Biodegradation of Chloroquine by a Fungus from Amazonian Soil, Penicillium guaibinense CBMAI 2758
Abstract
1. Introduction
2. Results and Discussion
2.1. Identification of Filamentous Fungi
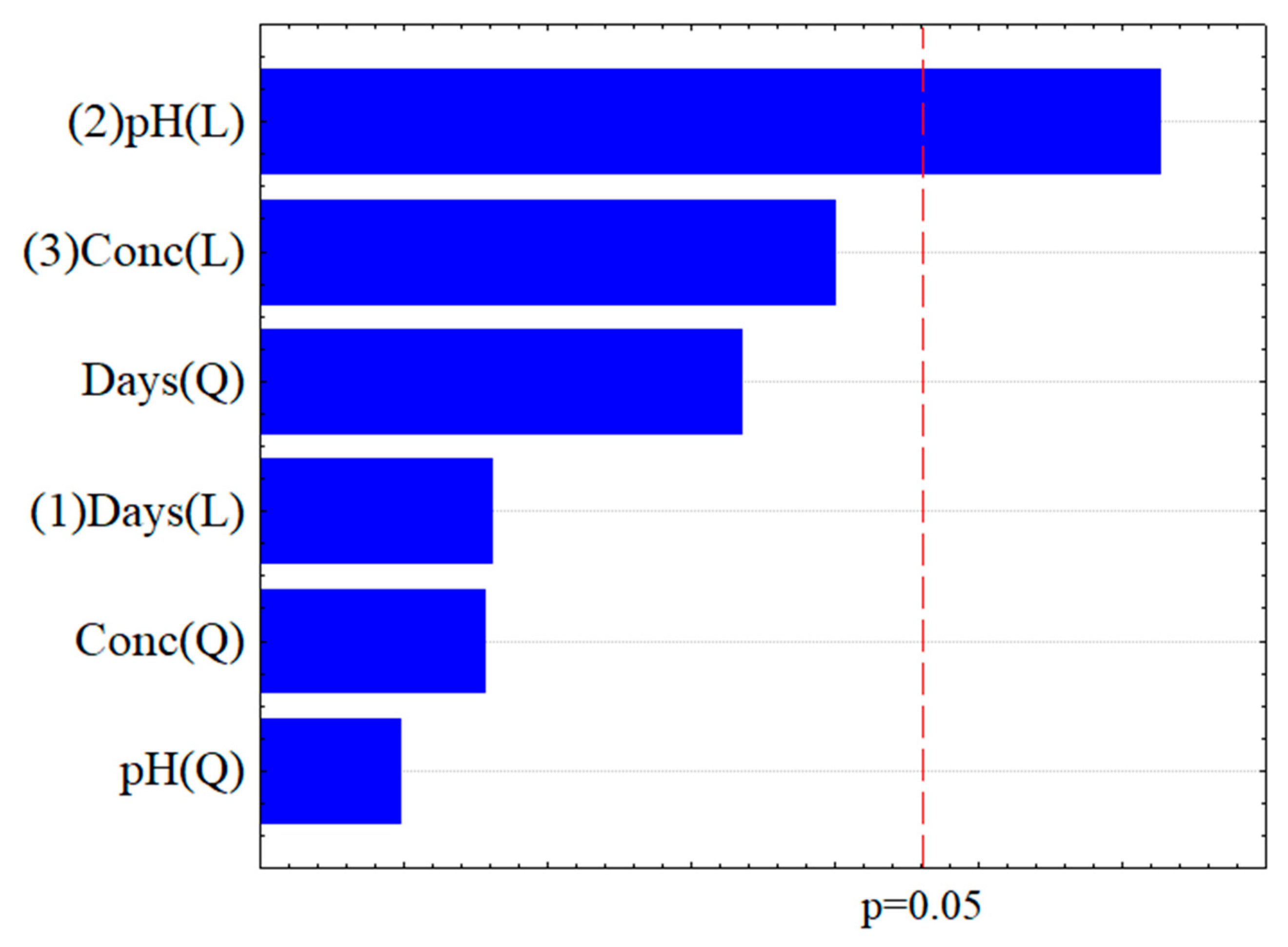
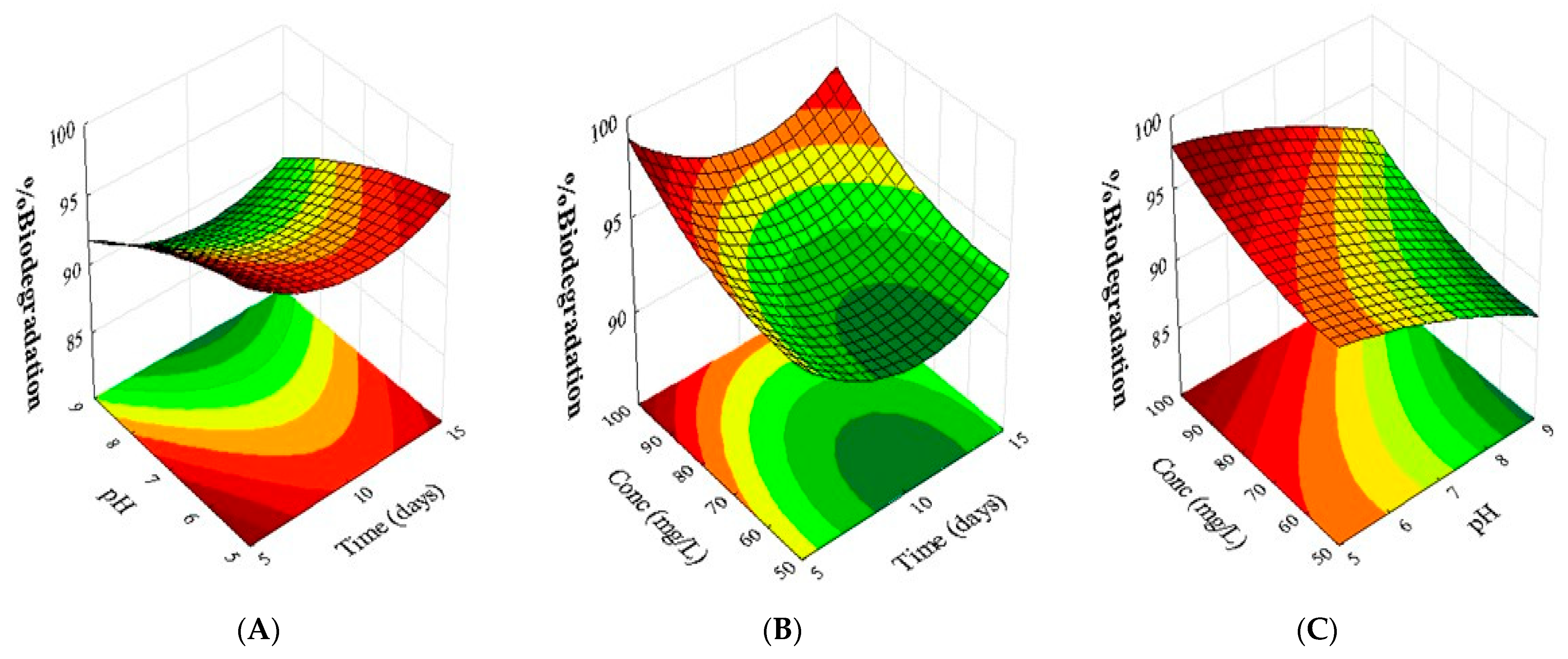
2.2. Evaluation of the Biodegradation of Chloroquine Diphosphate Through Experimental Design
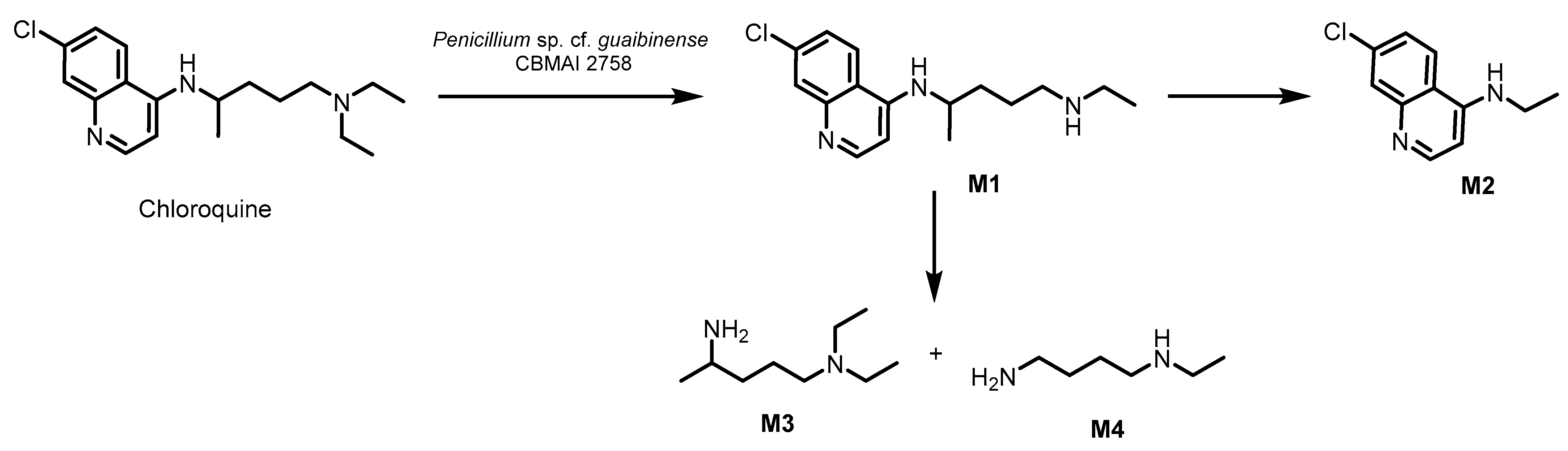
2.3. Identification of Biodegradation Metabolites by GC-MS
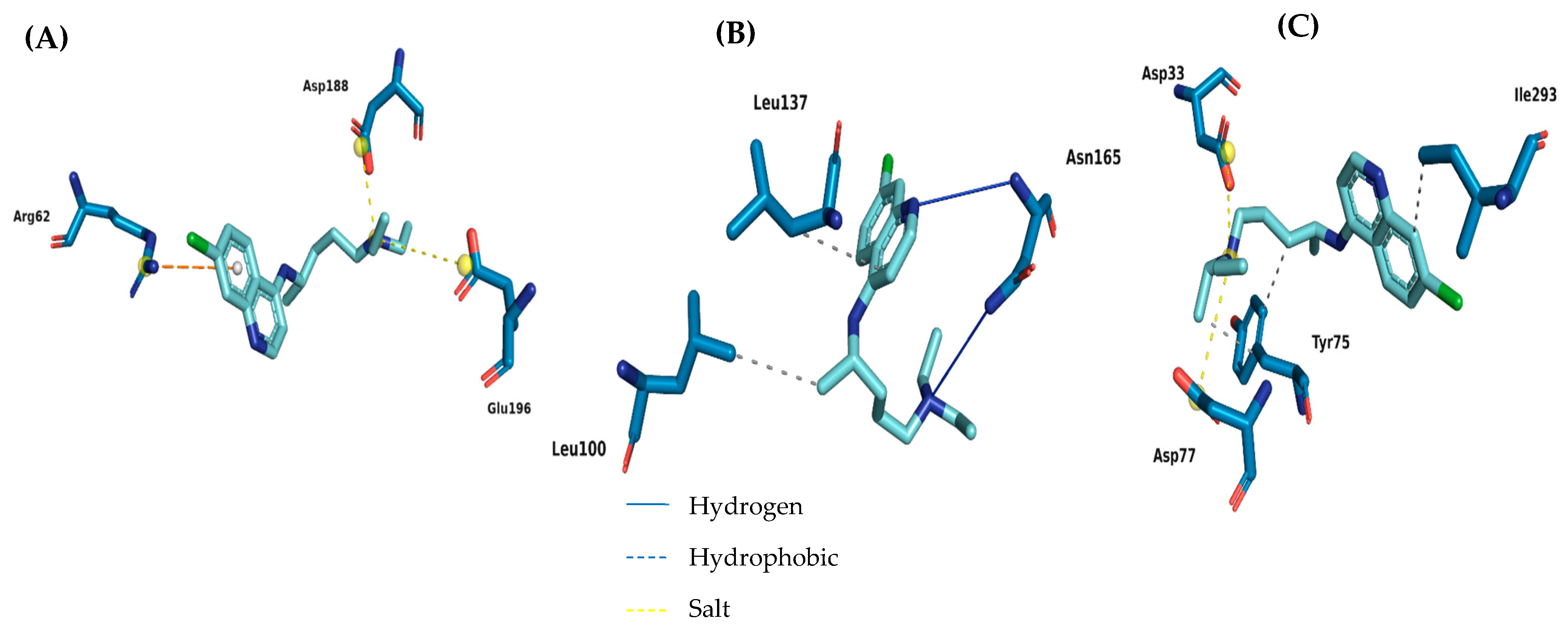
2.4. Toxicological Analysis
3. Materials and Methods
3.1. Reagents and Solvents
3.2. Isolation and Identification of Filamentous Fungi
3.3. Radial Growth of Strains in the Presence of Chloroquine Diphosphate (Solid)
3.4. Experimental Design and Statistical Model
3.5. Biodegradation Reaction of Chloroquine Diphosphate by the Fungus in Liquid Medium
3.6. Extraction of Chloroquine Diphosphate and Its Metabolites
3.7. Method Validation
3.8. Detection of Chloroquine Diphosphate and Metabolites Through Gas Chromatography–Mass Spectrometry (GC-MS) Analysis
3.9. Toxicity Predictions
3.10. Molecular Docking Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CBMAI | Brazilian Collection of Environmental and Industrial Microorganisms |
| pH | hydrogen of potential |
| EOMs | Emerging organic contaminants |
| GC-MS | Gas chromatography–mass spectrometry |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| COVID-19 | Coronavirus disease 2019 |
| RGR | Radial growth rate |
| EPS | Extracellular polymeric substances |
| CTC | Chlortetracycline |
| NSAIDs | Non-steroidal anti-inflammatory drugs |
| CAS | Chemical Abstracts Service |
| HPLC | High-performance liquid chromatography |
| CPQBA | Multidisciplinary Center for Chemical, Biological, and Agricultural Research |
| DNA | Deoxyribonucleic acid |
| PCR | Polymerase chain reaction |
| HCl | Hydrochloric acid |
| KOH | Sodium hydroxide |
| DMSO | Dimethyl sulfoxide |
| BD | Biodegradation rate |
| RDC | Collegiate Board Resolution |
| ANVISA | National Health Surveillance Agency |
| RSD | Relative standard deviation |
| CV | Coefficient of variation |
| SD | Standard deviation |
| DMC | Determined mean concentration |
| LD | Limit of detection |
| LQ | Limit of quantification |
References
- Serafin, M.B.; Bottega, A.; Foletto, V.S.; da Rosa, T.F.; Hörner, A.; Hörner, R. Drug Repositioning Is an Alternative for the Treatment of Coronavirus COVID-19. Int. J. Antimicrob. Agents 2020, 55, 105969. [Google Scholar] [CrossRef]
- Furlan, L.; Caramelli, B. The Regrettable Story of the “Covid Kit” and the “Early Treatment of COVID-19” in Brazil. Lancet Reg. Health Am. 2021, 4, 100089. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, R.; Xu, M.; Wang, X.; Zhang, H.; Hu, H.; Li, Y.; Hu, Z.; Zhong, W.; Wang, M. Hydroxychloroquine, a Less Toxic Derivative of Chloroquine, Is Effective in Inhibiting SARS-CoV-2 Infection in Vitro. Cell Discov. 