The Predicted Structure of S. cerevisiae Ssp1 Reveals Parallel Evolution in the Pil1 BAR Domain Family Proteins of Ascomycetes
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. Plasmids
2.3. Alphafold Predictions and Phylogenetic Analysis
2.4. Microsocopy
3. Results
3.1. Ssp1 Contains a Predicted BAR Domain with Strong Similarity to Pil1
3.2. The AlphaFold Model for the Ssp1 BAR Domain Predicts Amino Acids Critical for Function
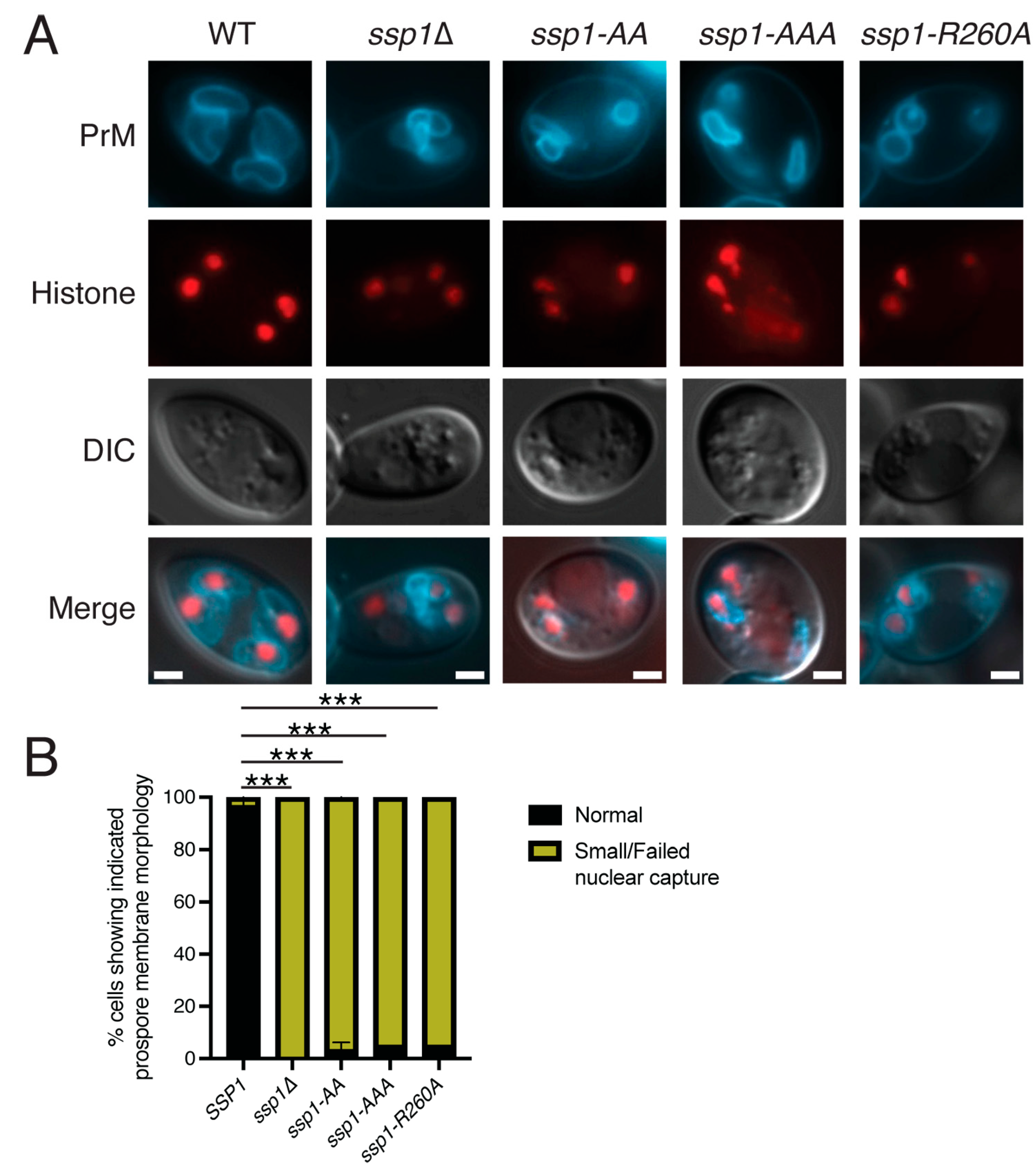
3.3. The Basic Patches on Ssp1 Are Required for Localization to the Leading Edge, but Not Prospore Membrane Association
3.4. The Ssp1R260A Protein Localizes to the Leading Edge
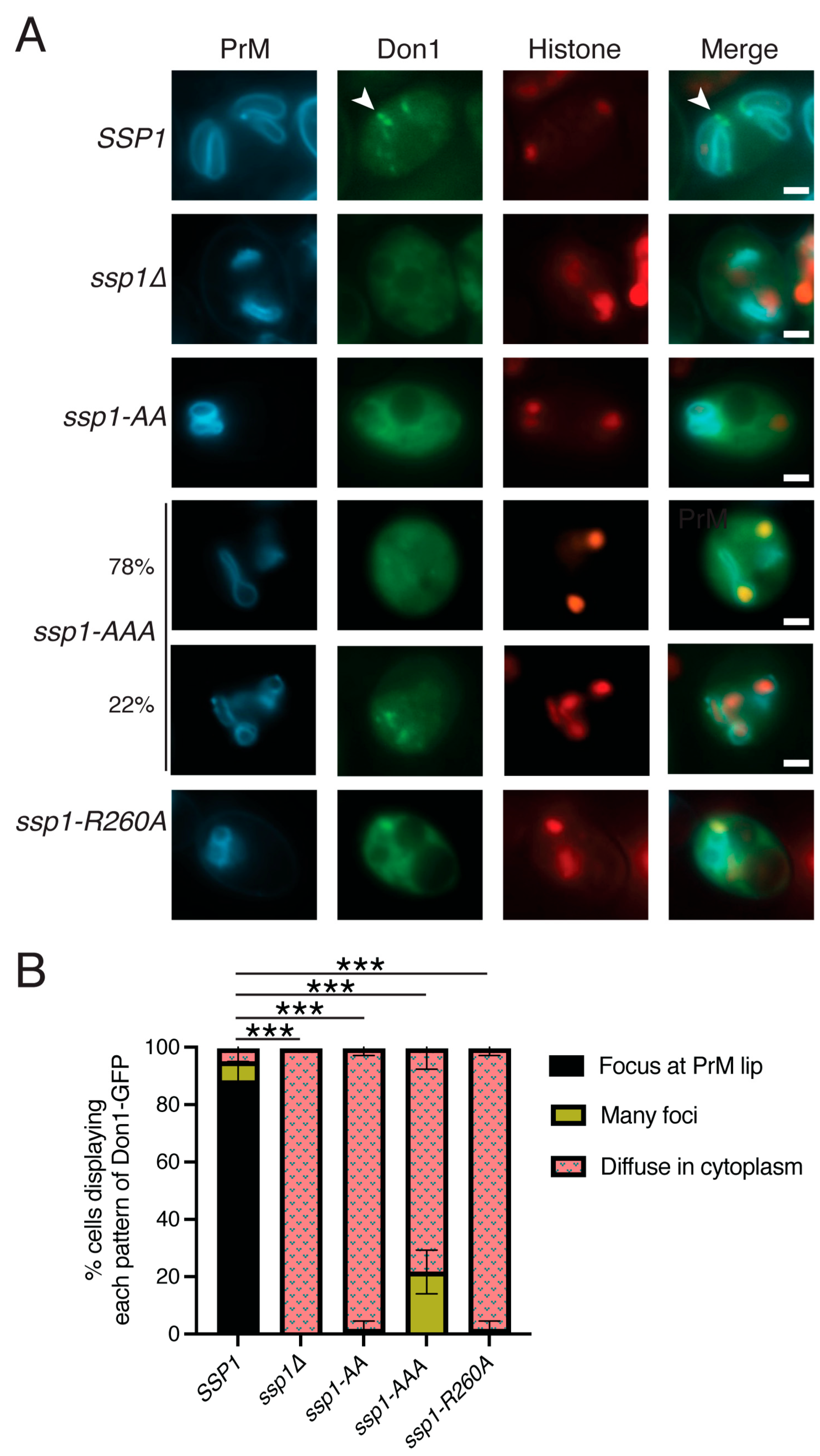
3.5. The BAR Domain Is Required for Leading Edge Complex Formation
3.6. Evolution of the Pil1 Family in Ascomycetes
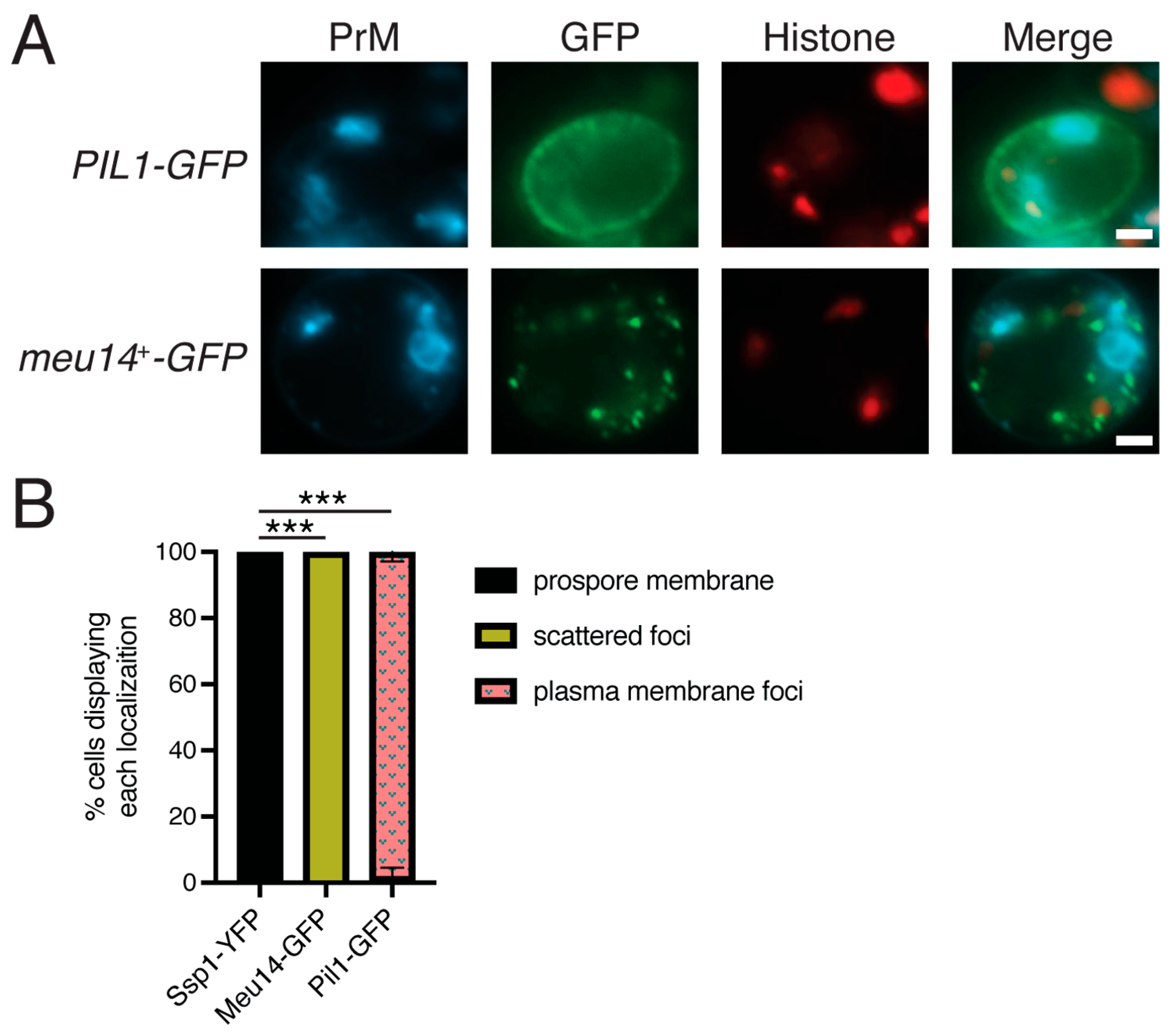
3.7. Neither Pil1 nor Meu14 Localize to the Prospore Membrane
4. Discussion
4.1. SSP1 Encodes a BAR Domain Protein of the Pil1 Family
4.2. Ssp1 Behavior Is Distinct from Pil1/Lsp1
4.3. Convergent Evolution of a Role for Pil1 Family Proteins in Prospore Membrane Assembly
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BAR | Bin/Amphiphysin/Rvs |
| PrM | Prospore Membrane |
| GFP | Green Fluorescent Protein |
| YFP | Yellow Fluorescent Protein |
| BFP | Blue Fluorescent Protein |
References
- Simunovic, M.; Evergren, E.; Callan-Jones, A.; Bassereau, P. Curving cells inside and out: Roles of BAR domain proteins in membrane shaping and its cellular implications. Annu. Rev. Cell Dev. Biol. 2019, 35, 111–129. [Google Scholar] [CrossRef]
- Zimmerberg, J.; McLaughlin, S. Membrane curvature: How BAR domains bend bilayers. Curr. Biol. 2004, 14, R250–R252. [Google Scholar] [CrossRef]
- Foderaro, J.E.; Douglas, L.M.; Konopka, J.B. MCC/Eisosomes regulate cell wall synthesis and stress responses in Fungi. J. Fungi 2017, 3, 61. [Google Scholar] [CrossRef]
- Ziolkowska, N.E.; Karotki, L.; Rehman, M.; Huiskonen, J.T.; Walther, T.C. Eisosome-driven plasma membrane organization is mediated by BAR domains. Nat. Struct. Mol. Biol. 2011, 18, 854–856. [Google Scholar] [CrossRef]
