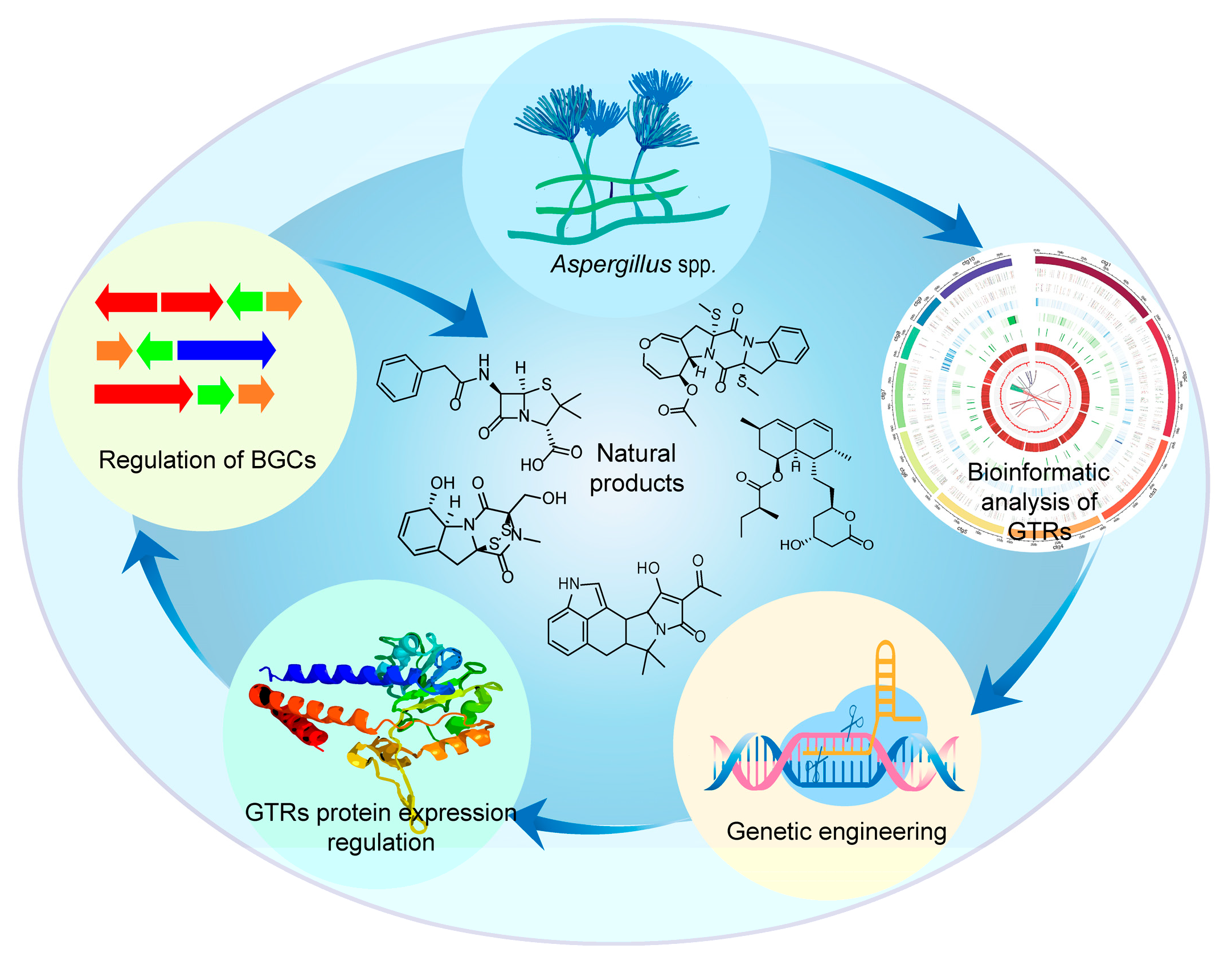
Engineering of Global Transcriptional Regulators (GTRs) in Aspergillus for Natural Product Discovery
