological potential of Beauveria bassiana as a source of novel biocatalysts and metabolites. Crit. Rev. Biotechnol. 2020, 40, 1019–1034. [Google Scholar] [CrossRef]
- Amobonye, A.; Bhagwat, P.; Singh, S.; Pillai, S. Enhanced xylanase and endoglucanase production from Beauveria bassiana SAN01, an entomopathogenic fungal endophyte. Fungal Biology 2021, 125, 39–48. [Google Scholar] [CrossRef]
- Martínez, Á.T.; Speranza, M.; Ruiz-Dueñas, F.J.; Ferreira, P.; Camarero, S.; Guillén, F.; Martínez, M.J.; Gutiérrez Suárez, A.; Río Andrade, J.C.D. Biodegradation of lignocellulosics: Microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int. Microbiol. 2005, 8, 195–204. [Google Scholar] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Woudenberg, J.H.C.; Groenewald, J.Z.; Binder, M.; Crous, P.W. Alternaria redefined. Stud. Mycol. 2009, 64, 171–212. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Komon, M.; Kubicek, C.P.; Druzhinina, I.S. Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 2006, 98, 106–115. [Google Scholar] [CrossRef]
- Samuels, G.J.; Dodd, S.L.; Lu, B.S.; Petrini, O.; Schroers , H.-J.; Druzhinina , I.S. The Trichoderma koningii aggregate species. Stud. Mycol. 2006, 56, 67–133. [Google Scholar] [CrossRef]
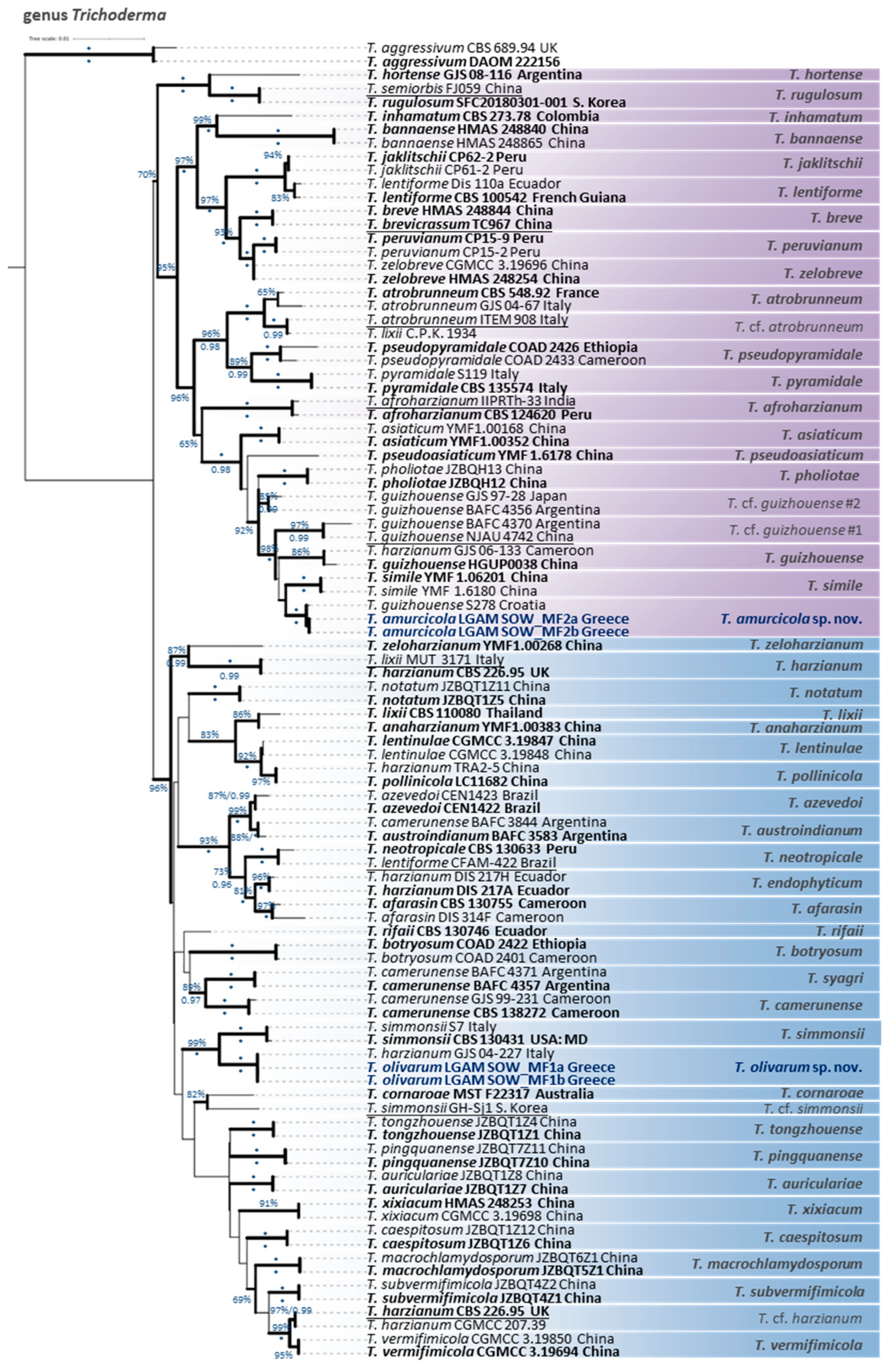

| Species | Strain Code | No. of MOTUs | ITS | tef1-α | tub2 | rpb2 | act | cal |
|---|---|---|---|---|---|---|---|---|
| ASCOMYCOTA | ||||||||
| Aspergillus (Aspergillaceae) | ||||||||
| A. filifer | A5 | 5 | PP766592 | PP768675 | ||||
| A. fumigatus | A1 | 3 | PP766593 | PP768676 | ||||
| A2 | 3 | PP766594 | PP768677 | |||||
| A3 | 3 | PP766595 | PP768693 | PP768678 | ||||
| A. novoparasiticus | T1 | 1 | PP766596 | PP768679 | ||||
| T2 | 1 | PP766597 | ||||||
| A. sydowii | PT1b | 4 | PP768694 | PP768680 | ||||
| A. tubingensis | A6 | 7 | PP766598 | |||||
| A7 | 7 | PP766599 | PP768681 | |||||
| PT4a | 7 | PP766600 | ||||||
| A. welwitschiae | A4 | 6 | PP766601 | |||||
| A8 | 6 | PP766602 | PP768682 | |||||