- Olivera-Couto, A.; Grana, M.; Harispe, L.; Aguilar, P.S. The eisosome core is composed of BAR domain proteins. Mol. Biol. Cell 2011, 22, 2360–2372. [Google Scholar] [CrossRef]
- Moreira, K.E.; Walther, T.C.; Aguilar, P.S.; Walter, P. Pil1 controls eisosome biogenesis. Mol. Biol. Cell 2009, 20, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Walther, T.C.; Brickner, J.H.; Aguilar, P.S.; Bernales, S.; Pantoja, C.; Walter, P. Eisosomes mark static sites of endocytosis. Nature 2006, 439, 998–1003. [Google Scholar] [CrossRef]
- Grossmann, G.; Malinsky, J.; Stahlschmidt, W.; Loibl, M.; Weig-Meckl, I.; Frommer, W.B.; Opekarova, M.; Tanner, W. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J. Cell Biol. 2008, 183, 1075–1088. [Google Scholar] [CrossRef]
- Lanze, C.E.; Gandra, R.M.; Foderaro, J.E.; Swenson, K.A.; Douglas, L.M.; Konopka, J.B. Plasma membrane MCC/Eisosome domains promote stress resistance in Fungi. Microbiol. Mol. Biol. Rev. 2020, 84, e00063-19. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.R.; Boxberger, J.; Colvin, R.; Lee, S.J.; Zahn, G.; Loor, F.; Kim, K. Pil1, an eisosome organizer, plays an important role in the recruitment of synaptojanins and amphiphysins to facilitate receptor-mediated endocytosis in yeast. Eur. J. Cell Biol. 2011, 90, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.Q.E.; Ng, A.Y.E.; Zhang, D. Plasma membrane furrows control plasticity of ER-PM contacts. Cell Rep. 2020, 30, 1434–1446 e1437. [Google Scholar] [CrossRef] [PubMed]
- Kabeche, R.; Baldissard, S.; Hammond, J.; Howard, L.; Moseley, J.B. The filament-forming protein Pil1 assembles linear eisosomes in fission yeast. Mol. Biol. Cell 2011, 22, 4059–4067. [Google Scholar] [CrossRef]
- Neiman, A.M. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 1998, 140, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Esposito, R.E.; Klapholz, S. Strathern, J.N., Jones, E.W., Broach, J.R., Eds.; Meiosis and ascospore development. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1981; pp. 211–287. [Google Scholar]
