Discovery and Identification of Four Novel Species of Distoseptispora (Distoseptisporaceae, Distoseptisporales) on Decaying Wood from Hainan and Fujian Provinces, China
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Treatment
2.2. Morphological and Cultural Characterization
2.3. DNA Extraction, PCR Amplification, and Sequencing
2.4. Phylogenetic Analysis
3. Results
3.1. Phylogenetic Analysis
3.2. Taxonomy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Su, H.Y.; Hyde, K.D.; Maharachchikumbura, S.S.N.; Ariyawansa, H.A.; Luo, Z.L.; Promputtha, I.; Tian, Q.; Lin, C.G.; Shang, Q.J.; Zhao, Y.C. The families Distoseptisporaceae fam. nov., Kirschsteiniotheliaceae, Sporormiaceae and Torulaceae, with new species from freshwater in Yunnan Province, China. Fungal Divers. 2016, 80, 375–409. [Google Scholar] [CrossRef]
- Sun, Y.R.; Hyde, K.D.; Liu, N.G.; Jayawardena, R.S.; Wijayawardene, N.N.; Ma, J.; Zhang, Q.; Al Otibi, F.; Wang, Y. Micro fungi in southern China and northern Thailand: Emphasis on medicinal plants. Fungal Divers. 2025, 130, 100. [Google Scholar] [CrossRef]
- Hyde, K.D.; Hongsanan, S.; Jeewon, R.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Phookamsak, R.; Ariyawansa, H.A.; Boonmee, S.; Zhao, Q.; et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 80, 1–270. [Google Scholar] [CrossRef]
- Hyde, K.D.; Tennakoon, D.S.; Jeewon, R.; Bhat, D.J.; Maharachchikumbura, S.S.N.; Rossi, W.; Leonardi, M.; Lee, H.B.; Mun, H.Y.; Houbraken, J.; et al. Fungal diversity notes 1036–1150: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2019, 96, 1–242. [Google Scholar] [CrossRef]
- Hyde, K.D.; Jeewon, R.; Chen, Y.J.; Bhunjun, C.S.; Calabon, M.S.; Jiang, H.-B.; Lin, C.-G.; Norphanphoun, C.; Sysouphanthong, P.; Pem, D.; et al. The numbers of fungi: Is the descriptive curve flattening? Fungal Divers. 2020, 103, 219–271. [Google Scholar] [CrossRef]
- Jayawardena, R.S.; Hyde, K.D.; Wang, S.; Sun, Y.R.; Suwannarach, N.; Sysouphanthong, P.; Abdel Wahab, M.A.; Abdel-Aziz, F.A.; Abeywickrama, P.D.; Abreu, V.P.; et al. Fungal diversity notes 1512–1610: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2022, 117, 1–272. [Google Scholar] [CrossRef]