2020, 6, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Cortegiani, A.; Ingoglia, G.; Ippolito, M.; Giarratano, A.; Einav, S. A systematic review on the efficacy and safety of chloroquine for the treatment of COVID-19. J. Crit. Care 2020, 57, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-Approved Drug Ivermectin Inhibits the Replication of SARS-CoV-2 in Vitro. Antivir. Res. 2020, 178, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Mirtaleb, M.S.; Mirtaleb, A.H.; Nosrati, H.; Heshmatnia, J.; Falak, R.; Zolfaghari Emameh, R. Potenciais agentes terapêuticos para COVID-19: Uma revisão atualizada sobre terapia antiviral, imunoterapia e terapia celular. Biomed. Pharmacother. 2021, 138, 111518. [Google Scholar] [CrossRef]
- Lou, Y.; Liu, L.; Yao, H.; Hu, X.; Su, J.; Xu, K.; Luo, R.; Yang, X.; He, L.; Lu, X.; et al. Clinical Outcomes and Plasma Concentrations of Baloxavir Marboxil and Favipiravir in COVID-19 Patients: An Exploratory Randomized, Controlled Trial. Eur. J. Pharm. Sci. 2021, 157, 105631. [Google Scholar] [CrossRef]
- Antinori, S.; Cossu, M.V.; Ridolfo, A.L.; Rech, R.; Bonazzetti, C.; Pagani, G.; Gubertini, G.; Coen, M.; Magni, C.; Castelli, A.; et al. Compassionate Remdesivir Treatment of Severe COVID-19 Pneumonia in Intensive Care Unit (ICU) and Non-ICU Patients: Clinical Outcome and Differences in Post-Treatment Hospitalisation Status. Pharmacol. Res. 2020, 158, 104899. [Google Scholar] [CrossRef]
- Khalili, J.S.; Zhu, H.; Mak, N.S.A.; Yan, Y.; Zhu, Y. Novel Coronavirus Treatment with Ribavirin: Groundwork for an Evaluation Concerning COVID-19. J. Med. Virol. 2020, 92, 740–746. [Google Scholar] [CrossRef]
- Dai, H.; Wang, C.; Yu, W.; Han, J. Tracing COVID-19 Drugs in the Environment: Are We Focusing on the Right Environmental Compartment? Environ. Pollut. 2023, 339, 122732. [Google Scholar] [CrossRef]
- Saadat, D.; Rawtani, C.M. Hussain. Environmental perspective of COVID-19. Sci. Total Environ. 2020, 728, 1138870. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, B.; Tscharke, P.M.; Bertsch, K.; Bibby, A.; Bivins, P.; Choi, L.; Clarke, J.; Dwyer, J.; Edson, T.M.H.; Nguyen, J.W.O.; et al. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community. Sci. Total Environ. 2020, 761, 144216. [Google Scholar] [CrossRef] [PubMed]
- Picó, Y.; Barceló, D. Microplastics and other emerging contaminants in the environment after COVID-19 pandemic: The need of global reconnaissance studies. Curr. Opin. Environ. Sci. Health 2023, 33, 100468. [Google Scholar] [CrossRef] [PubMed]
- Aus der Beek, T.; Weber, F.A.; Bergmann, A.; Hickmann, S.; Ebert, I.; Hein, A.; Küster, A. Pharmaceuticals in the Environment-Global Occurrences and Perspectives. Environ. Toxicol. Chem. 2016, 35, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Nannou, C.; Ofrydopoulou, A.; Evgenidou, E.; Heath, D.; Heath, E.; Lambropoulou, D. Antiviral Drugs in Aquatic Environment and Wastewater Treatment Plants: A Review on Occurrence, Fate, Removal and Ecotoxicity. Sci. Total Environ. 2020, 699, 134322. [Google Scholar] [CrossRef] [PubMed]