Abstract
1. Introduction
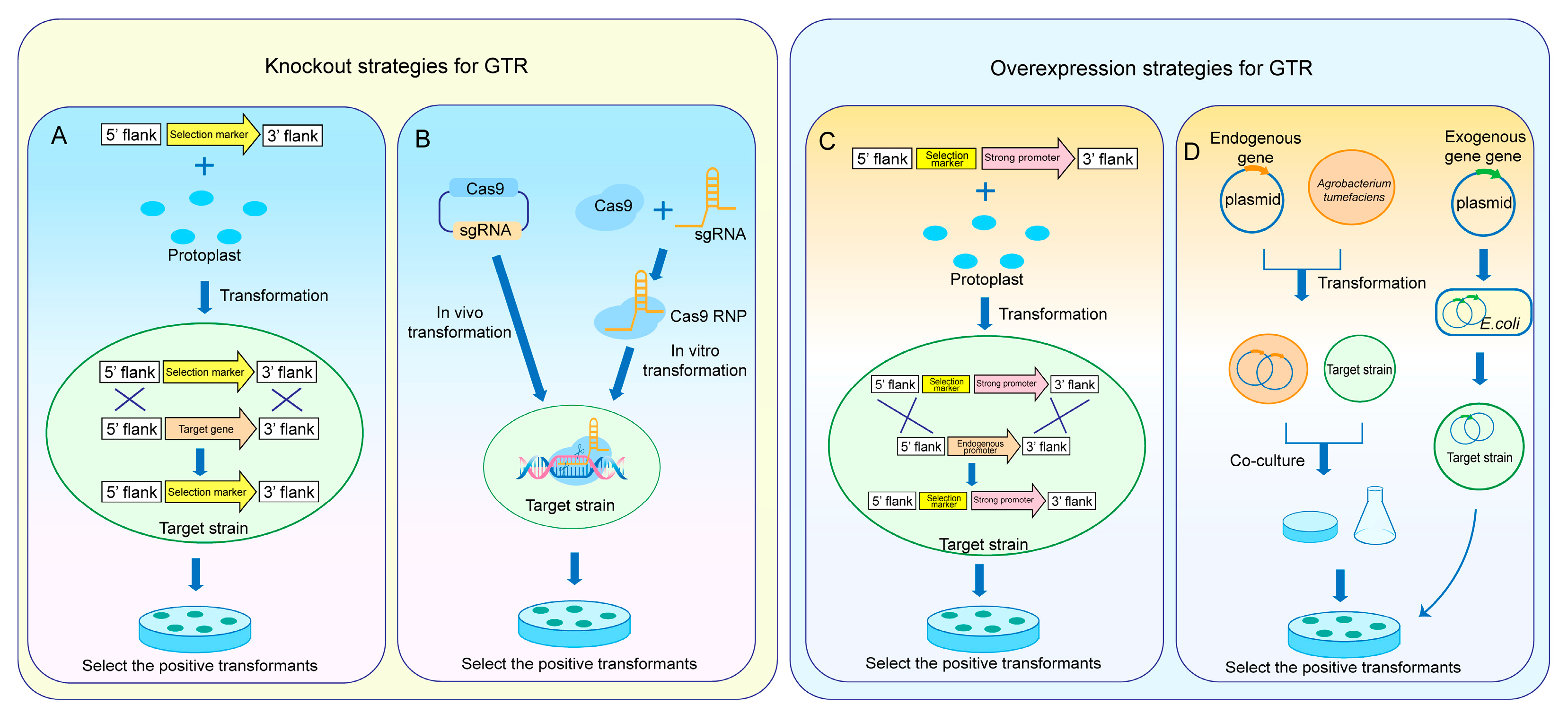
2. GTR Engineering Approaches
2.1. Knockout Strategies for GTR
2.1.1. Knockout of GTR Based on HR
2.1.2. Knockout of GTR Based on the CRISPR/Cas9 System
2.2. Overexpression Strategies for GTR
2.2.1. Overexpression of GTR Using Strong Promoters
2.2.2. Construction of Overexpression Vectors for GTR Expression
3. GTRs in Aspergillus spp.
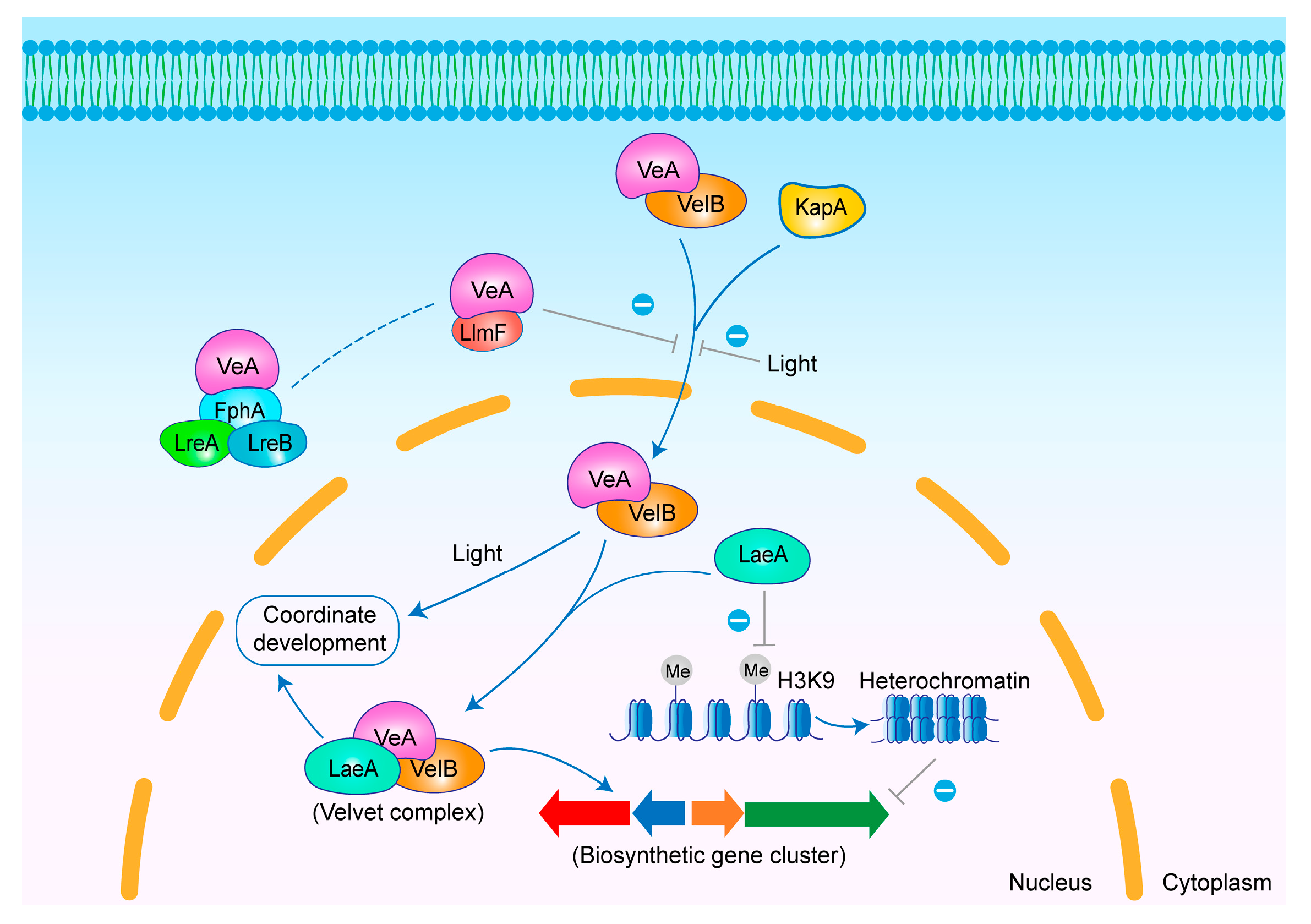
3.1. LaeA
3.1.1. Regulatory Mechanisms of LaeA
3.1.2. LaeA Regulation-Derived SMs
3.2. VeA
3.2.1. Regulatory Mechanisms of VeA
3.2.2. VeA Regulation-Derived SMs
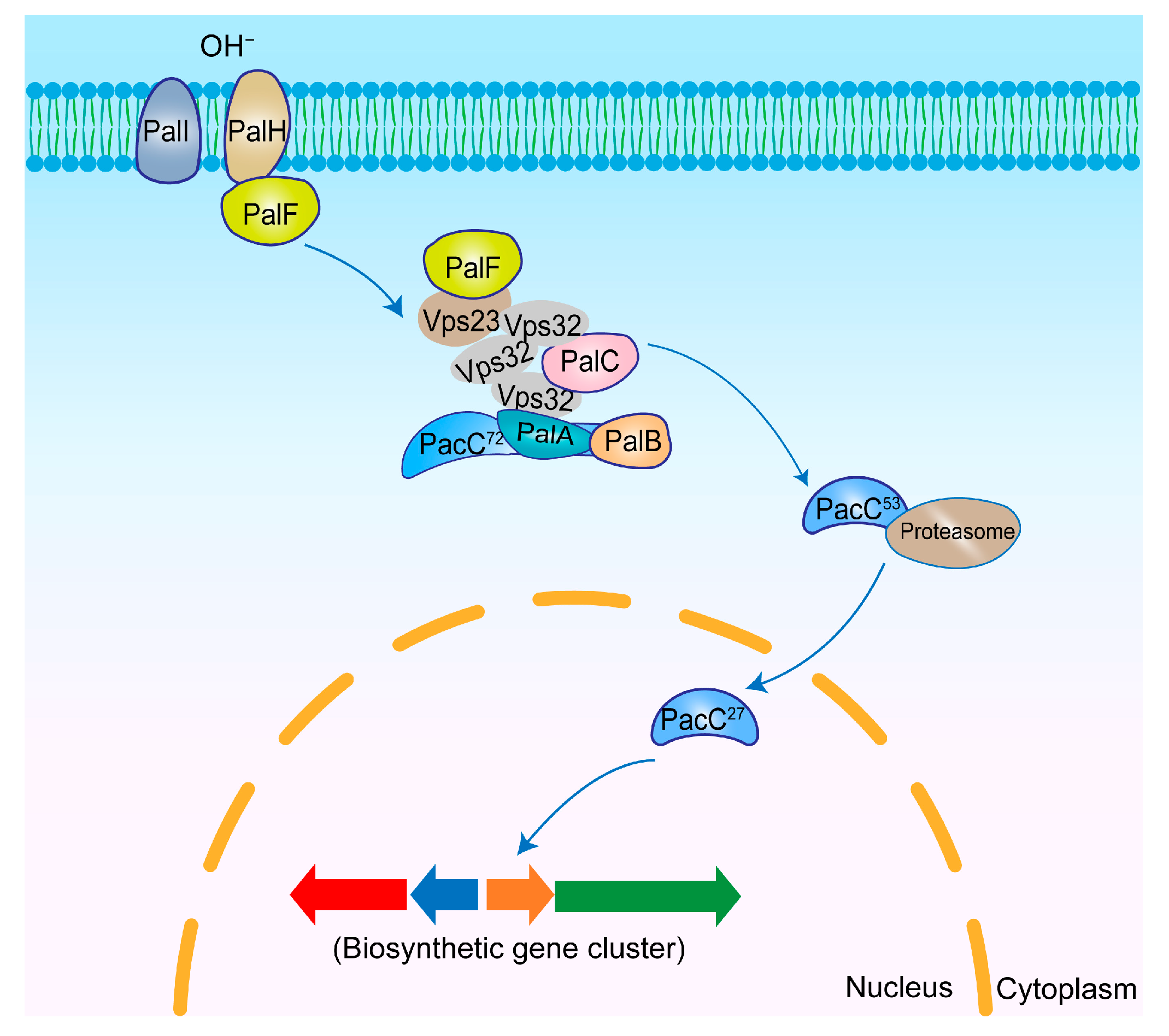
3.3. PacC
3.3.1. Regulatory Mechanisms of PacC
3.3.2. PacC Regulation-Derived SMs
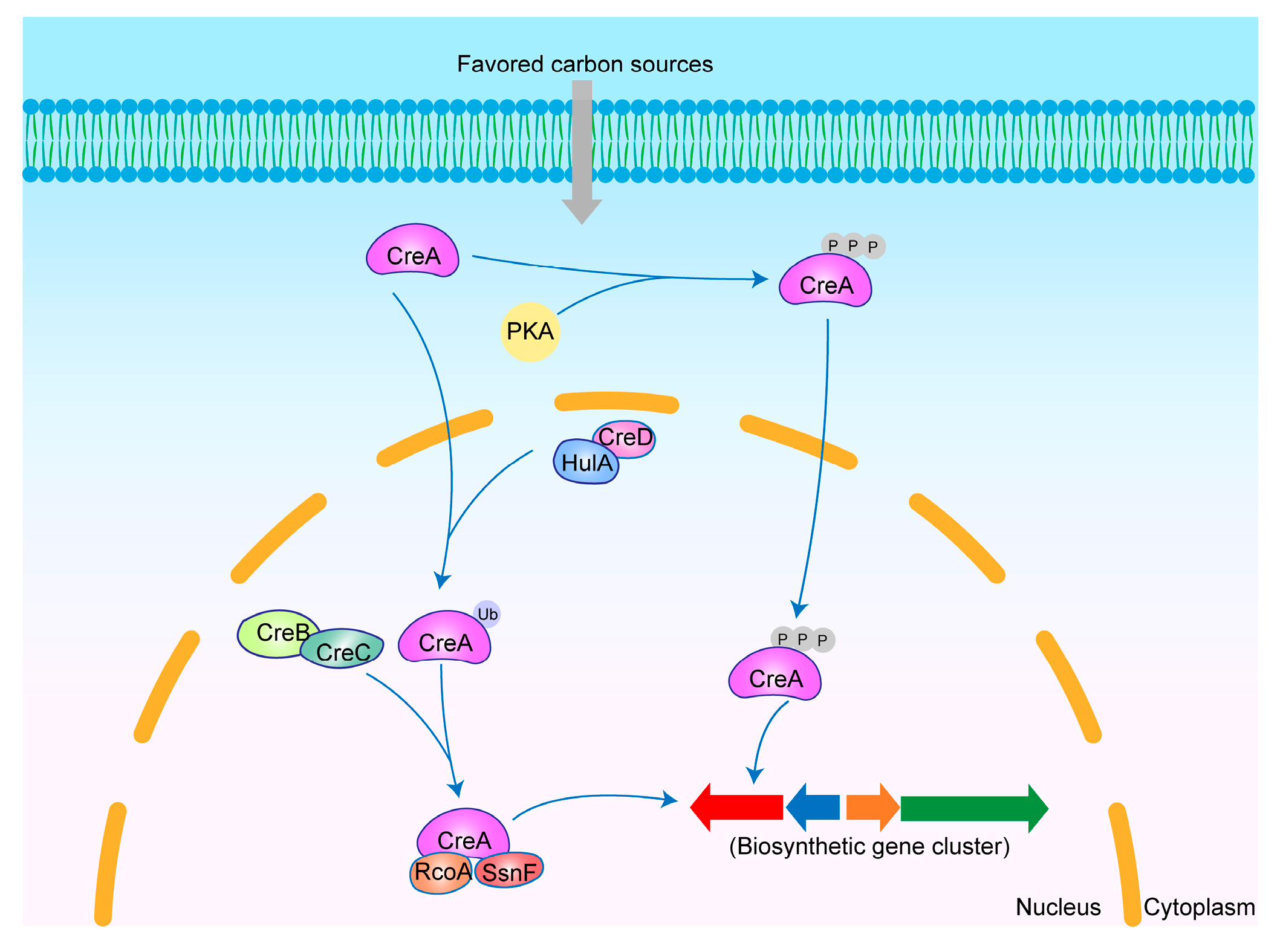
3.4. CreA
3.4.1. Regulatory Mechanisms of CreA
3.4.2. CreA Regulation-Derived SMs
3.5. AreA
3.5.1. Regulatory Mechanisms of AreA
3.5.2. AreA Regulation-Derived SMs
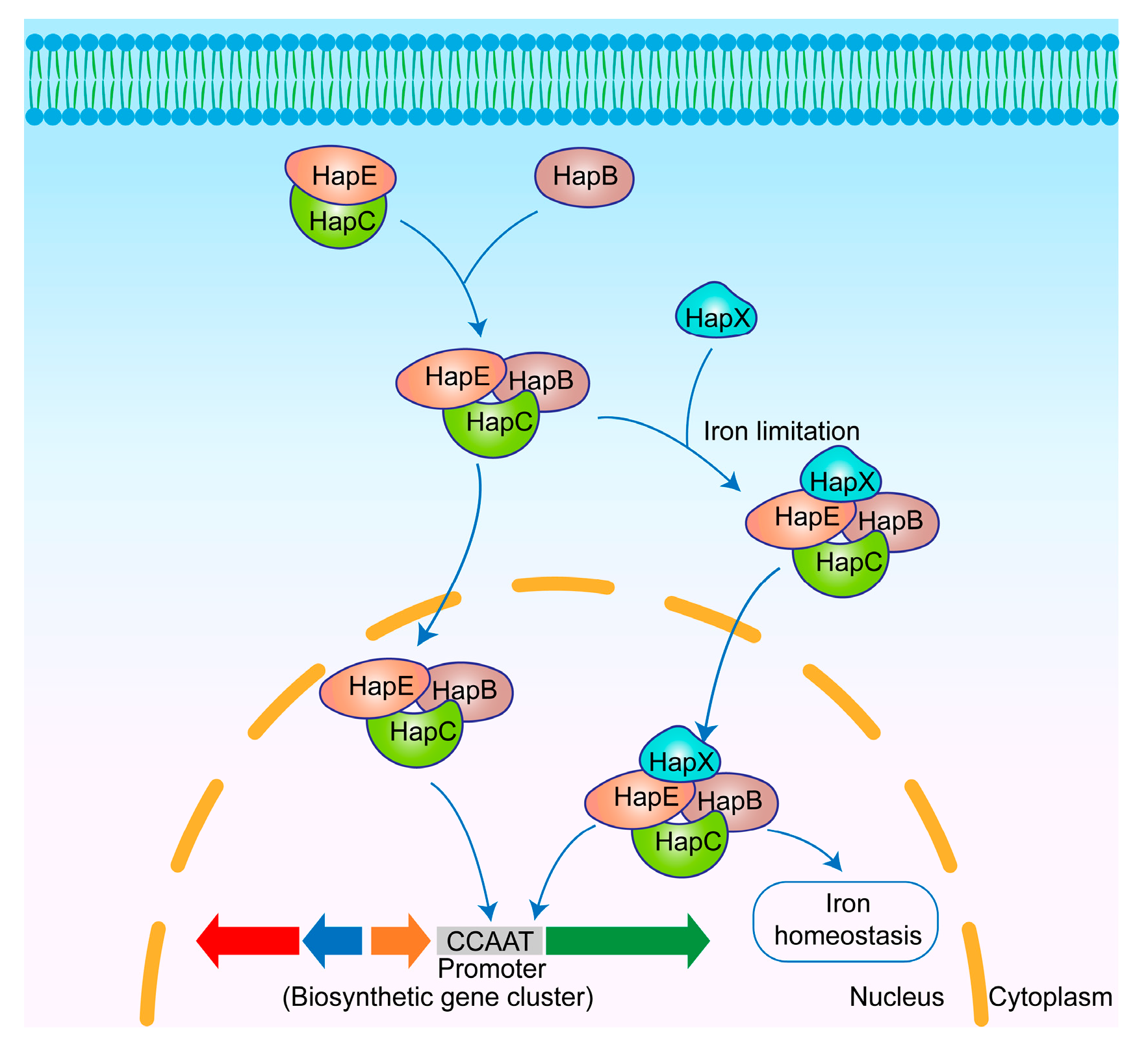
3.6. CBC
3.6.1. Regulatory Mechanisms of CBC
3.6.2. CBC Regulation-Derived SMs
3.7. Other GTRs
4. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ATMT | Agrobacterium tumefaciens-mediated transformation |
| BGCs | biosynthetic gene clusters |
| Cas9 | CRISPR-Associated Protein 9 |