| PT1 | 6 | PP766603 | PP768695 | |||||
| A. westerdijkiae | A9 | 2 | PP766604 | |||||
| Penicillium (Aspergillaceae) | ||||||||
| P. crustosum | M1b | 9 | PP766605 | PP768696 | ||||
| M2 | 10 | PP766606 | ||||||
| M7 | 10 | PP766607 | PP768697 | |||||
| M6 | 11 | PP766608 | PP768683 | |||||
| P1 | 11 | PP766609 | ||||||
| P2 | 12 | PP766610 | ||||||
| P3 | 12 | PP766611 | ||||||
| P4 | 13 | PP766612 | ||||||
| P5 | 13 | PP766613 | ||||||
| P. kongii | PT2 | 8 | PP766614 | |||||
| PT5 | 8 | PP766615 | ||||||
| P. paneum | M8 | 14 | PP766616 | PP768698 | ||||
| M15 | 14 | PP766617 | ||||||
| P. roqueforti | M5 | 15 | PP766618 | PP768699 | PP768684 | |||
| M12 | 15 | PP766619 | PP768700 | |||||
| M16 | 16 | PP766620 | ||||||
| M16a | 16 | PP766621 | ||||||
| M16b | 16 | PP766622 | ||||||
| M16c | 16 | PP766623 | ||||||
| OM27 | 17 | PP766624 | ||||||
| OM35 | 17 | PP766625 | PP768685 | |||||
| OM59 | 18 | PP766626 | ||||||
| OM73 | 18 | PP766627 | ||||||
| OM122 | 19 | PP766628 | ||||||
| OM125 | 19 | PP766629 | ||||||
| OM132 | 20 | PP766630 | ||||||
| OM147 | 20 | PP766631 | ||||||
| Cladosporium (Cladosporiaceae) | ||||||||
| C. cladosporioides | M4 | 27 | PP766643 | PP768710 | PP768725 | |||
| C. limoniforme | OM52 | 28 | PP766644 | PP768690 | PP768726 | |||
| C. ramotenellum | PT1a | 29 | PP766645 | PP768691 | ||||
| PT4 | 29 | PP766646 | PP768711 | PP768692 | PP768727 | |||
| Beauveria (Cordycipitaceae) | ||||||||
| B. pseudobassiana | BF1 | 24 | PP766639 | PP768706 | ||||
| Neocucurbitaria (Cucurbitariaceae) | ||||||||
| N. keratinophila | M4a | 30 | PP766647 | PP768712 | ||||
| Candida (Debaryomycetaceae) | ||||||||
| C. boidinii | Y1a | 32 | PP766649 | |||||
| Y2 | 33 | PP766650 | ||||||
| Y3 | 33 | PP766651 | ||||||
| Y8 | 34 | PP766652 | ||||||
| Y9 | 34 | PP766653 | ||||||
| Y5 | 35 | PP766654 | ||||||
| Y6 | 36 | PP766655 | ||||||
| Y7 | 37 | PP766656 | ||||||
| Y10 | 38 | PP766657 | PP768713 | |||||
| Y11 | 39 | PP766658 | ||||||
| Y12 | 40 | PP766659 | ||||||