- Morales-Paredes, C.A.; Rodríguez-Díaz, J.M.; Boluda-Botella, N. Pharmaceutical Compounds Used in the COVID-19 Pandemic: A Review of Their Presence in Water and Treatment Techniques for Their Elimination. Sci. Total Environ. 2022, 814, 152691. [Google Scholar] [CrossRef]
- Asif, M.B.; Li, C.; Ren, B.; Maqbool, T.; Zhang, X.; Zhang, Z. Elucidating the Impacts of Intermittent In-Situ Ozonation in a Ceramic Membrane Bioreactor: Micropollutant Removal, Microbial Community Evolution and Fouling Mechanisms. J. Hazard. Mater. 2021, 402, 123730. [Google Scholar] [CrossRef]
- Mohd Hanafiah, Z.; Wan Mohtar, W.H.M.; Wan-Mohtar, W.A.A.Q.I.; Bithi, A.S.; Rohani, R.; Indarto, A.; Yaseen, Z.M.; Sharil, S.; Binti Abdul Manan, T.S. Removal of Pharmaceutical Compounds and Toxicology Study in Wastewater Using Malaysian Fungal Ganoderma Lucidum. Chemosphere 2024, 358, 142209. [Google Scholar] [CrossRef]
- Williams, N.S.; Ray, M.B.; Gomaa, H.G. Removal of Ibuprofen and 4-Isobutylacetophenone by Non-Dispersive Solvent Extraction Using a Hollow Fibre Membrane Contactor. Sep. Purif. Technol. 2012, 88, 61–69. [Google Scholar] [CrossRef]
- Stachowiak, M.; Cegłowski, M.; Kurczewska, J. Hybrid Chitosan/Molecularly Imprinted Polymer Hydrogel Beads Doped with Iron for Selective Ibuprofen Adsorption. Int. J. Biol. Macromol. 2023, 251, 126356. [Google Scholar] [CrossRef]
- Magaela, B.N.; Ndlovu, K.S.; Tshangana, C.S.; Muleja, A.A.; Mamba, B.B.; Nyokong, T.; Managa, M. Photodegradation of Ibuprofen Using 5-10-15-20-Tetrakis(4-Bromophenyl) Porphyrin Conjugated to Graphene Quantum Dots. Opt. Mater. 2022, 134, 113147. [Google Scholar] [CrossRef]
- Danner, M.C.; Robertson, A.; Behrends, V.; Reiss, J. Antibiotic Pollution in Surface Fresh Waters: Occurrence and Effects. Sci. Total Environ. 2019, 664, 793–804. [Google Scholar] [CrossRef]
- Hasan, M.K.; Alfredo, S.; Murthy, R. Riffat. Biodegradation of salicylic acid, acetaminophen and ibuprofen by bacteria collected from a full-scale drinking water biofilter. J. Environ. Manag. 2021, 295, 113071. [Google Scholar] [CrossRef] [PubMed]
- 24. Holanda, F.H.e.; Birolli, W.G.; Morais, E.d.S.; Sena, I.S.; Ferreira, A.M.; Faustino, S.M.M.; Grace, d.S.; Solon, L.; Porto, A.M.; Ferreira, I.M. Study of Biodegradation of Chloramphenicol by Endophytic Fungi Isolated from Bertholletia excelsa (Brazil Nuts). Biocatal. Agric. Biotechnol. 2019, 20, 101200. [Google Scholar] [CrossRef]
- Midassi, S.; Bedoui, A.; Bensalah, N. Efficient Degradation of Chloroquine Drug by Electro-Fenton Oxidation: Effects of Operating Conditions and Degradation Mechanism. Chemosphere 2020, 260, 127558. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Huang, Y.; Wang, X.; Liu, Y.; Lin, W.; Zhang, F.; Chen, X. Degradation of Low Concentration Chloroquine Phosphate by Ozone Oxidation. Proc. J. Phys. Conf. Ser. Inst. Phys. 2024, 2679, 012050. [Google Scholar] [CrossRef]
- Birolli, W.G.; de A. Santos, D.; Alvarenga, N.; Garcia, A.C.F.S.; Romão, L.P.C.; Porto, A.L.M. Biodegradation of Anthracene and Several PAHs by the Marine-Derived Fungus Cladosporium Sp. CBMAI 1237. Mar. Pollut. Bull. 2018, 129, 525–533. [Google Scholar] [CrossRef]