| CBC | the CCAAT-binding complex |
| CCR | carbon catabolite repression |
| ChIP | chromatin immunoprecipitation |
| CPA | cyclopiazonic acid |
| CRISPR | clustered Regularly Interspaced Short Palindromic Repeats |
| DPPH | 2,2-diphenyl-1-picrylhydrazyl |
| ECB | echinocandin B |
| ESCRT | endosomal sorting complex required for transport |
| GTRs | global transcriptional regulators |
| HR | homologous recombination |
| H3K4 | histone 3 lysine 4 |
| H3K9 | histone 3 lysine 9 |
| IC50 | half maximal inhibitory concentration |
| MIC | minimum Inhibitory Concentration |
| MONJ | monacolin J |
| NHEJ | non-homologous end joining |
| NLS | nuclear localization sequence |
| NMR | nitrogen metabolite repression |
| NRPS | non-ribosomal peptide synthetase |
| OSMAC | one strain many compounds |
| OTA | ochratoxin A |
| PCR | polymerase chain reaction |
| PKA | protein kinase A |
| PKS | polyketide synthase |
| qRT-PCR | quantitative reverse transcription polymerase chain reaction |
| RNA-seq | RNA sequencing |
| SirE | sirtuin E |
| SMs | secondary metabolites |
| ST | sterigmatocystin |
References
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef] [PubMed]
- The Dictionary of Natural Products. Available online: https://dnp.chemnetbase.com (accessed on 27 March 2025).