| Y14 | 41 | PP766660 | ||||||
| Stagonosporopsis (Didymellaceae) | ||||||||
| S. ailanthicola | OM34 | 31 | PP766648 | |||||
| Geotrichum (Dipodascaceae) | ||||||||
| G. candidum | OM5 | 43 | PP766662 | |||||
| OM58 | 43 | PP766663 | ||||||
| OM62 | 43 | PP766664 | ||||||
| OM84 | 43 | PP766665 | ||||||
| GZ3 | 44 | PP766666 | ||||||
| G1 | 45 | PP766667 | ||||||
| G2 | 45 | PP766668 | ||||||
| Trichoderma (Hypocreaceae) | ||||||||
| T. amurcicola sp. nov. | MF2a | 23 | PP766637 | PP768704 | PP768717 | PP768723 | PP768730 | |
| MF2b | 23 | PP766638 | PP768705 | PP768718 | PP768724 | PP768731 | ||
| T. olivarum sp. nov. | MF1a | 22 | PP766635 | PP768702 | PP768715 | PP768721 | PP768728 | |
| MF1b | 22 | PP766636 | PP768703 | PP768716 | PP768722 | PP768729 | ||
| Pleurostoma (Pleurostomataceae) | ||||||||
| P. richardsiae | M3 | 26 | PP766641 | PP768708 | PP768688 | PP768719 | ||
| M3a | 26 | PP766642 | PP768709 | PP768689 | PP768720 | |||
| Barnettozyma (Phaffomycetaceae) | ||||||||
| B. californica | Y4 | 42 | PP766661 | PP768714 | ||||
| Sarocladium (Sarocladiaceae) | ||||||||
| S. kiliense | M10 | 25 | PP766640 | PP768707 | ||||
| Talaromyces (Trichocomaceae) | ||||||||
| T. nanjingensis | M13 | 21 | PP766632 | PP768701 | PP768686 | |||
| Μ13a | 21 | PP766633 | PP768687 | |||||
| Μ13c | 21 | PP766634 | ||||||
| BASIDIOMYCOTA | ||||||||
| Fuscoporia (Hymenochaetaceae) | ||||||||
| F. ferrea | M9a | 48 | PP766673 | |||||
| Pseudophlebia (Meruliaceae) | ||||||||
| P. setulosa | OM106 | 46 | PP766669 | |||||
| OM141 | 46 | PP766670 | ||||||
| PT3N | 47 | PP766671 | ||||||
| PT5N | 47 | PP766672 | ||||||
| Peniophora (Peniophoraceae) | ||||||||
| P. lycii | Μ11 | 49 | PP766676 | |||||
| M14 | 49 | PP766677 | ||||||
| MUCOROMYCOTA | ||||||||
| Mucor (Mucoraceae) | ||||||||
| M. circinelloides | Z3 | 54 | PP766678 | |||||
| M. pseudolusitanicus | Z4 | 55 | PP766679 | |||||
| Z6 | 55 | PP766680 | ||||||
| Z7 | 56 | PP766681 | ||||||
| Z9 | 50 | PP766682 | ||||||