- Alvarenga, N.; Birolli, W.G.; Nitschke, M.; Rezende, M.O.O.; Seleghim, M.H.R.; Porto, A.L.M. Biodegradation of Chlorpyrifos by Whole Cells of Marine-Derived Fungi Aspergillus sydowii and Trichoderma sp. J. Microb. Biochem. Technol. 2015, 7, 133–139. [Google Scholar] [CrossRef]
- Li, Z.; Wang, J.; Gu, C.; Guo, Y.; Wu, S. Marine Bacteria-Mediated Abiotic-Biotic Coupling Degradation Mechanism of Ibuprofen. J. Hazard. Mater. 2022, 435, 128960. [Google Scholar] [CrossRef]
- Murshid, S.; Dhakshinamoorthy, G.P. Biodegradation of Sodium Diclofenac and Mefenamic Acid: Kinetic Studies, Identification of Metabolites and Analysis of Enzyme Activity. Int. Biodeterior. Biodegrad. 2019, 144, 104756. [Google Scholar] [CrossRef]
- Arun, K.B.; Madhavan, A.; Tarafdar, A.; Sirohi, R.; Anoopkumar, A.N.; Kuriakose, L.L.; Awasthi, M.K.; Binod, P.; Varjani, S.; Sindhu, R. Filamentous fungi for pharmaceutical compounds degradation in the environment: A sustainable approach. Environ. Technol. Innov. 2023, 31, 103182. [Google Scholar] [CrossRef]
- dos Santos, V.H.P.; dos Santos, W.T.; Ionta, M.; de Paula, A.C.C.; Silva, E.O. Biotransformation of hydroxychloroquine to evaluate the cytotoxicity of its metabolites and mimic mammalian metabolism. Results Chem. 2023, 5, 100761. [Google Scholar] [CrossRef] [PubMed]
- Correa, L.O.; Bezerra, A.F.M.; Honorato, L.R.S.; Cortez, A.C.A.; Souza, J.V.B.; Souza, E.S. Amazonian soil fungi are efficient degraders of glyphosate herbicide; novel isolates of Penicillium, Aspergillus, and Trichoderma. Braz. J. Biol. 2023, 83, e242830. [Google Scholar] [CrossRef] [PubMed]
- Ramos, M.M.; Dos SMorais, E.; da SSena, I.; Lima, A.L.; de Oliveira, F.R.; de Freitas, C.M.; Fernandes, C.P.; de Carvalho, J.C.T.; Ferreira, I.M. Silver nanoparticle from whole cells of the fungi Trichoderma spp. isolated from Brazilian Amazon. Biotechnol. Lett. 2020, 42, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Nóbrega, P.A.; Ferreira, B.L.; Sá, L.S.; Braga, F.C.; Bezerra, R.M.; Ferreira, M.I. Isolation and Identification of Iron, Chromium, and Manganese Tolerant Filamentous Fungi from Mining Soil. Curr. Biotechnol. 2024, 13, 174–184. [Google Scholar] [CrossRef]
- Oliveira, A.N.; Oliveira, L.A.; Andrade, J.S. Chagas júnior, A.F. Atividade enzimática de isolados de rizóbia nativos da amazônia central crescendo em diferentes níveis de acidez. Ciênc. Tecnol. Aliment. Camp. 2006, 26, 204–210. [Google Scholar] [CrossRef]
- Colla, L.M.; Primaz, A.L.; de Lima, M.; Bertolin, T.E.; Costa, J.A.V. Isolation and Screening of Fungi to Bioremediation from Triazine Herbicide Contaminated Soil. Cienc. Agrotecnologia 2008, 32, 809–813. [Google Scholar] [CrossRef]
- Tortora, G.J.; Case, C.L.; Funke, B.R. Microbiologia; Artmed: Porto Alegre, Brazil, 2016; pp. 1–964. ISBN 8582713541/9788582713549. [Google Scholar]
- Palanivel, T.M.; Pracejus, B.; Novo, L.A. Bioremediation of copper using indigenous fungi Aspergillus species isolated from an abandoned copper mine soil. Chemosphere 2023, 314, 37688. [Google Scholar] [CrossRef]