- Bai, X.; Sheng, Y.; Tang, Z.; Pan, J.; Wang, S.; Tang, B.; Zhou, T.; Shi, L.; Zhang, H. Polyketides as secondary metabolites from the genus Aspergillus. J. Fungi 2023, 9, 261. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Yuan, X.L.; Li, C.; Li, X.D. Recent discovery of heterocyclic alkaloids from marine-derived Aspergillus species. Mar. Drugs 2020, 18, 54. [Google Scholar] [CrossRef]
- Yu, R.; Liu, J.; Wang, Y.; Wang, H.; Zhang, H. Aspergillus niger as a secondary metabolite factory. Front. Chem. 2021, 9, 701022. [Google Scholar] [CrossRef]
- Bladt, T.T.; Frisvad, J.C.; Knudsen, P.B.; Larsen, T.O. Anticancer and antifungal compounds from Aspergillus, penicillium and other filamentous fungi. Molecules 2013, 18, 11338–11376. [Google Scholar] [CrossRef]
- Andersen, M.R.; Nielsen, J.B.; Klitgaard, A.; Petersen, L.M.; Zachariasen, M.; Hansen, T.J.; Blicher, L.H.; Gotfredsen, C.H.; Larsen, T.O.; Nielsen, K.F.; et al. Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc. Natl. Acad. Sci. USA 2013, 110, E99–E107. [Google Scholar] [CrossRef]
- Bergmann, S.; Schümann, J.; Scherlach, K.; Lange, C.; Brakhage, A.A.; Hertweck, C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat. Chem. Biol. 2007, 3, 213–217. [Google Scholar] [CrossRef]
- Xu, S.; Li, M.; Hu, Z.; Shao, Y.; Ying, J.; Zhang, H. The potential use of fungal co-culture strategy for discovery of new secondary metabolites. Microorganisms 2023, 11, 464. [Google Scholar] [CrossRef]
- Zhao, F.; Sun, C.; Liu, Z.; Cabrera, A.; Escobar, M.; Huang, S.; Yuan, Q.; Nie, Q.; Luo, K.L.; Lin, A.; et al. Multiplex base-editing enables combinatorial epigenetic regulation for genome mining of fungal natural products. J. Am. Chem. Soc. 2023, 145, 413–421. [Google Scholar] [CrossRef]
- Wei, Q.; Bai, J.; Yan, D.; Bao, X.; Li, W.; Liu, B.; Zhang, D.; Qi, X.; Yu, D.; Hu, Y. Genome mining combined metabolic shunting and OSMAC strategy of an endophytic fungus leads to the production of diverse natural products. Acta Pharm. Sin. B 2021, 11, 572–587. [Google Scholar] [CrossRef]
- Hautbergue, T.; Jamin, E.L.; Debrauwer, L.; Puel, O.; Oswald, I.P. From genomics to metabolomics, moving toward an integrated strategy for the discovery of fungal secondary metabolites. Nat. Prod. Rep. 2018, 35, 147–173. [Google Scholar] [CrossRef] [PubMed]
- Lyu, H.N.; Liu, H.W.; Keller, N.P.; Yin, W.B. Harnessing diverse transcriptional regulators for natural product discovery in fungi. Nat. Prod. Rep. 2020, 37, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32. [Google Scholar] [CrossRef]
- Yu, W.; Pei, R.; Zhou, J.; Zeng, B.; Tu, Y.; He, B. Molecular regulation of fungal secondary metabolism. World J. Microbiol. Biotechnol. 2023, 39, 204. [Google Scholar] [CrossRef]
- Macheleidt, J.; Mattern, D.J.; Fischer, J.; Netzker, T.; Weber, J.; Schroeckh, V.; Valiante, V.; Brakhage, A.A. Regulation and role of fungal secondary metabolites. Annu. Rev. Genet. 2016, 50, 371–392. [Google Scholar] [CrossRef]
- Wang, S.; Chen, H.; Tang, X.; Zhang, H.; Chen, W.; Chen, Y.Q. Molecular tools for gene manipulation in filamentous fungi. Appl. Microbiol. Biotechnol. 2017, 101, 8063–8075. [Google Scholar] [CrossRef]
- Mei, Y.Z.; Zhu, Y.L.; Huang, P.W.; Yang, Q.; Dai, C.C. Strategies for gene disruption and expression in filamentous fungi. Appl. Microbiol. Biotechnol. 2019, 103, 6041–6059. [Google Scholar] [CrossRef]
- Yu, J.H.; Hamari, Z.; Han, K.H.; Seo, J.A.; Reyes-Domínguez, Y.; Scazzocchio, C. Double-joint PCR: A PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004, 41, 973–981. [Google Scholar] [CrossRef]
- Szewczyk, E.; Nayak, T.; Oakley, C.E.; Edgerton, H.; Xiong, Y.; Taheri-Talesh, N.; Osmani, S.A.; Oakley, B.R. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 2006, 1, 3111–3120. [Google Scholar] [CrossRef]
- Son, Y.E.; Park, H.S. Genetic Manipulation and transformation methods for Aspergillus spp. Mycobiology 2021, 49, 95–104. [Google Scholar] [CrossRef]
- Shrivastav, M.; De Haro, L.P.; Nickoloff, J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008, 18, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, N.D.S.P.; Arentshorst, M.; Jin Kwon, M.; Meyer, V.; Ram, A.F.J. Expanding the ku70 toolbox for filamentous fungi: Establishment of complementation vectors and recipient strains for advanced gene analyses. Appl. Microbiol. Biotechnol. 2010, 87, 1463–1473. [Google Scholar] [CrossRef]
- Nayak, T.; Szewczyk, E.; Oakley, C.E.; Osmani, A.; Ukil, L.; Murray, S.L.; Hynes, M.J.; Osmani, S.A.; Oakley, B.R. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 2006, 172, 1557–1566. [Google Scholar] [CrossRef]
- Meyer, V.; Arentshorst, M.; El-Ghezal, A.; Drews, A.C.; Kooistra, R.; van den Hondel, C.A.M.J.J.; Ram, A.F.J. Highly efficient gene targeting in the Aspergillus niger kusA mutant. J. Biotechnol. 2007, 128, 770–775. [Google Scholar] [CrossRef]
- Da Silva Ferreira, M.E.; Kress, M.R.V.Z.; Savoldi, M.; Goldman, M.H.S.; Härtl, A.; Heinekamp, T.; Brakhage, A.A.; Goldman, G.H. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 2006, 5, 207–211. [Google Scholar] [CrossRef]
- Krappmann, S.; Sasse, C.; Braus, G.H. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end- joining-deficient genetic background. Eukaryot. Cell 2006, 5, 212–215. [Google Scholar] [CrossRef]
- Fairhead, C.; Llorente, B.; Denis, F.; Soler, M.; Dujon, B. New vectors for combinatorial deletions in yeast chromosomes and for gap-repair cloning using ‘split-marker’ recombination. Yeast 1996, 12, 1439–1457. [Google Scholar] [CrossRef]
- Goswami, R.S. Targeted gene replacement in fungi using a split-marker approach. In Plant Fungal Pathogens: Methods and Protocols; Bolton, M.D., Thomma, B.P.H.J., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 255–269. [Google Scholar]
- Nielsen, M.L.; Albertsen, L.; Lettier, G.; Nielsen, J.B.; Mortensen, U.H. Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet. Biol. 2006, 43, 54–64. [Google Scholar] [CrossRef]
- Li, D.; Tang, Y.; Lin, J.; Cai, W. Methods for genetic transformation of filamentous fungi. Microb. Cell Fact. 2017, 16, 168. [Google Scholar] [CrossRef]