- Neiman, A.M. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 2011, 189, 737–765. [Google Scholar] [CrossRef]
- Lynn, R.R.; Magee, P.T. Development of the spore wall during ascospore formation in Saccharomyces cerevisiae. J. Cell Biol. 1970, 44, 688–692. [Google Scholar] [CrossRef]
- Moens, P.B. Fine structure of ascospore development in the yeast Saccharomyces cerevisiae. Can. J. Microbiol. 1971, 17, 507–510. [Google Scholar] [CrossRef]
- Raju, N.B. Meiosis and ascospore genesis in Neurospora. Eur. J. Cell Biol. 1980, 23, 208–223. [Google Scholar]
- Reeves, F., Jr. The fine structure of ascospore formation in Pyronema domesticum. Mycologia 1967, 59, 1018–1033. [Google Scholar]
- Tanaka, K.; Hirata, A. Ascospore development in the fission yeasts Schizosaccharomyces pombe and S. japonicus. J. Cell Sci. 1982, 56, 263–279. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuka, H.; Imada, K.; Shimasaki, T.; Aiba, H. Sporulation: A response to starvation in the fission yeast Schizosaccharomyces pombe. Microbiologyopen 2022, 11, e1303. [Google Scholar] [CrossRef]
- Shimoda, C. Forespore membrane assembly in yeast: Coordinating SPBs and membrane trafficking. J. Cell Sci. 2004, 117, 389–396. [Google Scholar] [CrossRef]
- Knop, M.; Strasser, K. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. Embo J. 2000, 19, 3657–3667. [Google Scholar] [CrossRef]
- Lam, C.; Santore, E.; Lavoie, E.; Needleman, L.; Fiacco, N.; Kim, C.; Neiman, A.M. A visual screen of protein localization during sporulation identifies new components of prospore membrane-associated complexes in budding yeast. Eukaryot. Cell 2014, 13, 383–391. [Google Scholar] [CrossRef]
- Moreno-Borchart, A.C.; Strasser, K.; Finkbeiner, M.G.; Shevchenko, A.; Shevchenko, A.; Knop, M. Prospore membrane formation linked to the leading edge protein (LEP) coat assembly. Embo J. 2001, 20, 6946–6957. [Google Scholar] [CrossRef]