- Rigo, D.; Luana, G.; Tres, G.A.; Camera, F.D.; Zeni, J. Microbiological Production of Enzymes: A Review. Braz. J. Dev. 2021, 7, 9232–9254. [Google Scholar] [CrossRef]
- Sena, I.S.; Ferreira, A.M.; Marinho, V.H.; e Holanda, F.H.; Borges, S.F.; de Souza, A.A.; de Carvalho, R.; Koga, R.; Lima, A.L.; Florentino, A.C.; et al. Euterpe oleracea Mart (Açaizeiro) from the Brazilian Amazon: A Novel Font of Fungi for Lipase Production. Microorganisms 2022, 10, 2394. [Google Scholar] [CrossRef]
- Daneshfar, A.; Vafafard, S. Solubility of Chloroquine Diphosphate and 4,7-Dichloroquinoline in Water, Ethanol, Tetrahydrofuran, Acetonitrile, and Acetone from (298.2 to 333.2) K. J. Chem. Eng. Data 2009, 54, 2170–2174. [Google Scholar] [CrossRef]
- Tomasini, A.; Leon-Santiesteban, H.H. Fungal Bioremediation: Fundamentals and Applications; Tomasini, A., Leâon-Santiesteban, H.H., Eds.; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Rodriguez-Rodrıíguez, C.E.; Caminal, G.; Vicent, T.; Diaz-Cruz, M.S.; Eljarrat, E.; Farre, M.; López de Alda, M.J.; Petrovic, M.; Barcelo, D. Fungal-Mediated Degradation of Emerging Pollutants in Sewage Sludge. Handb. Environ. Chem. 2013, 24, 137–164. [Google Scholar] [CrossRef]
- Akerman-Sanchez, G.; Rojas-Jimenez, K. Fungi for the Bioremediation of Pharmaceutical-Derived Pollutants: A Bioengineering Approach to Water Treatment. J. Fungi 2021, 4, 100071. [Google Scholar] [CrossRef]
- Park, H.; Choi, I.G. Genomic and transcriptomic perspectives on mycoremediation of polycyclic. aromatic hydrocarbons. Appl. Microbiol. Biotechnol. 2020, 104, 6919–6928. [Google Scholar] [CrossRef]
- Cai, Y.; Yu, H.; Ren, L.; Ou, Y.; Jiang, S.; Chai, Y.; Chen, A.; Yan, B. Treatment of amoxicillin-containing wastewater by Trichoderma strains selected from activated sludge. Sci. Total Environ. 2023, 867, 161565. [Google Scholar] [CrossRef]
- Li, Y.; Chen, H.; Wang, Y.; Yang, Z.; Zhang, H. Efficient biodegradation of chlortetracycline in high concentration from strong-acidity pharmaceutical residue with degrading fungi. J. Hazard. Mater. 2022, 424, 127671. [Google Scholar] [CrossRef] [PubMed]
- Bankole, P.O.; Adekunle, A.A.; Jeon, B.H.; Govindwar, S.P. Novel cobiomass degradation of NSAIDs by two wood rot fungi, Ganoderma applanatum and Laetiporus sulphureus: Ligninolytic enzymes induction, isotherm and kinetic studies. Ecotoxicol. Environ. Saf. Ecotoxicol. 2020, 203, 110997. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Z.; Wang, G.; Zhang, G.; Wang, Q.; Wang, W. New insight into biodegradation mechanism of phenylurea herbicides by cytochrome P450 enzymes: Successive N-demethylation. Environ. Int. 2023, 182, 108332. [Google Scholar] [CrossRef] [PubMed]
- Marmor, M.F.; Kellner, U.; Lai, T.Y.Y.; Melles, R.B.; Mieler, W.F. Recommendations on Screening for Chloroquine and Hydroxychloroquine Retinopathy (2016 Revision). Ophthalmology 2016, 123, 1386–1394. [Google Scholar] [CrossRef]