- De Groot, M.J.A.; Bundock, P.; Hooykaas, P.J.J.; Beijersbergen, A.G.M. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 1998, 16, 839–842. [Google Scholar] [CrossRef]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef] [PubMed]
- Wassano, N.S.; da Silva, G.B.; Reis, A.H.; Gerhardt, J.A.; Antoniel, E.P.; Akiyama, D.; Rezende, C.P.; Neves, L.X.; Vasconcelos, E.J.R.; de Figueiredo, F.L.; et al. Sirtuin E deacetylase is required for full virulence of Aspergillus fumigatus. Commun. Biol. 2024, 7, 704. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Keller, N.P.; Oakley, B.R.; Stajich, J.E.; Wang, C.C.C. Manipulation of the global regulator mcrA upregulates secondary metabolite production in Aspergillus wentii using CRISPR-Cas9 with in vitro assembled ribonucleoproteins. ACS Chem. Biol. 2022, 17, 2828–2835. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Grau, M.F.; Murata, R.M.; Torok, T.; Venkateswaran, K.; Stajich, J.E.; Wang, C.C.C. Identification of the neoaspergillic acid biosynthesis gene cluster by establishing an in vitro CRISPR-Ribonucleoprotein genetic system in Aspergillus melleus. ACS Omega 2023, 8, 16713–16721. [Google Scholar] [CrossRef]
- Juven-Gershon, T.; Hsu, J.-Y.; Theisen, J.W.; Kadonaga, J.T. The RNA polymerase II core promoter—The gateway to transcription. Curr. Opin. Cell Biol. 2008, 20, 253–259. [Google Scholar] [CrossRef]
- Punt, P.J.; Dingemanse, M.A.; Kuyvenhoven, A.; Soede, R.D.M.; Pouwels, P.H.; van den Hondel, C.A.M.J.J. Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene 1990, 93, 101–109. [Google Scholar] [CrossRef]
- Fowler, T.; Berka, R.M.; Ward, M. Regulation of the glaA gene of Aspergillus niger. Curr. Genet. 1990, 18, 537–545. [Google Scholar] [CrossRef]
- Wei, P.L.; Fan, J.; Yu, J.; Ma, Z.; Guo, X.; Keller, N.P.; Li, E.; Lou, C.; Yin, W.B. Quantitative characterization of filamentous fungal promoters on a single-cell resolution to discover cryptic natural products. Sci. China Life Sci. 2023, 66, 848–860. [Google Scholar] [CrossRef]
- Nishikawa, R.; Yoshida, M.; Noda, T.; Okuhara, T.; Taguchi, G.; Inatomi, S.; Shimosaka, M. pFungiway: A series of plasmid vectors used for gene manipulation in fungi. Ann. Microbiol. 2016, 66, 825–832. [Google Scholar] [CrossRef]
- Roux, I.; Woodcraft, C.; Sbaraini, N.; Pepper, A.; Wong, E.; Bracegirdle, J.; Chooi, Y.H. Next-generation AMA1-based plasmids for enhanced heterologous expression in filamentous fungi. Microb. Biotechnol. 2024, 17, e70010. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Zhang, J.; Zhang, C. Overexpression of llm1 affects the synthesis of secondary metabolites of Aspergillus cristatus. Microorganisms 2022, 10, 1707. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Liang, C.; Xing, W.; Wu, W.; Hou, Y.; Zhang, L.; Xiao, S.; Xu, H.; An, Y.; Zheng, M.; et al. Transcriptional factor engineering in microbes for industrial biotechnology. J. Chem. Technol. Biotechnol. 2020, 95, 3071–3078. [Google Scholar] [CrossRef]
- Khan, I.; Xie, W.L.; Yu, Y.C.; Sheng, H.; Xu, Y.; Wang, J.Q.; Debnath, S.C.; Xu, J.Z.; Zheng, D.Q.; Ding, W.J.; et al. Heteroexpression of Aspergillus nidulans laeA in marine-derived fungi triggers upregulation of secondary metabolite biosynthetic genes. Mar. Drugs 2020, 18, 652. [Google Scholar] [CrossRef]
- Bok, J.W.; Keller, N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 2004, 3, 527–535. [Google Scholar]
- Jin, W.B.; Balajee, S.A.; Marr, K.A.; Andes, D.; Nielsen, K.F.; Frisvad, J.C.; Keller, N.P. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell 2005, 4, 1574–1582. [Google Scholar]
- Keller, N.; Bok, J.; Chung, D.; Perrin, R.M.; Keats Shwab, E. LaeA, a global regulator of Aspergillus toxins. Med. Mycol. 2006, 44, 83–85. [Google Scholar] [CrossRef]
- Reyes-Dominguez, Y.; Bok, J.W.; Berger, H.; Shwab, E.K.; Basheer, A.; Gallmetzer, A.; Scazzocchio, C.; Keller, N.; Strauss, J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Mol. Microbiol. 2010, 76, 1376–1386. [Google Scholar] [CrossRef]
- Kadooka, C.; Nakamur, E.; Mori, K.; Okutsu, K.; Yoshizaki, Y.; Takamine, K.; Goto, M.; Tamaki, H.; Futagami, T. LaeA controls citric acid production through regulation of the citrate exporter-encoding cexA gene in Aspergillus luchuensis mut. kawachii. Appl. Environ. Microbiol. 2020, 86, e01950-19. [Google Scholar] [CrossRef]
- Lind, A.L.; Lim, F.Y.; Soukup, A.A.; Keller, N.P.; Rokas, A. An LaeA- and BrlA-dependent cellular network governs tissue-specific secondary metabolism in the human pathogen Aspergillus fumigatus. mSphere 2018, 3, e00050-18. [Google Scholar] [CrossRef]
- Bayram, Ö.; Krappmann, S.; Ni, M.; Bok, J.W.; Helmstaedt, K.; Valerius, O.; Braus-Stromeyer, S.; Kwon, N.-J.; Keller, N.P.; Yu, J.H.; et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 2008, 320, 1504–1506. [Google Scholar] [CrossRef]
- Bok, J.W.; Noordermeer, D.; Kale, S.P.; Keller, N.P. Secondary metabolic gene cluster silencing in Aspergillus nidulans. Mol. Microbiol. 2006, 61, 1636–1645. [Google Scholar] [CrossRef] [PubMed]
- Bok, J.W.; Jr., R.A.C.; Wortman, J.R.; Kim, H.S.; Nierman, W.C.; Keller, N.P. Transcriptional regulation of chemical diversity in Aspergillus fumigatus by LaeA. PLoS Pathog. 2007, 3, e50. [Google Scholar]
- Sugui, J.A.; Pardo, J.; Chang, Y.C.; Müllbacher, A.; Zarember, K.A.; Galvez, E.M.; Brinster, L.; Zerfas, P.; Gallin, J.I.; Simon, M.M.; et al. Role of laeA in the regulation of alb1, gliP, conidial morphology, and virulence in Aspergillus fumigatus. Eukaryot. Cell 2007, 6, 1552–1561. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.K.; Scharfenstein, L.L.; Ehrlich, K.C.; Wei, Q.; Bhatnagar, D.; Ingber, B.F. Effects of laeA deletion on Aspergillus flavus conidial development and hydrophobicity may contribute to loss of aflatoxin production. Fungal Biol. 2012, 116, 298–307. [Google Scholar] [CrossRef]
- Lim, F.Y.; Hou, Y.; Chen, Y.; Oh, J.H.; Lee, I.; Bugni, T.S.; Keller, N.P. Genome-based cluster deletion reveals an endocrocin biosynthetic pathway in Aspergillus fumigatus. Appl. Environ. Microbiol. 2012, 78, 4117–4125. [Google Scholar] [CrossRef]