- Nickas, M.E.; Neiman, A.M. Ady3p links spindle pole body function to spore wall synthesis in Saccharomyces cerevisiae. Genetics 2002, 160, 1439–1450. [Google Scholar] [CrossRef]
- Okuzaki, D.; Satake, W.; Hirata, A.; Nojima, H. Fission yeast meu14+ is required for proper nuclear division and accurate forespore membrane formation during meiosis II. J. Cell Sci. 2003, 116, 2721–2735. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.D.; Fink, G.R. Methods in Yeast Genetics; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1990. [Google Scholar]
- Boeke, J.D.; Trueheart, J.; Natsoulis, G.; Fink, G.R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzym. 1987, 154, 164–175. [Google Scholar]
- Neiman, A.M.; Katz, L.; Brennwald, P.J. Identification of domains required for developmentally regulated SNARE function in Saccharomyces cerevisiae. Genetics 2000, 155, 1643–1655. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar] [CrossRef]
- Christianson, T.W.; Sikorski, R.S.; Dante, M.; Shero, J.H.; Hieter, P. Multifunctional yeast high-copy-number shuttle vectors. Gene 1992, 110, 119–122. [Google Scholar] [CrossRef]
- Niedenthal, R.K.; Riles, L.; Johnston, M.; Hegemann, J.H. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast 1996, 12, 773–786. [Google Scholar] [CrossRef]
- Nunez, G.; Zhang, K.; Moghbeli, K.; Hollingsworth, N.M.; Neiman, A.M. Recruitment of the lipid kinase Mss4 to the meiotic spindle pole promotes prospore membrane formation in Saccharomyces cerevisiae. Mol. Biol. Cell 2023, 34, ar33. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.P.; Kim, C.; Smith, S.O.; Neiman, A.M. A highly redundant gene network controls assembly of the outer spore wall in S. cerevisiae. PLoS Genet. 2013, 9, e1003700. [Google Scholar] [CrossRef]
- Shen, X.X.; Steenwyk, J.L.; LaBella, A.L.; Opulente, D.A.; Zhou, X.; Kominek, J.; Li, Y.; Groenewald, M.; Hittinger, C.T.; Rokas, A. Genome-scale phylogeny and contrasting modes of genome evolution in the fungal phylum Ascomycota. Sci. Adv. 2020, 6, eabd0079. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Hassabis, D. Protein structure predictions to atomic accuracy with AlphaFold. Nat. Methods 2022, 19, 11–12. [Google Scholar] [CrossRef]