| M. racemosus | Z1 | 51 | PP766683 | |||||
| Z5 | 51 | PP766684 | ||||||
| Z2 | 52 | PP766685 | ||||||
| Z8 | 53 | PP766686 |
| Species | Strain (Tested/Total) | Isolation Conditions | CEA | XEA | LEA | PDA-G | MS-RB |
|---|---|---|---|---|---|---|---|
| Ascomycota | |||||||
| Aspergillus filifer | 1/1 | PDA/EC-SD/25 | 1 | 1 | 0 | 0 | 1 |
| Aspergillus fumigatus | 2/3 | CEA, LEA/WU/45 | 1 | 1 | 0 | 0 | 0 |
| Aspergillus novoparasiticus | 2/2 | PDA/EL-SD/25, 45 | 1–2 | 2 | 0 | 0 | 0–1 |
| Aspergillus sydowii | 1/1 | CEA/SD/25 | 3 | 1 | 0 | 1 | 0 |
| Aspergillus tubingensis | 2/3 | PDA, CEA/EC-SD, EL-SD, SD/25 | 2 | 1 | 0 | 0 | 1 |
| Aspergillus welwitschiae | 2/3 | PDA-RB/EC-SD, EL-SD, SD/25, 45 | 0 | 1 | 0 | 0 | 1 |
| Aspergillus westerdijkiae | 1/1 | PDA/EL-SD/25 | 1 | 2 | 0 | 0 | 1 |
| Penicillium crustosum | 5/9 | CEA, CEA-EC25, LEA/WU, EL-SD, SD/25 | 1–3 | 1–2 | 0 | 0 | 0–1 |
| Penicillium kongii | 2/2 | CEA/SD/25 | 2–3 | 1 | 0 | 0 | 1 |
| Penicillium paneum | 2/2 | YPDA-EC25, LEA/WU/25 | 3 | 2 | 0 | 0 | 1 |
| Penicillium roqueforti | 9/14 | LEA, CEA, PDA/SD, EL-SD, EY-SD, WU/25 | 1–3 | 1 | 0 | 0 | 0–2 |
| Cladosporium cladosporioides | 1/1 | LEA/EL-SD/25 | 2 | 2 | 0 | 0 | 1 |
| Cladosporium limoniforme | 1/1 | CEA/SD/25 | 2 | 1 | 3 | 2 | 2 |
| Cladosporium ramotenellum | 2/2 | CEA/SD/25 | 2–3 | 1–2 | 3 | 3 | 2 |
| Beauveria pseudobassiana | 1/1 | CEA/SD/25 | 3 | 3 | 0 | 3 | 3 |
| Neocucurbitaria keratinophila | 1/1 | YPDA/SD/25 | 2 | 3 | 0 | 0 | 1 |
| Candida boidinii | 2/12 | YPDA, YPDA-EC10, YPDA-EC25, CEA, CEA-EC10, LEA-EC25, PDA-EC25/EC-SD, EL-SD, EY-SD, WU/25 | 3 | 2–3 | 0 | 0 | 3 |
| Stagonosporopsis ailanthicola | 1/1 | CEA/SD/25 | 0 | 0 | 0 | 0 | 0 |
| Geotrichum candidum | 6/7 | CEA, YPDA, PDA-RB/SD, EC-SD, EL-SD/25 | 1–3 | 3 | 0 | 0 | 0–1 |
| Trichoderma amurcicola sp. nov. | 1/2 | CEA/SD/25 | 1 | 1 | 0 | 1 | 1 |
| Trichoderma olivarum sp. nov. | 1/2 | CEA/SD/25 | 1 | 1 | 0 | 0 | 0 |
| Pleurostoma richardsiae | 2/2 | CEA, PDA/EC-SD/25 | 1–2 | 1–2 | 0 | 1 | 1–2 |