- Oda, K.; Kobayashi, A.; Ohashi, S.; Sano, M. Aspergillus oryzae laeA regulates kojic acid synthesis genes. Biosci. Biotechnol. Biochem. 2011, 75, 1832–1834. [Google Scholar] [CrossRef]
- Niu, J.; Arentshorst, M.; Nair, P.D.S.; Dai, Z.; Baker, S.E.; Frisvad, J.C.; Nielsen, K.F.; Punt, P.J.; Ram, A.F.J. Identification of a classical mutant in the industrial host Aspergillus niger by systems genetics: LaeA is required for citric acid production and regulates the formation of some secondary metabolites. G3 2016, 6, 193–204. [Google Scholar] [CrossRef]
- Crespo-Sempere, A.; Marín, S.; Sanchis, V.; Ramos, A.J. VeA and LaeA transcriptional factors regulate ochratoxin a biosynthesis in Aspergillus carbonarius. Int. J. Food Microbiol. 2013, 166, 479–486. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, H.; Wang, Y.; Liu, F.; Li, E.; Ma, J.; Yang, B.; Zhang, C.; Li, L.; Liu, Y. Requirement of LaeA, VeA, and VelB on asexual development, ochratoxin A biosynthesis, and fungal virulence in Aspergillus ochraceus. Front. Microbiol. 2019, 10, 2759. [Google Scholar] [CrossRef]
- Iliadi, M.K.; Varveri, M.; Kapetanakou, A.E.; Skandamis, P.N.; Tsitsigiannis, D.I. The global secondary metabolite regulator AcLaeA modulates Aspergillus carbonarius virulence, ochratoxin biosynthesis, and the mode of action of biopesticides and essential oils. Toxins 2025, 17, 2. [Google Scholar] [CrossRef]
- Pomraning, K.R.; Dai, Z.; Munoz, N.; Kim, Y.M.; Gao, Y.; Deng, S.; Lemmon, T.; Swita, M.S.; Zucker, J.D.; Kim, J.; et al. Itaconic acid production is regulated by LaeA in Aspergillus pseudoterreus. Metab. Eng. Commun. 2022, 15, e00203. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, P.; Qi, C.; Zha, Z.; Meng, J.; Liu, C.; Han, J.; Zhou, Q.; Luo, Z.; Wang, J.; et al. Cryptic piperazine derivatives activated by knocking out the global regulator LaeA in Aspergillus flavipes. Bioorg. Med. Chem. 2024, 103, 117685. [Google Scholar] [CrossRef] [PubMed]
- Hong, E.J.; Kim, N.K.; Lee, D.; Kim, W.G.; Lee, I. Overexpression of the laeA gene leads to increased production of cyclopiazonic acid in Aspergillus fumisynnematus. Fungal Biol. 2015, 119, 973–983. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Wang, Y.; Du, L.; Li, F.; Hu, Q.; Cheng, G.; Wang, W. Overexpression of global regulator LaeA induced secondary metabolite production in Aspergillus versicolor 0312. Rec. Nat. Prod. 2020, 14, 387–394. [Google Scholar] [CrossRef]
- Wang, B.; Li, X.; Tabudravu, J.; Wang, S.; Deng, H.; Pan, L. The chemical profile of activated secondary metabolites by overexpressing LaeA in Aspergillus niger. Microbiol. Res. 2021, 248, 126735. [Google Scholar] [CrossRef]
- Bingxin, Z.; Wen, S.; Qin, X.Y.; Wang, Z.H.; Yao, G.S. Activating biosynthesis by overexpressing LaeA and anti-Vibrio activity of dihydroisoflavipucines of marine-derived Aspergillus terreus. Mycosystema 2023, 42, 562–569. [Google Scholar]
- Zhou, W.; Li, M.; Wang, W.; Bai, X.; Zhang, H. Overexpression of the global transcriptional regulator LaeA leads to production of cyclic lipopeptides in marine-derived Aspergillus niger L14. Chem. Biodivers. 2025, 22, e202402704. [Google Scholar] [CrossRef]
- Käfer, E. Origins of translocations in Aspergillus nidulans. Genetics 1965, 52, 217–232. [Google Scholar] [CrossRef]
- Rauscher, S.; Pacher, S.; Hedtke, M.; Kniemeyer, O.; Fischer, R. A phosphorylation code of the Aspergillus nidulans global regulator Velvet A (VeA) determines specific functions. Mol. Microbiol. 2016, 99, 909–924. [Google Scholar] [CrossRef]
- Stinnett, S.M.; Espeso, E.A.; Cobeño, L.; Araújo-Bazán, L.; Calvo, A.M. Aspergillus nidulans VeA subcellular localization is dependent on the importin α carrier and on light. Mol. Microbiol. 2007, 63, 242–255. [Google Scholar] [CrossRef]
- Kim, H.S.; Han, K.Y.; Kim, K.J.; Han, D.M.; Jahng, K.Y.; Chae, K.S. The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet. Biol. 2002, 37, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Purschwitz, J.; Müller, S.; Kastner, C.; Schöser, M.; Haas, H.; Espeso, E.A.; Atoui, A.; Calvo, A.M.; Fischer, R. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr. Biol. 2008, 18, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.M.; Theisen, J.M.; Duran, R.M.; Grayburn, W.S.; Calvo, A.M.; Keller, N.P. Secondary metabolism and development is mediated by LlmF control of VeA subcellular localization in Aspergillus nidulans. PLoS Genet. 2013, 9, e1003193. [Google Scholar] [CrossRef]
- Kato, N.; Brooks, W.; Calvo, A.M. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot. Cell 2003, 2, 1178–1186. [Google Scholar] [CrossRef]
- Spröte, P.; Brakhage, A.A. The light-dependent regulator velvet A of Aspergillus nidulans acts as a repressor of the penicillin biosynthesis. Arch. Microbiol. 2007, 188, 69–79. [Google Scholar] [CrossRef]
- Marui, J.; Ohashi-Kunihiro, S.; Ando, T.; Nishimura, M.; Koike, H.; Machida, M. Penicillin biosynthesis in Aspergillus oryzae and its overproduction by genetic engineering. J. Biosci. Bioeng. 2010, 110, 8–11. [Google Scholar] [CrossRef]
- Duran, R.M.; Cary, J.W.; Calvo, A.M. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 2007, 73, 1158–1168. [Google Scholar] [CrossRef]
- Cary, J.W.; OBrian, G.R.; Nielsen, D.M.; Nierman, W.; Harris-Coward, P.; Yu, J.; Bhatnagar, D.; Cleveland, T.E.; Payne, G.A.; Calvo, A.M. Elucidation of veA-dependent genes associated with aflatoxin and sclerotial production in Aspergillus flavus by functional genomics. Appl. Microbiol. Biotechnol. 2007, 76, 1107–1118. [Google Scholar] [CrossRef]
- Dhingra, S.; Andes, D.; Calvo, A.M. VeA regulates conidiation, gliotoxin production, and protease activity in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot. Cell 2012, 11, 1531–1543. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, H.; Sumarah, M.W.; Gao, Q.; Wang, D.; Zhang, Y. veA gene acts as a positive regulator of conidia production, ochratoxin A biosynthesis, and oxidative stress tolerance in Aspergillus niger. J. Agric. Food Chem. 2018, 66, 13199–13208. [Google Scholar] [CrossRef]