- van Kempen, M.; Kim, S.S.; Tumescheit, C.; Mirdita, M.; Lee, J.; Gilchrist, C.L.M.; Soding, J.; Steinegger, M. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 2024, 42, 243–246. [Google Scholar] [CrossRef]
- Peter, B.J.; Kent, H.M.; Mills, I.G.; Vallis, Y.; Butler, P.J.; Evans, P.R.; McMahon, H.T. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science 2004, 303, 495–499. [Google Scholar] [CrossRef]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.W.; Green, T.; Zidek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv 2022. [Google Scholar] [CrossRef]
- Karotki, L.; Huiskonen, J.T.; Stefan, C.J.; Ziolkowska, N.E.; Roth, R.; Surma, M.A.; Krogan, N.J.; Emr, S.D.; Heuser, J.; Grunewald, K.; et al. Eisosome proteins assemble into a membrane scaffold. J. Cell Biol. 2011, 195, 889–902. [Google Scholar] [CrossRef]
- Diamond, A.; Park, J.S.; Inoue, I.; Tachikawa, H.; Neiman, A.M. The APC targeting subunit Ama1 links meiotic exit to cytokinesis during sporulation in Saccharomyces cerevisiae. Mol. Biol. Cell 2008, 20, 134–145. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maier, P.; Rathfelder, N.; Finkbeiner, M.G.; Taxis, C.; Mazza, M.; Le Panse, S.; Haguenauer-Tsapis, R.; Knop, M. Cytokinesis in yeast meiosis depends on the regulated removal of Ssp1p from the prospore membrane. Embo J. 2007, 26, 1843–1852. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.S.; Suda, Y.; Muneshige, K.; Fujieda, Y.; Okumura, Y.; Inoue, I.; Tanaka, T.; Takahashi, T.; Nakanishi, H.; Gao, X.D.; et al. Suppression of Vps13 adaptor protein mutants reveals a central role for PI4P in regulating prospore membrane extension. PLoS Genet. 2021, 17, e1009727. [Google Scholar] [CrossRef]