| Barnettozyma californica | 1/1 | CEA/EL-SD/25 | 3 | 3 | 0 | 0 | 2 |
| Sarocladium kiliense | 1/1 | CEA/EL-SD/25 | 1 | 2 | 0 | 0 | 1 |
| Talaromyces nanjingensis | 2/3 | YPDA-EC10, YPDA-EC25/WU, EL-SD/25 | 1 | 1 | 0 | 0 | 0 |
| Basidiomycota | |||||||
| Fuscoporia ferrea | 1/1 | LEA/WU/25 | 3 | 1 | 1 | 1 | 1 |
| Pseudophlebia setulosa | 2/4 | CEA/SD/25 | 1–2 | 1 | 1–2 | 2–3 | 1–2 |
| Peniophora lycii | 2/2 | CEA-EC25/WU/25 | 1 | 3 | 2 | 0 | 2 |
| Mucoromycota | |||||||
| Mucor circinelloides | 1/1 | LEA/SD/25 | 1 | 2 | 0 | 0 | 1 |
| Mucor pseudolusitanicus | 4/4 | PDA-RB, LEA, CEA/WU, EC-SD/25 | 1 | 1–2 | 0 | 0 | 1 |
| Mucor racemosus | 4/4 | CEA, LEA, PDA/WU, EC-SD/25 | 1 | 0 | 0 | 0 | 0–1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fryssouli, V.; Kefalogianni, I.; Polemis, E.; Typas, M.A.; Zervakis, G.I. Diversity of Culturable Fungi in Two-Phase Olive Mill Waste, a Preliminary Evaluation of Their Enzymatic Potential, and Two New Trichoderma Species. J. Fungi 2025, 11, 687. https://doi.org/10.3390/jof11090687
Fryssouli V, Kefalogianni I, Polemis E, Typas MA, Zervakis GI. Diversity of Culturable Fungi in Two-Phase Olive Mill Waste, a Preliminary Evaluation of Their Enzymatic Potential, and Two New Trichoderma Species. Journal of Fungi. 2025; 11(9):687. https://doi.org/10.3390/jof11090687
Chicago/Turabian StyleFryssouli, Vassiliki, Io Kefalogianni, Elias Polemis, Milton A. Typas, and Georgios I. Zervakis. 2025. "Diversity of Culturable Fungi in Two-Phase Olive Mill Waste, a Preliminary Evaluation of Their Enzymatic Potential, and Two New Trichoderma Species" Journal of Fungi 11, no. 9: 687. https://doi.org/10.3390/jof11090687
APA StyleFryssouli, V., Kefalogianni, I., Polemis, E., Typas, M. A., & Zervakis, G. I. (2025). Diversity of Culturable Fungi in Two-Phase Olive Mill Waste, a Preliminary Evaluation of Their Enzymatic Potential, and Two New Trichoderma Species. Journal of Fungi, 11(9), 687. https://doi.org/10.3390/jof11090687