- Sun, Y.; Goonasekara, I.D.; Thambugala, K.M.; Jayawardena, R.; Wang, Y.; Hyde, K. Distoseptispora bambusae sp. nov. (Distoseptisporaceae) on bamboo from China and Thailand. Biodivers. Data J. 2020, 8, 6. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; Mckenzie, E.H.C.; Bhat, D.J.; Phillips, A.J.L.; Wanasinghe, D.N.; Samarakoon, M.C.; Jayawardena, R.S.; Dissanayake, A.J.; Tennakoon, D.S.; et al. Fungal diversity notes 840–928: Micro-fungi associated with Pandanaceae. Fungal Divers. 2018, 93, 1–160. [Google Scholar] [CrossRef]
- Yang, J.; Maharachchikumbura, S.S.N.; Liu, J.K.; Hyde, K.D.; Jones, E.B.G.; Al-Sadi, A.M.; Liu, Z.Y. Pseudostanjehughesia aquitropica gen. et sp. nov. and Sporidesmium sensu lato species from freshwater habitats. Mycol. Prog. 2018, 17, 591–616. [Google Scholar] [CrossRef]
- Yang, J.; Liu, L.L.; Jones, E.B.G.; Li, W.L.; Hyde, K.D.; Liu, Z.Y. Morphological variety in Distoseptispora and introduction of six novel species. J. Fungi. 2021, 7, 945. [Google Scholar] [CrossRef] [PubMed]
- Monkai, J.; Boonmee, S.; Ren, G.; Wei, D.-P.; Phookamsak, R.; Mortimer, P.E. Distoseptispora hydei sp. nov. (Distoseptisporaceae), a novel lignicolous fungus on decaying bamboo in Thailand. Phytotaxa 2020, 459, 093–107. [Google Scholar] [CrossRef]
- Konta, S.; Tibpromma, S.; Karunarathna, S.; Samarakoon, M.; Steven, L.; Mapook, A.; Boonmee, S.; Senwanna, C.; Balasuriya, A.; Eungwanichayapant, P.; et al. Morphology and multigene phylogeny reveal ten novel taxa in Ascomycota from terrestrial palm substrates (Arecaceae) in Thailand. Mycosphere 2023, 14, 107–152. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Ma, J.; Yang, H.D.; Zhang, J.Y.; Du, T.Y.; Gao, Y.; de Farias, A.G.; He, S.; He, Y.; et al. Mycosphere notes 387–412—Novel species of fungal taxa from around the world. Mycosphere 2023, 14, 663–744. [Google Scholar] [CrossRef]
- Karimi, O.; Chethana, K.W.T.; De Farias, A.R.G.; Asghari, R.; Kaewchai, S.; Hyde, K.D.; Li, Q. Morphology and multigene phylogeny reveal three new species of Distoseptispora (Distoseptisporales, Distoseptisporaceae) on palms (Arecaceae) from peat swamp areas in southern Thailand. MycoKeys 2024, 102, 55–81. [Google Scholar] [CrossRef]