- Santiago-Tirado, F.H.; Legesse-Miller, A.; Schott, D.; Bretscher, A. PI4P and Rab inputs collaborate in myosin-V-dependent transport of secretory compartments in yeast. Dev. Cell 2011, 20, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Heuser, J.E.; Roth, R.; Goodenough, U. Eisosome Ultrastructure and Evolution in Fungi, Microalgae, and Lichens. Eukaryot. Cell 2015, 14, 1017–1042. [Google Scholar] [CrossRef]
- Athanasopoulos, A.; Boleti, H.; Scazzocchio, C.; Sophianopoulou, V. Eisosome distribution and localization in the meiotic progeny of Aspergillus nidulans. Fungal Genet. Biol. 2013, 53, 84–96. [Google Scholar] [CrossRef]



| Strain | Genotype | Source |
|---|---|---|
| AN117-4B | MATα ura3 his3∆SK leu2 trp1::hisG lys2 arg4-NspI rme1∆::LEU2 ho∆::LYS2 | [30] |
| AN117-16D | MATa ura3 his3∆SK leu2 trp1::hisG lys2 ho∆::LYS2 | [30] |
| NY66 | same as AN117-4B except ssp1∆::kanMX6 | [24] |
| NY67 | same as AN117-16D, except ssp1∆::kanMX6 | [24] |
| AN1159 | same as AN117-4B, except ssp1-K276A K277A K280A | this study |
| AN1160 | same as AN117-16D, except ssp1-K276A K277A K280A | this study |
| AN1161 | same as AN117-4B, except ssp1-K185A R186A | this study |
| AN1162 | same as AN117-16D, except ssp1-K185A R186A | this study |
| AN1163 | same as AN117-16D, except ssp1-R260A | this study |
| AN1164 | same as AN117-4B, except ssp1-R260A | this study |
| AN120 | MATα ura3 his3∆SK leu2 trp1::hisG lys2 arg4-NspI rme1∆::LEU2 ho∆::LYS2 MATa ura3 his3∆SK leu2 trp1::hisG lys2 ARG4 RME1 ho∆::LYS2 | [30] |
| NY551 | same as AN120 except ssp1∆::kanMX6 | [24] |
| AN617 | same as AN120 except ssp1-K276A K277A K280A | this study |
| AN618 | same as AN120 except ssp1-K185A R186A | this study |
| AN619 | same as AN120 except ssp1-R260A | this study |
| Plasmid | Relevant Features | Source |
|---|---|---|
| pRS306 | URA3 | [31] |
| pRS314 | TRP1 CEN ARS | [31] |
| pRS313 | HIS3 CEN ARS | [31] |
| pRS424 | 2µ TRP1 | [32] |
| pRS426 | 2µ URA3 | [32] |
| pGFP-C-FUS | GFP | [33] |
| pRS424-PSPO20-GFP | 2µ TRP1 PSPO20 | H. Nakanishi |
| pRS424-PSPO20 | 2µ TRP1 PSPO20 GFP | H. Nakanishi |
| pRS306-SSP1 | URA3 SSP1 | This study |
| pRS306-SSP1-AA | URA3 | this study |
| pRS306-SSP1-AAA | URA3 | this study |