- Chorin, E.; Dai, M.; Shulman, E.; Wadhwani, L.; Bar-Cohen, R.; Barbhaiya, C.; Aizer, A.; Holmes, D.; Bernstein, S.; Spinelli, M.; et al. The QT Interval in Patients with COVID-19 Treated with Hydroxychloroquine and Azithromycin. Nat. Med. 2020, 26, 808–809. [Google Scholar] [CrossRef]
- Van Eaton, K.M.; Gustafson, D.L. Pharmacokinetic and Pharmacodynamic Assessment of Hydroxychloroquine in Breast Cancer. J. Pharmacol. Exp. Ther. 2021, 379, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Rynes, R.I. Antimalarial drugs in the treatment of rheumatological diseases. Rheumatology 1997, 36, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Matúš, P.; Littera, P.; Farkas, B.; Urík, M. Review on Performance of Aspergillus and Penicillium Species in Biodegradation of Organochlorine and Organophosphorus Pesticides. Microorganisms 2023, 11, 1485. [Google Scholar] [CrossRef] [PubMed]
- Sowmya, H.V.; Ramalingappa; Krishnappa, M.; Thippeswamy, B. Degradation of Polyethylene by Penicillium simplicissimum Isolated from Local Dumpsite of Shivamogga District. Environ. Dev. Sustain. 2015, 17, 731–745. [Google Scholar] [CrossRef]
- Boucherit, Z.; Flahaut, S.; Djoudi, B.; Mouas, T.N.; Mechakra, A.; Ameddah, S. Potential of Halophilic Penicillium chrysogenum Isolated from Algerian Saline Soil to Produce Laccase on Olive Oil Wastes. Curr. Microbiol. 2022, 79, 178. [Google Scholar] [CrossRef] [PubMed]
- Selim, S.; Al-Sanea, M.M.; Alhejely, A.; Moawad, H.; Masmali, I.; Hendawy, O.M. Degradative potential of laccase and manganese peroxidase to mycotoxins on infected maize grains by fungi with docking interaction studies. Bioresources 2024, 19, 9773–9787. [Google Scholar] [CrossRef]
- Sundaramoorthy, M.; Kishi, K.; Gold, M.H.; Poulos, T.L. Preliminary crystallographic analysis of manganese peroxidase from Phanerochaete chrysosporium. J. Mol. Biol. 1994, 238, 845–848. [Google Scholar] [CrossRef]
- Ducros, V.; Brzozowski, A.M.; Wilson, K.S.; Østergaard, P.; Schneider, P.; Svendson, A.; Davies, G.J. Structure of the laccase from Coprinus cinereus at 1.68 Å resolution: Evidence for different ‘type 2 Cu-depleted’ isoforms. Acta Crystallogr. D Biol. Crystallogr. 2001, 57 Pt 3, 333–336. [Google Scholar] [CrossRef]
- Oakley, A.J. The Structure of Aspergillus niger Phytase PhyA in Complex with a Phytate Mimetic. Biochem. Biophys. Res. Commun. 2010, 397, 745–749. [Google Scholar] [CrossRef]
- Koszelak, S.; Ng, J.D.; Day, J.; Ko, T.P.; Greenwood, A.; McPherson, A. The Crystallographic Structure of the Subtilisin Protease from Penicillium cyclopium. Biochemistry 1997, 36, 6597–6604. [Google Scholar] [CrossRef]
- Vacondio, B.W.G.; Birolli, I.M.; Ferreira, M.H.; Seleghim, S.; Gonçalves, S.P.; Vasconcellos, A.L. Biodegradation of pentachlorophenol by marine-derived fungus Trichoderma harzianum CBMAI 1677 isolated from ascidian Didemnun ligulum. Biocatal. Agric. Biotechnol. 2015, 4, 266–275. [Google Scholar] [CrossRef]