- Wang, S.; Liu, X.M.; Xiong, C.L.; Gao, S.S.; Xu, W.M.; Zhao, L.L.; Song, C.Y.; Liu, X.Y.; James, T.Y.; Li, Z.; et al. ASF1 regulates asexual and sexual reproduction in Stemphylium eturmiunum by DJ-1 stimulation of the PI3K/AKT signaling pathway. Fungal Divers. 2023, 123, 159–176. [Google Scholar] [CrossRef]
- Zhang, Z.X.; Shang, Y.X.; Liu, Q.Y.; Li, D.H.; Yin, C.Z.; Liu, X.Y.; Tao, M.F.; Jiang, Y.; Wang, Y.X.; Zhang, M.Y.; et al. Deciphering the evolutionary and taxonomic complexity of Diaporthales (Sordariomycetes, Ascomycota) through integrated phylogenomic and divergence time estimation. Fungal Divers. 2025, 132, 1–125. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Guo, L.D.; Hyde, K.D.; Liew, E.C.Y. Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytol. 2000, 147, 617–630. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: New York, NY, USA, 1990; Volume 18, pp. 315–322. [Google Scholar] [CrossRef]
- Rehner, S.A.; Samuels, G.J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 1994, 98, 625–634. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Sung, G.H.; Sung, J.M.; Hywel-Jones, N.L.; Spatafora, J.W. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenetics Evol. 2007, 44, 1204–1223. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Zhao, N.; Luo, Z.L.; Hyde, K.D.; Su, H.Y.; Bhat, D.J.; Liu, J.K.; Bao, D.F.; Hao, Y.E. Helminthosporium submersum sp. nov. (Massarinaceae) from submerged wood in north-western Yunnan Province, China. Phytotaxa 2018, 348, 269–278. [Google Scholar] [CrossRef]
- Zhang, Z.X.; Liu, R.Y.; Liu, S.B.; Mu, T.C.; Zhang, X.G.; Xia, J.W. Morphological and phylogenetic analyses reveal two new species of Sporocadaceae from Hainan, China. MycoKeys 2022, 88, 171–192. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.G.; Luo, X.X.; Hu, Y.F.; Castaeda-Ruíz, R.F.; Xu, Z.H.; Ma, J. Morphological and phylogenetic analyses reveal four novel species of Distoseptispora (Distoseptisporaceae, Distoseptisporales) from southern China. MycoKeys 2025, 113, 31–55. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES science gateway: Enabling high-impact science for phylogenetics researchers with limited resources. ACM 2012, 39, 1–8. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Nylander, J.A.A. MrModelTest v. 2. Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; Volume 7. [Google Scholar] [CrossRef]
- Dissanayake, L.S.; Samarakoon, M.C.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Tang, X.; Li, Q.R.; Mortimer, P.E.; Faraj, T.; Xu, J.C.; Kang, J.C.; et al. Exploring the taxonomy and phylogeny of Sordariomycetes taxa emphasizing Xylariomycetidae in Southwestern China. Mycosphere 2024, 15, 1675–1793. [Google Scholar] [CrossRef]
- Hyde, K.D.; Suwannarach, N.; Jayawardena, R.S.; Manawasinghe, I.S.; Liao, C.F.; Doilom, M.; Cai, L.; Zhao, P.; Buyck, B.; Phukhamsakda, C.; et al. My cosphere notes 325–344–Novel species and records of fungal taxa from around the world. Mycosphere 2021, 12, 1101–1156. [Google Scholar] [CrossRef]
- Hu, Y.F.; Liu, J.W.; Luo, X.X.; Xu, Z.H.; Xia, J.W.; Zhang, X.-G.; Castañeda-Ruíz, R.F.; Ma, J.; Anderson, M.Z. Multi-locus phylogenetic analyses reveal eight novel species of Distoseptispora from southern China. Microbiol. Spectr. 2023, 11, e0246823. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.W.; Hu, Y.F.; Luo, X.X.; Xu, Z.H.; Castañeda-Ruíz, R.F.; Xia, J.; Zhang, X.; Zhang, L.; Cui, R.; Ma, J. Morphological and Phylogenetic Analyses Reveal Three New Species of Distoseptispora (Distoseptisporaceae, Distoseptisporales) from Yunnan, China. J. Fungi. 2023, 9, 470. [Google Scholar] [CrossRef]
- Delgado, G.; Maciá-Vicente, J.G.; Colbert, W.; Piepenbring, M. Redefining Ellisembia sensu stricto with a reassessment of related taxa in Sordariomycetes. Mycol. Prog. 2024, 23, 32. [Google Scholar] [CrossRef]
- Xu, R.J.; Hyde, K.D.; Li, J.N.; Boonmee, S.; Liu, N.J.; Yang, J.; Li, Y.; Bao, D.-F.; Shen, H.-W.; Zhu, X.-T.; et al. Lignicolous freshwater fungi of the pan Qinghai-Xizang Plateau, China. Fungal Divers. 2025, 153, 1–212. [Google Scholar] [CrossRef]