| pRS306-SSP1-R260A | URA3 | this study |
| pRS314-SSP1-YFP | TRP1 CEN ARS SSP1-YFP | [24] |
| pRS314-SSP1-AA-YFP | TRP1 CEN ARS ssp1-K185A R186A-YFP | this study |
| pRS314-SSP1-AAA-YFP | TRP1 CEN ARSssp1-K276A K277A K280A-YFP | this study |
| pRS314-SSP1-R260A-YFP | TRP1 CEN ARS ssp1-R260A-YFP | this study |
| pRS313-HTB1-mOrange2 | HIS3 CEN ARS HTB1-mOrange2 | [34] |
| pRS426-Spo2051-91-mTagBFP | URA3 2µ spo2051-91-mTagBFP | [35] |
| pRS424-PSPO20-Pil1-GFP | TRP1 2µ PSPO20-PIL1-GFP | this study |
| pRS424-PSPO20-Meu14-GFP | TRP1 2µ PSPO20-meu14+-GFP | this study |
| pSB9 | TRP1 2µ DON1-GFP | [24] |
| Genotype 1 | Gene Expressed from Plasmid 2 | % Sporulation (±SD) 3 |
|---|---|---|
| SSP1 | - | 73.8 (±4.9) |
| ssp1∆ | - | 0 (±0) |
| ssp1-AA | - | 0 (±0) |
| ssp1-AA | SSP1 | 68 (±6.5) |
| ssp1-AAA | - | 0 (±0) |
| ssp1-AAA | SSP1 | 70.3 (±8) |
| ssp1-R260A | - | 0 (±0) |
| ssp1-R260A | SSP1 | 71 (±4.4) |
| ssp1∆ | SSP1-YFP | 32.5 (±6.6) |
| ssp1∆ | ssp1-AA-YFP | 0 (±0) |
| ssp1∆ | ssp1-AAA-YFP | 15.5 (±1.8) |
| ssp1∆ | ssp1-R260A-YFP | 0 (±0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suda, Y.; Neiman, A.M. The Predicted Structure of S. cerevisiae Ssp1 Reveals Parallel Evolution in the Pil1 BAR Domain Family Proteins of Ascomycetes. J. Fungi 2025, 11, 661. https://doi.org/10.3390/jof11090661
Suda Y, Neiman AM. The Predicted Structure of S. cerevisiae Ssp1 Reveals Parallel Evolution in the Pil1 BAR Domain Family Proteins of Ascomycetes. Journal of Fungi. 2025; 11(9):661. https://doi.org/10.3390/jof11090661
Chicago/Turabian StyleSuda, Yasuyuki, and Aaron M. Neiman. 2025. "The Predicted Structure of S. cerevisiae Ssp1 Reveals Parallel Evolution in the Pil1 BAR Domain Family Proteins of Ascomycetes" Journal of Fungi 11, no. 9: 661. https://doi.org/10.3390/jof11090661
APA StyleSuda, Y., & Neiman, A. M. (2025). The Predicted Structure of S. cerevisiae Ssp1 Reveals Parallel Evolution in the Pil1 BAR Domain Family Proteins of Ascomycetes. Journal of Fungi, 11(9), 661. https://doi.org/10.3390/jof11090661