- Soares, P.R.S.; Birolli, W.G.; Ferreira, I.M.; Porto, A.L.M. Biodegradation pathway of the organophosphate pesticides chlorpyrifos, methyl parathion and profenofos by the marine-derived fungus Aspergillus sydowii CBMAI 935 and its potential for methylation reactions of phenolic compounds. Mar. Pollut. Bull. 2021, 166, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Ramos, R.S.; Borges, R.S.; de Souza, J.S.N.; Araujo, I.F.; Chaves, M.H.; Santos, C.B.R. Identification of Potential Antiviral Inhibitors from Hydroxychloroquine and 1,2,4,5-Tetraoxanes Analogues and Investigation of the Mechanism of Action in SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 1781. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, E.S.R.; De Faveri Moreira, R.; da Silva Ramos, R.; de Souza, S.A.; Filho, J.W.M.S.; da Silva, B.J.P.; Jumbo, L.O.V.; de Oliveira, E.E.; Lima, E.S.; de Souza Aguiar, R.W. Safety Assessment of the Ethanolic Extract of Siparuna guianensis: Cell Viability, Molecular Risk Predictions and Toxicity Risk for Acute and Sub-Chronic Oral Ingestion. J. Ethnopharmacol. 2025, 347, 119751. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.R.; Parrish, J.C.; Fraser, M.E.; Smith, W.W.; Bartlett, P.A.; James, M.N.G. Lowering the Entropic Barrier for Binding Conformationally Flexible Inhibitors to Enzymes. Biochemistry 1998, 37, 16839–16845. [Google Scholar] [CrossRef]
- Guedes, I.A.; Pereira da Silva, M.M.; Galheigo, M.; Krempser, E.; de Magalhães, C.S.; Correa Barbosa, H.J.; Dardenne, L.E. DockThor-VS: A Free Platform for Receptor-Ligand Virtual Screening. J. Mol. Biol. 2024, 436, 168548. [Google Scholar] [CrossRef]
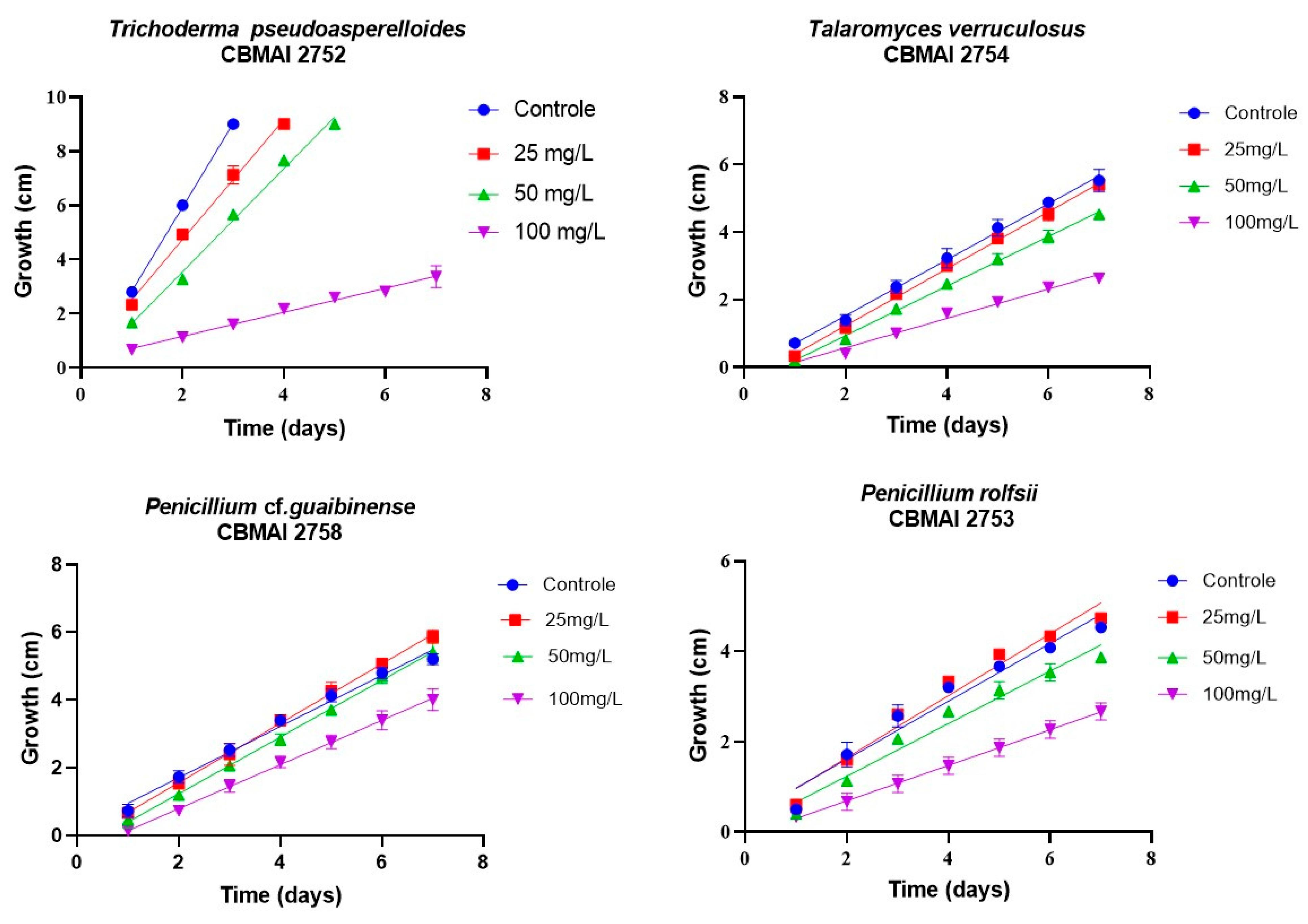
| Fungus | Growth Medium | Time (Days) | Growth Speed (cm/d) | |
|---|---|---|---|---|
| 3 | 6 | |||
| Trichoderma pseudoasperelloides CBMAI 2752 | Control |  | 3.10 | |
| Chloroquine 25 mg·L−1 |  | 2.22 | ||
| Chloroquine 50 mg·L−1 |  | 1.9 | ||
| Chloroquine 100 mg·L−1 |  | 0.44 | ||
| Penicillium rolfsii CBMAI 2753 | Control |  | 0.82 | |
| Chloroquine 25 mg·L−1 |  | 0.84 | ||
| Chloroquine 50 mg·L−1 |  | 0.733 | ||
| Chloroquine 100 mg·L−1 |  | 0.435 | ||
| Talaromyces verruculosus CBMAI 2754 | Control |  | 0.75 | |
| Chloroquine 25 mg·L−1 | 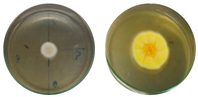 | 0.87 | ||
| Chloroquine 50 mg·L−1 | 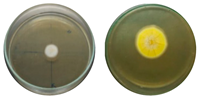 | 0.83 | ||
| Chloroquine 100 mg·L−1 |  | 0.65 | ||
| Penicillium cf. guaibinense CBMAI 2758 | Control |  | 0.64 | |
| Chloroquine 25 mg·L−1 |  | 0.68 | ||
| Chloroquine 50 mg·L−1 |  | 0.58 | ||
| Chloroquine 100 mg·L−1 |  | 0.39 | ||
| Factors | Variables | Levels | ||
|---|---|---|---|---|
| −1 | 0 | +1 | ||
| X1 | Time (days) | 5 | 10 | 15 |
| X2 | pH | 5 | 7 | 9 |
| X3 | Chloroquine diphosphate concentration (mg·L−1) | 50 | 75 | 100 |
| Run | Levels of Uncoded and Coded Variables | Response (%BD) | |||||
|---|---|---|---|---|---|---|---|
| X1 | X2 | X3 | |||||
| T1 | 5 | −1 | 5 | −1 | 75 | 0 | >99.0 |
| T2 | 15 | 1 | 5 | −1 | 75 | 0 | 98.2 |
| T3 | 5 | −1 | 9 | 1 | 75 | 0 | 88.2 |
| T4 | 15 | 1 | 9 | 1 | 75 | 0 | 91.2 |
| T5 | 5 | −1 | 7 | 0 | 50 | −1 | 97.0 |
| T6 | 15 | 1 | 7 | 0 | 50 | −1 | 89.0 |