- Yang, J.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Bhat, D.J.; McKenzie, E.H.C.; Bahkali, A.H.; Gareth Jones, E.B.G.; Liu, Z.Y. Aquapteridospora lignicola gen. et sp. nov., a new hyphomycetous taxon (Sordariomycetes) from wood submerged in a freshwater stream. Cryptogam. Mycol. 2015, 36, 469–478. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C. Outline of fungi and fungus-like taxa. Mycosphere J. Fungal Biol. 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Yin, C.Z.; Zhang, Z.X.; Wang, S.; Liu, W.W.; Zhang, X.G. A taxonomic and phylogenetic study of anamorphic strains of Daldinia (hypoxylaceae, xylariales) in southern china. J. Fungi 2024, 10, 700. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.W.; Yin, C.Z.; Zhang, Z.X.; Wang, X.S.; Meng, Z.; Zhang, X.G.; Wang, S. Four new species of Beltraniella (Amphisphaeriales, Beltraniaceae) revealed by morphology and phylogenetic analyses from china. MycoKeys 2025, 116, 125–144. [Google Scholar] [CrossRef]
- Chen, X.M.; Tang, X.; Ma, J.; Liu, N.-G.; Tibpromma, S.; Karunarathna, S.C.; Xiao, Y.-P.; Lu, Y.-Z. Identification of two new species and a new host record of Distoseptispora (Distoseptisporaceae, Distoseptisporales, Sordariomycetes) from terrestrial and freshwater habitats in Southern China. MycoKeys. 2024, 102, 83–105. [Google Scholar] [CrossRef]

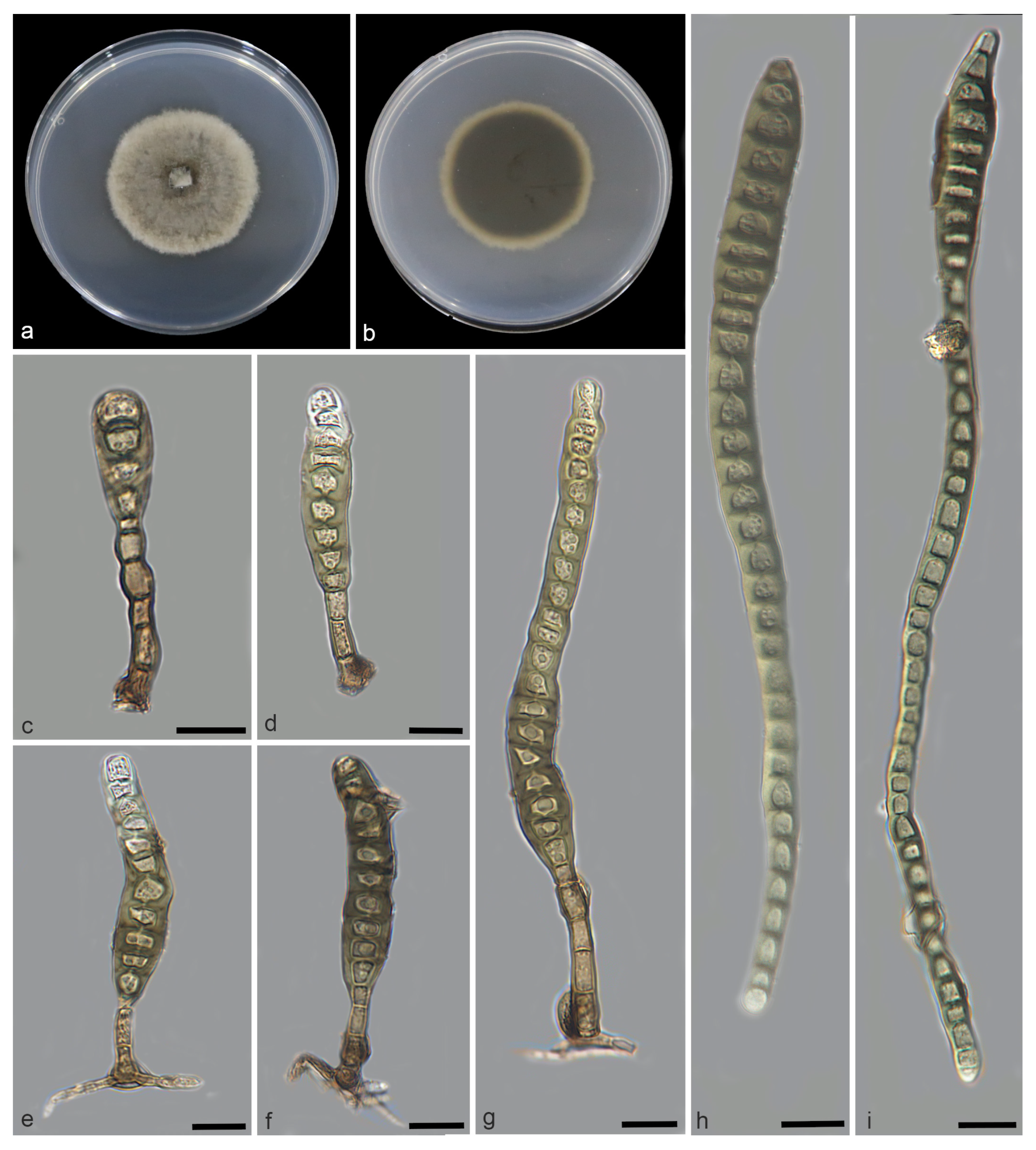
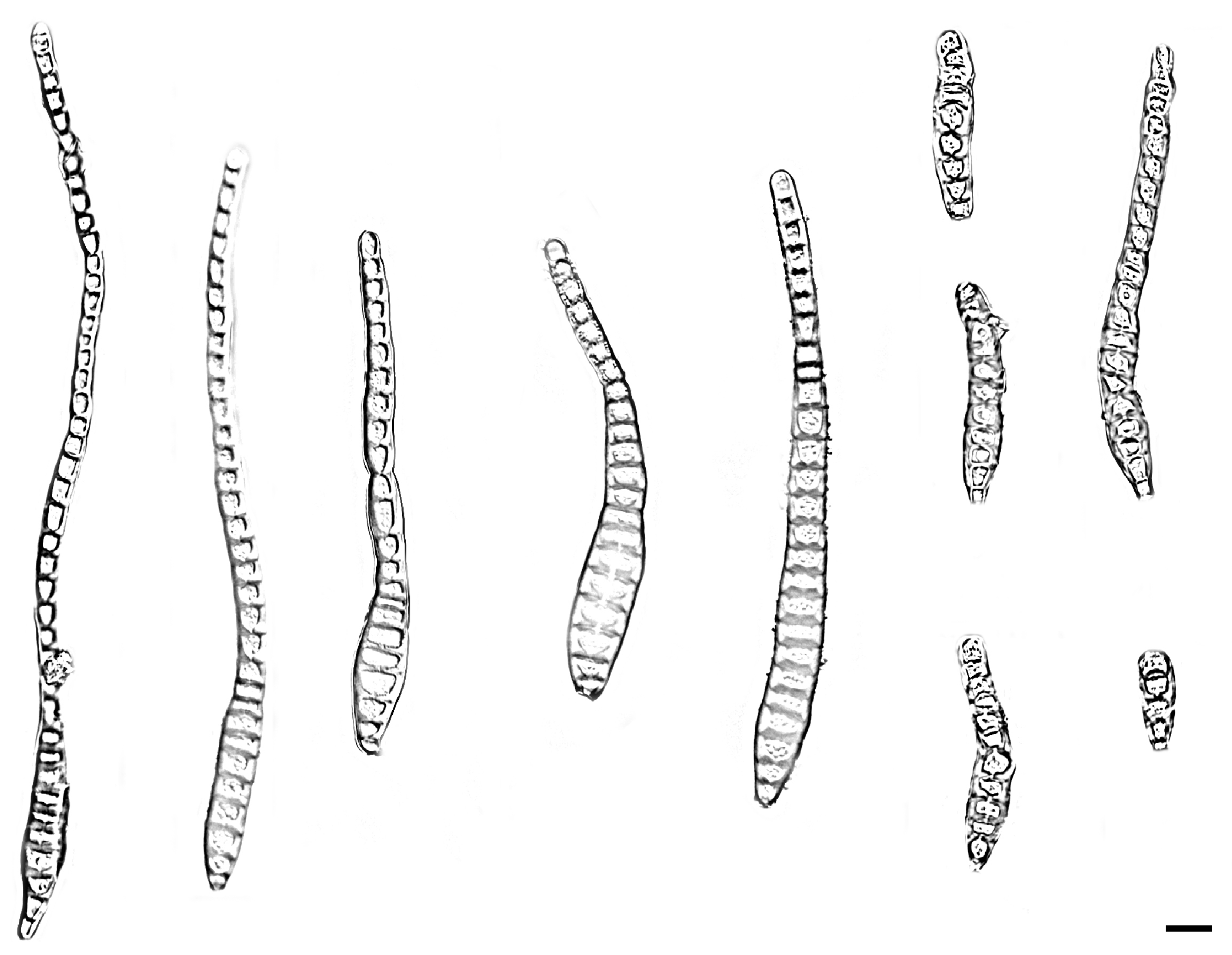

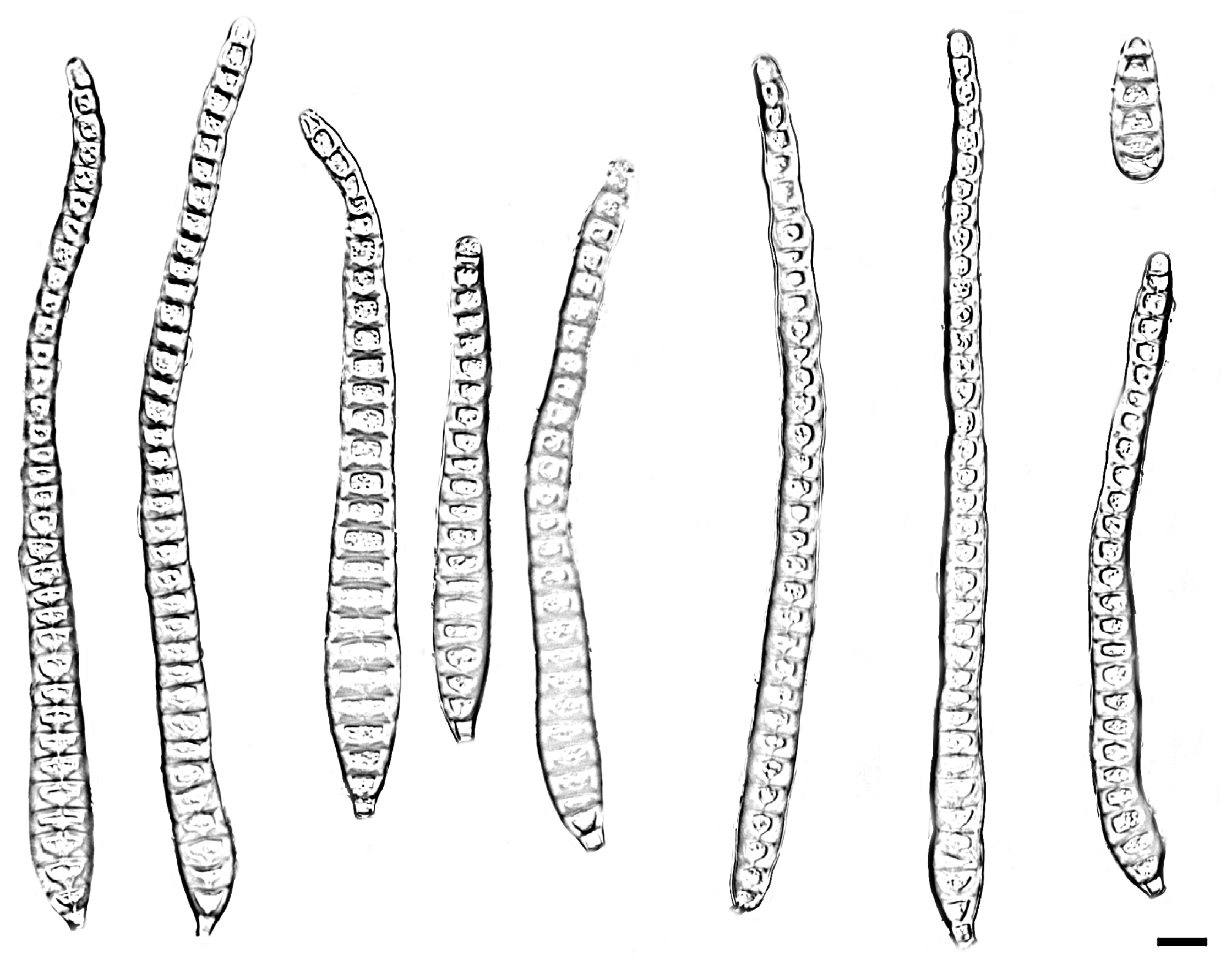
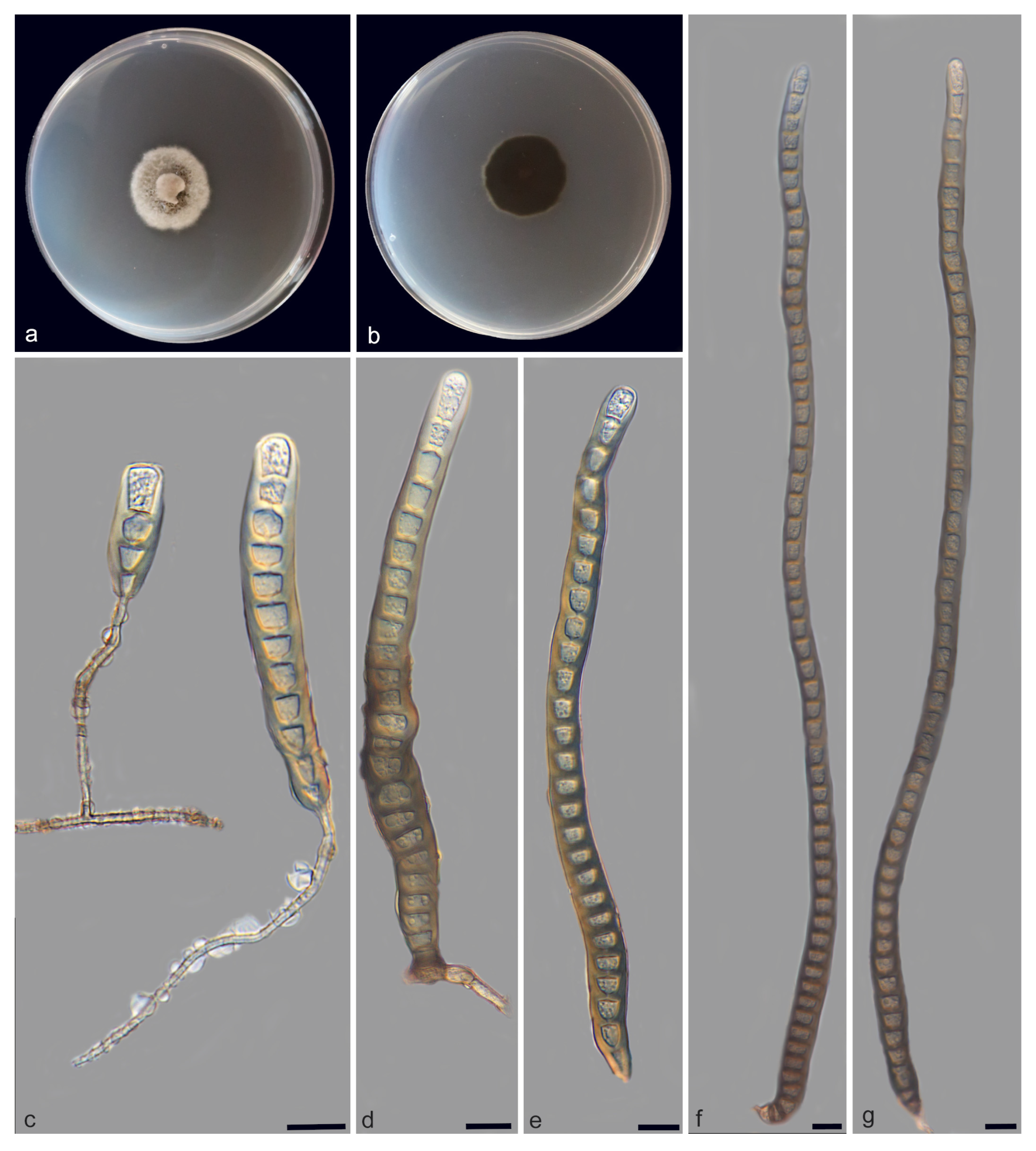
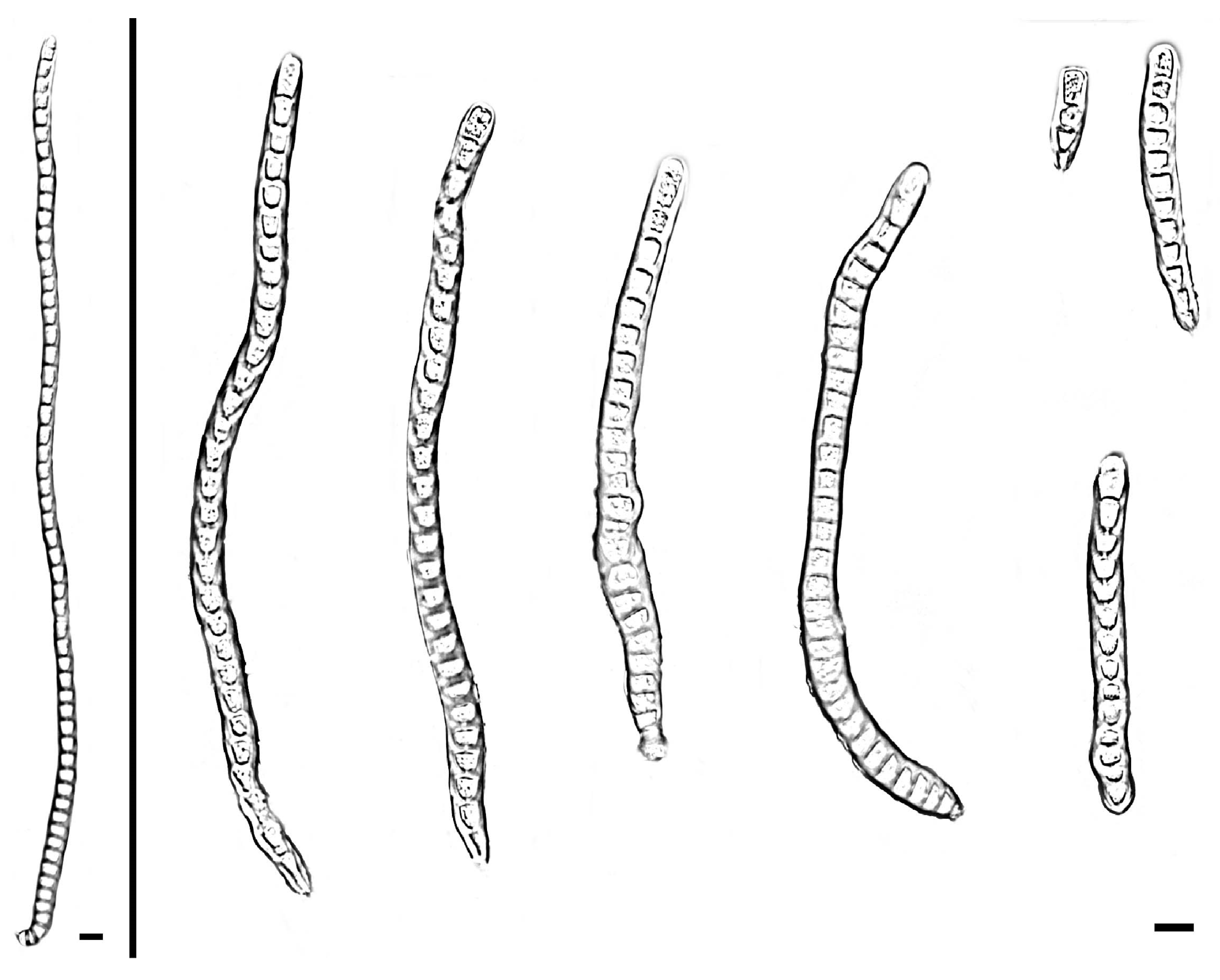
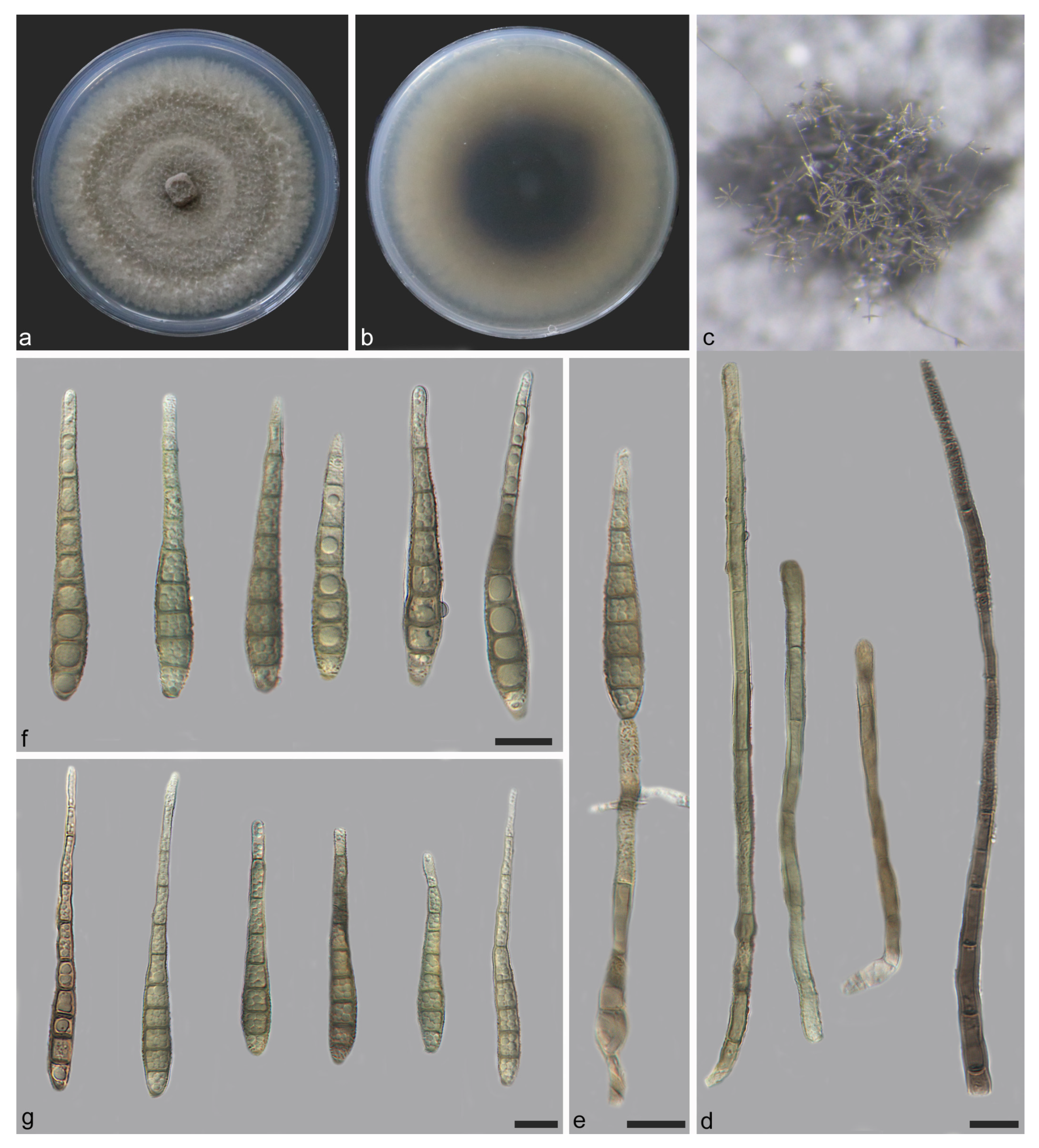
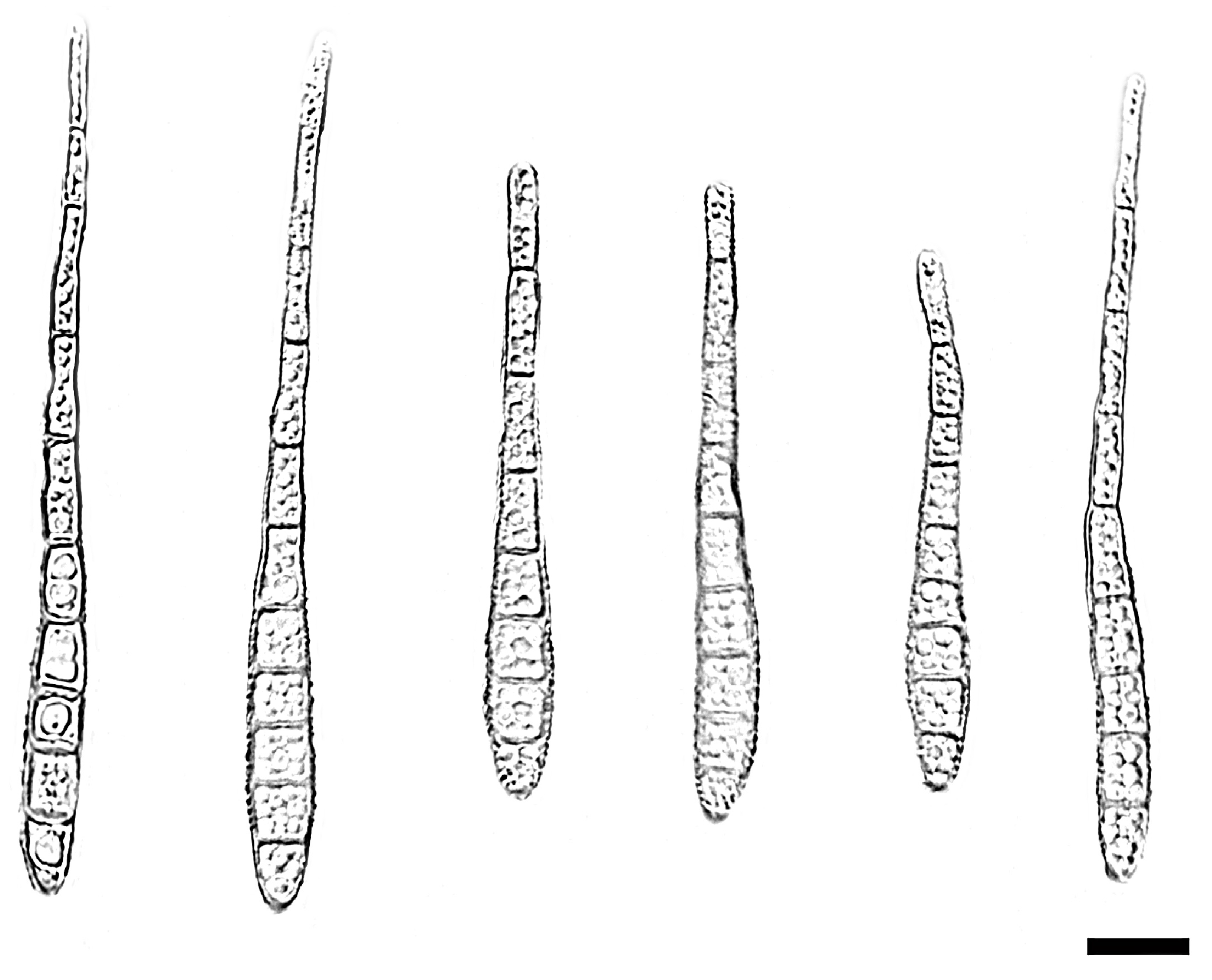
| Species | Colonies | Conidiphores (μm) | Conida | Habitat | Locality | Host | ||
|---|---|---|---|---|---|---|---|---|
| Size (μm) | Morphology | Septation | ||||||
| Distoseptispora bawanglingensis vs. D. sichuanensis | raised, circular, velutinous edge, grayish-brown, pale edge, surface velvety, felted, dense, reverse concentric, dark brown inner ring, outer pale brown halo vs. effuse, dark brown to black, hairy. | = 22.5 × 5.2, n = 20) = 20 × 5, n = 5) | = 141.7 × 10.4, n = 25) = 114 × 12.8, n = 20) | acrogenous or obclavate, solitary, straight or slightly curved, brown to pale brown vs. solitary, obclavate, elongated, straight or slightly curved, truncate at the base, rounded at the apex and hyaline, straight or slightly curved | 3–45 vs. 12–20 | Peat swamp forest vs. terrestrial | China | decaying Wood vs. dead branches |