- Dhingra, S.; Lind, A.L.; Lin, H.C.; Tang, Y.; Rokas, A.; Calvo, A.M. The fumagillin gene cluster, an example of hundreds of genes under veA control in Aspergillus fumigatus. PLoS ONE 2013, 8, e77147. [Google Scholar] [CrossRef] [PubMed]
- Lan, N.; Yue, Q.; An, Z.; Bills, G.F. Apc.LaeA and Apc.VeA of the velvet complex govern secondary metabolism and morphological development in the echinocandin-producing fungus Aspergillus pachycristatus. J. Ind. Microbiol. Biotechnol. 2020, 47, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Bok, J.W.; Soukup, A.A.; Chadwick, E.; Chiang, Y.-M.; Wang, C.C.C.; Keller, N.P. VeA and MvlA repression of the cryptic orsellinic acid gene cluster in Aspergillus nidulans involves histone 3 acetylation. Mol. Microbiol. 2013, 89, 963–974. [Google Scholar] [CrossRef]
- Selvig, K.; Alspaugh, J.A. pH Response Pathways in Fungi: Adapting to host-derived and environmental signals. Mycobiology 2011, 39, 249–256. [Google Scholar] [CrossRef]
- Tilburn, J.; Sarkar, S.; Widdick, D.A.; Espeso, E.A.; Orejas, M.; Mungroo, J.; Peñalva, M.A.; Arst, H.N. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995, 14, 779–790. [Google Scholar] [CrossRef]
- Espeso, E.A.; Tilburn, J.; Sánchez-Pulido, L.; Brown, C.V.; Valencia, A.; Arst, H.N.; Peñalva, M.A. Specific DNA recognition by the Aspergillus nidulans three zinc finger transcription factor PacC. J. Mol. Biol. 1997, 274, 466–480. [Google Scholar] [CrossRef]
- Fernández-Martínez, J.; Brown, C.V.; Díez, E.; Tilburn, J.; Arst, H.N.; Peñalva, M.Á.; Espeso, E.A. Overlap of nuclear localisation signal and specific DNA-binding residues within the zinc finger domain of PacC. J. Mol. Biol. 2003, 334, 667–684. [Google Scholar] [CrossRef]
- Ke, R.; Haynes, K.; Stark, J. Modelling the activation of alkaline pH response transcription factor PacC in Aspergillus nidulans: Involvement of a negative feedback loop. J. Theor. Biol. 2013, 326, 11–20. [Google Scholar] [CrossRef]
- Díez, E.; Álvaro, J.; Espeso, E.A.; Rainbow, L.; Suárez, T.; Tilburn, J.; Arst, H.N.; Peñalva, M.Á. Activation of the Aspergillus PacC zinc finger transcription factor requires two proteolytic steps. EMBO J. 2002, 21, 1350–1359. [Google Scholar] [CrossRef]
- Galindo, A.; Calcagno-Pizarelli, A.M.; Arst, H.N., Jr.; Peñalva, M.Á. An ordered pathway for the assembly of fungal ESCRT-containing ambient pH signalling complexes at the plasma membrane. J. Cell Sci. 2012, 125, 1784–1795. [Google Scholar] [CrossRef]
- Hervás-Aguilar, A.; Galindo, A.; Peñalva, M.A. Receptor-independent Ambient pH Signaling by Ubiquitin Attachment to Fungal Arrestin-like PalF. J. Biol. Chem. 2010, 285, 18095–18102. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Galán, O.; Galindo, A.; Hervás-Aguilar, A.; Arst, H.N.; Peñalva, M.A. Physiological Involvement in pH Signaling of Vps24-mediated Recruitment of Aspergillus PalB Cysteine Protease to ESCRT-III. J. Biol. Chem. 2009, 284, 4404–4412. [Google Scholar] [CrossRef] [PubMed]
- Orejas, M.; Espeso, E.A.; Tilburn, J.; Sarkar, S.; Arst, H.N.; Peñalva, M.A. Activation of the Aspergillus PacC transcription factor in response to alkaline ambient pH requires proteolysis of the carboxy-terminal moiety. Genes Dev. 1995, 9, 1622–1632. [Google Scholar] [CrossRef] [PubMed]
- Hervás-Aguilar, A.; RodrÁguez, J.M.; Tilburn, J.; Arst, H.N.; Peñalva, M.A. Evidence for the direct involvement of the proteasome in the proteolytic processing of the Aspergillus nidulans zinc finger transcription factor PacC. J. Biol. Chem. 2007, 282, 34735–34747. [Google Scholar] [CrossRef]
- Arst, H.N.; Peñalva, M.A. pH regulation in Aspergillus and parallels with higher eukaryotic regulatory systems. Trends Genet. 2003, 19, 224–231. [Google Scholar] [CrossRef]
- Brakhage, A.A. Molecular regulation of penicillin biosynthesis in Aspergillus (Emericella) nidulans. FEMS Microbiol. Lett. 1997, 148, 1–10. [Google Scholar] [CrossRef]
- Espeso, E.A.; Tilburn, J.; Arst, H.N.; Peñalva, M.A. pH regulation is a major determinant in expression of a fungal penicillin biosynthetic gene. EMBO J. 1993, 12, 3947–3956. [Google Scholar] [CrossRef]
- Cary, J.W.; Ehrlich, K.C.; Wright, M.; Chang, P.K.; Bhatnagar, D. Generation of aflR disruption mutants of Aspergillus parasiticus. Appl. Microbiol. Biotechnol. 2000, 53, 680–684. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, F.; Wang, L.; Wang, Q.; Selvaraj, J.N.; Zhao, Y.; Wang, Y.; Xing, F.; Liu, Y. pH-signaling transcription factor AopacC regulates ochratoxin A biosynthesis in Aspergillus ochraceus. J. Agric. Food Chem. 2018, 66, 4394–4401. [Google Scholar] [CrossRef]
- Barda, O.; Maor, U.; Sadhasivam, S.; Bi, Y.; Zakin, V.; Prusky, D.; Sionov, E. The pH-responsive transcription factor PacC governs pathogenicity and ochratoxin A biosynthesis in Aspergillus carbonarius. Front. Microbiol. 2020, 11, 210. [Google Scholar] [CrossRef]
- Fasoyin, O.E.; Wang, B.; Qiu, M.; Han, X.; Chung, K.R.; Wang, S. Carbon catabolite repression gene creA regulates morphology, aflatoxin biosynthesis and virulence in Aspergillus flavus. Fungal Genet. Biol. 2018, 115, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, Y.; Yang, B.; Zhang, C.; Zhang, H.; Xing, F.; Liu, Y. Carbon catabolite repression gene AoCreA regulates morphological development and ochratoxin A biosynthesis responding to carbon sources in Aspergillus ochraceus. Toxins 2020, 12, 697. [Google Scholar] [CrossRef] [PubMed]
- Gressler, M.; Meyer, F.; Heine, D.; Hortschansky, P.; Hertweck, C.; Brock, M. Phytotoxin production in Aspergillus terreus is regulated by independent environmental signals. eLife 2015, 4, e07861. [Google Scholar] [CrossRef] [PubMed]
- Fasoyin, O.E.; Yang, K.; Qiu, M.; Wang, B.; Wang, S.; Wang, S. Regulation of morphology, aflatoxin production, and virulence of Aspergillus flavus by the major nitrogen regulatory gene areA. Toxins 2019, 11, 718. [Google Scholar] [CrossRef]
- Litzka, O.; Papagiannopolous, P.; Davis, M.A.; Hynes, M.J.; Brakhage, A.A. The penicillin regulator PENR1 of Aspergillus nidulans is a HAP-like transcriptional complex. Eur. J. Biochem. 1998, 251, 758–767. [Google Scholar] [CrossRef]