| T7 | 5 | −1 | 7 | 0 | 100 | 1 | >99.4 |
| T8 | 15 | 1 | 7 | 0 | 100 | 1 | 98.6 |
| T9 | 10 | 0 | 5 | −1 | 50 | −1 | 95.5 |
| T10 | 10 | 0 | 9 | 1 | 50 | −1 | 87.9 |
| T11 | 10 | 0 | 5 | −1 | 100 | 1 | 93.9 |
| T12 | 10 | 0 | 9 | 1 | 100 | 1 | 93.8 |
| T13 | 10 | 0 | 7 | 0 | 75 | 0 | 92.8 |
| T14 | 10 | 0 | 7 | 0 | 75 | 0 | 92.5 |
| T15 | 10 | 0 | 7 | 0 | 75 | 0 | 91.7 |
| Computational Toxicological Parameters | Prediction of Toxicity Risks | Tolerated Doses of Potential Carcinogen | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecules | Pimephales promelas | Daphnia magna | Rat | Rat | Mouse | Rat | ||||||||
| Female/ Male | Ames | Skin | Ocular | Oral | Chronic | Inhalation | (Body Weight/Day) | |||||||
| Mouse | Rat | Mutagenicity | Irritancy | Sensitization | Irritation | LC50 (g/L) | EC50 (mg/L) | LD50 (g/kg) | LOAEL (g/kg) | LC50 (mg/m3/h) | TD50 (mg/kg) | RMTD (g/kg) | ||
| Chloroquine | C− | C− | M+ | − | − | ++++ | 0.009 | 11.40 | 1.068 | 0.062 | 24,400.2 | 0.376 | 12.73 | 0.016 |
| M1 | C− | C− | M− | − | − | +++ | 0.001 | 0.22 | 1.465 | 0.092 | 1511.87 | 3.394 | 28.74 | 0.093 |
| M2 | C | C− | M− | ++ | − | +++ | 2.56 × 10−5 | 0.32 | 0.853 | 0.007 | 2230.09 | 94.806 | 16.00 | 0.103 |
| M3 | C− | C− | M− | ++ | − | − | 0.001 | 0.72 | 0.138 | 0.065 | 8395.07 | 3.020 | 27.90 | 0.392 |
| M4 | C− | C− | M− | − | + | +++ | 0.001 | 0.33 | 6.877 | 0.070 | 771.57 | 0.798 | 45.80 | 0.736 |
| Ligand | Binding Affinity (kcal/mol) | ||
|---|---|---|---|
| Phytase | Protease | Penicillopepsin-1 | |
| HIS | −7.208 | -- | -- |
| PMS | -- | −7.475 | -- |
| PP7 | -- | -- | −9.547 |
| Chloroquine | −7.097 | −7.262 | −7.923 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Almeida Nóbrega, P.; Lopes, S.Q.; Sá, L.S.; da Silva Ramos, R.; Holanda, F.H.e.; de Araújo, I.F.; Porto, A.L.M.; Birolli, W.G.; Ferreira, I.M. Biodegradation of Chloroquine by a Fungus from Amazonian Soil, Penicillium guaibinense CBMAI 2758. J. Fungi 2025, 11, 579. https://doi.org/10.3390/jof11080579
de Almeida Nóbrega P, Lopes SQ, Sá LS, da Silva Ramos R, Holanda FHe, de Araújo IF, Porto ALM, Birolli WG, Ferreira IM. Biodegradation of Chloroquine by a Fungus from Amazonian Soil, Penicillium guaibinense CBMAI 2758. Journal of Fungi. 2025; 11(8):579. https://doi.org/10.3390/jof11080579
Chicago/Turabian Stylede Almeida Nóbrega, Patrícia, Samuel Q. Lopes, Lucas S. Sá, Ryan da Silva Ramos, Fabrício H. e Holanda, Inana F. de Araújo, André Luiz M. Porto, Willian G. Birolli, and Irlon M. Ferreira. 2025. "Biodegradation of Chloroquine by a Fungus from Amazonian Soil, Penicillium guaibinense CBMAI 2758" Journal of Fungi 11, no. 8: 579. https://doi.org/10.3390/jof11080579
APA Stylede Almeida Nóbrega, P., Lopes, S. Q., Sá, L. S., da Silva Ramos, R., Holanda, F. H. e., de Araújo, I. F., Porto, A. L. M., Birolli, W. G., & Ferreira, I. M. (2025). Biodegradation of Chloroquine by a Fungus from Amazonian Soil, Penicillium guaibinense CBMAI 2758. Journal of Fungi, 11(8), 579. https://doi.org/10.3390/jof11080579