| D. bawanglingensis vs. D. xinpingensis | raised, circular, velutinous edge, grayish-brown, pale edge, surface velvety, felted, dense, reverse concentric, dark brown inner ring, outer pale brown halo vs. effuse, brown to dark brown, solitary or gregarious | = 22.5 × 5.2, n = 20) vs. (97–)105–149(–175) × 4–5 μm ( = 127 × 5 μm, n = 40) | = 141.7 × 10.4, n = 25) = 123 × 8 μm, n = 40) | acrogenous or obclavate, solitary, acrogenous, straight or slightly curved, brown to pale brown vs. acrogenous, solitary, obclavate, truncate at base, tapering towards the apex, straight or slightly curved, brown, smooth, thin-wall | 3–45 vs. 8–12 | Peat swamp forest vs. freshwater stream | China | decaying Wood |
| D. changjiangensis vs. D. olivaceoviridis | circular, velutinous edge, low convex, concentric, light gray inner ring, outer dark gray, grayish-brown at the central parts, surface felted, dense, reverse dark brown, outer grayish white halo vs. single or in groups, numerous, hairy, dark brown | = 132.1 × 5.1, n = 20) vs. 34–78 × 5–7 | = 153.6 × 14.1, n = 25) = 80 × 9 μm, n = 15) | acrogenous or obclavate, solitary, straight or slightly curved, brown to pale brown vs. acrogenous or obclavate, solitary, straight or slightly curved, brown to pale brown | 6–40 vs. 3–45 | Peat swamp forest vs. unknown | China vs. Thailand | decaying wood vs. Clerodendrum quadriloculare |
| D. daanyuanensis vs. D. aquatica | circular, white, with gray exudates, velutinous edge, dull surface, felted, dense, reverse concentric, blackish-brown inner ring, outer grayish white halo vs. effuse, scattered, hairy, dark brown, gray or black | = 38.4 × 1.8, n = 20) = 35 × 8 μm, n = 10) | = 302.4 × 309 11.3, n = 25) vs. 29–41 × 7–9 | acrogenous, obclavate, solitary, straight or slightly curved, brown to pale brown vs. acrogenous, solitary, dry, obclavate, elongated, straight or slightly curved, truncate at the base, smooth, dark brown with bluish, green to malachite green tinge, paler towards the apex, thick- walled | 4–92 vs. 15–28 | Peat swamp forest vs. aquatic | China | decaying wood |
| D. daanyuanensis vs. D. nanchangensis | circular, white, with gray exudates, velutinous edge, dull surface, felted, dense, reverse con-centric, blackish-brown inner ring, outer grayish white halo vs. effuse, scattered, dark brown to black, and hairy | = 38.4 × 1.8, n = 20) = 40.6 × 7.2, n = 20) | = 302.4 × 309 11.3, n = 25) = 203.5 ×15.4, n = 30) | acrogenous, obclavate, solitary, straight or slightly curved, brown to pale brown vs. acrogenous, solitary, obclavate, straight or curved, brown to dark brown, smooth, thick-walled | 4–92 vs. (17–)21–43 | Peat swamp forest vs. unknown | China | decaying wood vs. dead branches |
| D. daanyuanensis vs. D. nanpingensis | circular, white, with gray exudates, velutinous edge, dull surface, felted, dense, reverse con-centric, blackish-brown inner ring, outer grayish white halo vs. effuse, scattered, dark brown to black, hairy | = 38.4 × 1.8, n = 20) = 18.1 × 6.1, n = 10) | = 302.4 × 309 11.3, n = 25) = 227.1 × 14.8, n = 20) | acrogenous, obclavate, solitary, straight or slightly curved, brown to pale brown vs. solitary, acrogenous, dry, obclavate, straight or curved, sometimes with a swollen cell, reddish-brown and slightly paler towards the apex, sometimes constricted at the septa, smooth | 4–92 vs. 28–41 | Peat swamp forest vs. terrestrial | China | decaying wood vs. dead branches |
| D. jianfenglingensis vs. D. liupanshuiensis | circular, dense aerial mycelia, velutinous edge, dull surface, concentric, dark brown at the central parts, brown inner ring, outer pale brown halo, felted, dense, reverse concentric, blackish-brown inner ring, outer light brown halo. vs. effuse, brown to dark-brown, hairy | = 4.9 × 4, n = 20) = 179 × 7.8, n = 30) | = 53.0 × 6.5, n = 25) = 74 × 8.8, n = 30) | acrogenous, solitary, straight or slightly curved, brown to pale brown, rostrate vs. acrogenous, solitary, straight, obpyriform, thick walled, light brown below, hyaline towards apex, rounded at the apex, truncate at base, tapering towards the apex | 5–10 vs. 8–10 | Peat swamp forest vs. unknown | China | decaying wood vs. dead culms of bamboo |
| Loci | PCR Primers | Sequence (5′–3′) | PCR Cycles |
|---|---|---|---|
| ITS | ITS4 | GGA AGT AAA AGT CGT AAC AAG G | (95 °C at 30 s, 55 °C at 30 s, 72 °C at 1 min) × 35 cycles |
| ITS5 | TCC TCC GCT TAT TGA TAT GC | ||
| LSU | LR0R | GTA CCC GCT GAA CTT AAG C | (95 °C at 30 s, 55 °C at 50 s, 72 °C at 1 min) × 35 cycles |
| LR5 | TCC TGA GGG AAA CTT CG | ||
| RPB2 | fRPB2-5F | CAT CGA GAA GTT CGA GAA GG | (95 °C at 45 s, 57 °C at 50 s, 72 °C at 90 s) × 40 cycles |
| fRPB2-7cR | GGA RGT ACC AGT SAT CAT GTT | ||
| TEF1 | TEF1-983F | GCY CCY GGH CAY CGT GAY TTY AT | (94 °C at 30 s, 55 °C at 50 s, 72 °C at 1 min) × 35 cycles |
| TEF1-2218R | AT GAC ACC RAC RGC RAC RGT YTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Yin, C.; Jiang, Y.; Yan, X.; Wang, X.; Zhang, X.; Wang, S. Discovery and Identification of Four Novel Species of Distoseptispora (Distoseptisporaceae, Distoseptisporales) on Decaying Wood from Hainan and Fujian Provinces, China. J. Fungi 2025, 11, 667. https://doi.org/10.3390/jof11090667
Liu W, Yin C, Jiang Y, Yan X, Wang X, Zhang X, Wang S. Discovery and Identification of Four Novel Species of Distoseptispora (Distoseptisporaceae, Distoseptisporales) on Decaying Wood from Hainan and Fujian Provinces, China. Journal of Fungi. 2025; 11(9):667. https://doi.org/10.3390/jof11090667
Chicago/Turabian StyleLiu, Wenwen, Changzhun Yin, Yang Jiang, Xigang Yan, Xingsheng Wang, Xiuguo Zhang, and Shi Wang. 2025. "Discovery and Identification of Four Novel Species of Distoseptispora (Distoseptisporaceae, Distoseptisporales) on Decaying Wood from Hainan and Fujian Provinces, China" Journal of Fungi 11, no. 9: 667. https://doi.org/10.3390/jof11090667
APA StyleLiu, W., Yin, C., Jiang, Y., Yan, X., Wang, X., Zhang, X., & Wang, S. (2025). Discovery and Identification of Four Novel Species of Distoseptispora (Distoseptisporaceae, Distoseptisporales) on Decaying Wood from Hainan and Fujian Provinces, China. Journal of Fungi, 11(9), 667. https://doi.org/10.3390/jof11090667