- Gsaller, F.; Hortschansky, P.; Furukawa, T.; Carr, P.D.; Rash, B.; Capilla, J.; Müller, C.; Bracher, F.; Bowyer, P.; Haas, H.; et al. Sterol biosynthesis and azole tolerance is governed by the opposing actions of SrbA and the CCAAT binding complex. PLoS Pathog. 2016, 12, e1005775. [Google Scholar]
- Dowzer, C.E.A.; Kelly, J.M. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol. Cell. Biol. 1991, 11, 5701–5709. [Google Scholar]
- Kulmburg, P.; Mathieu, M.; Dowzer, C.; Kelly, J.; Felenbok, B. Specific binding sites in the alcR and alcA promoters of the ethanol regulon for the CreA repressor mediating carbon cataboiite repression in Aspergillus nidulans. Mol. Microbiol. 1993, 7, 847–857. [Google Scholar] [CrossRef]
- Tamayo, E.N.; Villanueva, A.; Hasper, A.A.; de Graaff, L.H.; Ramón, D.; Orejas, M. CreA mediates repression of the regulatory gene xlnR which controls the production of xylanolytic enzymes in Aspergillus nidulans. Fungal Genet. Biol. 2008, 45, 984–993. [Google Scholar] [CrossRef]
- Strauss, J.; Horvath, H.K.; Abdallah, B.M.; Kindermann, J.; Mach, R.L.; Kubicek, C.P. The function of CreA, the carbon catabolite repressor of Aspergillus nidulans, is regulated at the transcriptional and post-transcriptional level. Mol. Microbiol. 1999, 32, 169–178. [Google Scholar] [CrossRef]
- Prathumpai, W.; McIntyre, M.; Nielsen, J. The effect of CreA in glucose and xylose catabolism in Aspergillus nidulans. Appl. Microbiol. Biotechnol. 2004, 63, 748–753. [Google Scholar] [CrossRef] [PubMed]
- De Assis, L.J.; Ries, L.N.A.; Savoldi, M.; dos Reis, T.F.; Brown, N.A.; Goldman, G.H. Aspergillus nidulans protein kinase A plays an important role in cellulase production. Biotechnol. Biofuels 2015, 8, 213. [Google Scholar] [CrossRef] [PubMed]
- Boase, N.A.; Kelly, J.M. A role for creD, a carbon catabolite repression gene from Aspergillus nidulans, in ubiquitination. Mol. Microbiol. 2004, 53, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.A.; Kelly, J.M. Proteins interacting with CreA and CreB in the carbon catabolite repression network in Aspergillus nidulans. Curr. Genet. 2017, 63, 669–683. [Google Scholar] [CrossRef]
- Ma, X.; Jiang, Y.; Ma, L.; Luo, S.; Du, H.; Li, X.; Xing, F. Corepressors SsnF and RcoA regulate development and aflatoxin B1 biosynthesis in Aspergillus flavus NRRL 3357. Toxins 2022, 14, 174. [Google Scholar] [CrossRef]
- Adnan, M.; Zheng, W.; Islam, W.; Arif, M.; Abubakar, Y.S.; Wang, Z.; Lu, G. Carbon catabolite repression in filamentous fungi. Int. J. Mol. Sci. 2018, 19, 48. [Google Scholar] [CrossRef]
- Ribeiro, L.F.C.; Chelius, C.; Boppidi, K.R.; Naik, N.S.; Hossain, S.; Ramsey, J.J.J.; Kumar, J.; Ribeiro, L.F.; Ostermeier, M.; Tran, B.; et al. Comprehensive analysis of Aspergillus nidulans PKA phosphorylome identifies a novel mode of CreA regulation. mBio 2019, 10, e02825-18. [Google Scholar] [CrossRef]
- De Assis, L.J.; Silva, L.P.; Bayram, O.; Dowling, P.; Kniemeyer, O.; Krüger, T.; Brakhage, A.A.; Chen, Y.; Dong, L.; Tan, K.; et al. Carbon catabolite repression in filamentous fungi is regulated by phosphorylation of the transcription factor CreA. mBio 2021, 12, e03146-20. [Google Scholar] [CrossRef]
- Qin, L.; Guo, S.; Li, A.; Fan, L.; Tan, K.; Wong, K.H. An effective strategy for identifying autogenous regulation of transcription factors in filamentous fungi. Microbiol. Spectr. 2023, 11, e02347-23. [Google Scholar] [CrossRef]
- Chen, Y.; Dong, L.; Alam, M.A.; Pardeshi, L.; Miao, Z.; Wang, F.; Tan, K.; Hynes, M.J.; Kelly, J.M.; Wong, K.H. Carbon catabolite repression governs diverse physiological processes and development in Aspergillus nidulans. mBio 2022, 13, e03734-21. [Google Scholar] [CrossRef]
- Caddick, M.X.; Peters, D.; Platt, A. Nitrogen regulation in fungi. Antonie Van Leeuwenhoek 1994, 65, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.A.; Arst, H.N. Mutational analysis of AreA, a transcriptional activator mediating nitrogen metabolite repression in Aspergillus nidulans and a member of the “Streetwise” GATA family of transcription factors. Microbiol. Mol. Biol. Rev. 1998, 62, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Chant, A.; Provatopoulou, X.; Manfield, I.W.; Kneale, G.G. Structural and functional characterisation of the DNA binding domain of the Aspergillus nidulans gene regulatory protein AreA. Biochim. Biophys. Acta 2003, 1648, 84–89. [Google Scholar] [CrossRef]
- Muro-Pastor, M.I.; Gonzalez, R.; Strauss, J.; Narendja, F.; Scazzocchio, C. The GATA factor AreA is essential for chromatin remodelling in a eukaryotic bidirectional promoter. EMBO J. 1999, 18, 1584–1597. [Google Scholar] [CrossRef]
- Berger, H.; Basheer, A.; Böck, S.; Reyes-Dominguez, Y.; Dalik, T.; Altmann, F.; Strauss, J. Dissecting individual steps of nitrogen transcription factor cooperation in the Aspergillus nidulans nitrate cluster. Mol. Microbiol. 2008, 69, 1385–1398. [Google Scholar] [CrossRef]
- Small, J.A.; Todd, R.B.; Zanker, M.C.; Delimitrou, S.; Hynes, M.J.; Davis, M.A. Functional analysis of TamA, a coactivator of nitrogen-regulated gene expression in Aspergillus nidulans. Mol. Genet. Genom. 2001, 265, 636–646. [Google Scholar] [CrossRef]
- Chudzicka-Ormaniec, P.; Macios, M.; Koper, M.; Weedall, G.D.; Caddick, M.X.; Weglenski, P.; Dzikowska, A. The role of the GATA transcription factor AreB in regulation of nitrogen and carbon metabolism in Aspergillus nidulans. FEMS Microbiol. Lett. 2019, 366, fnz066. [Google Scholar] [CrossRef]
- Kotaka, M.; Johnson, C.; Lamb, H.K.; Hawkins, A.R.; Ren, J.; Stammers, D.K. Structural analysis of the recognition of the negative regulator NmrA and DNA by the zinc finger from the GATA-type transcription factor AreA. J. Mol. Biol. 2008, 381, 373–382. [Google Scholar] [CrossRef]
- Wong, K.H.; Hynes, M.J.; Todd, R.B.; Davis, M.A. Transcriptional control of nmrA by the bZIP transcription factor MeaB reveals a new level of nitrogen regulation in Aspergillus nidulans. Mol. Microbiol. 2007, 66, 534–551. [Google Scholar] [CrossRef]
- Hunter, C.C.; Siebert, K.S.; Downes, D.J.; Wong, K.H.; Kreutzberger, S.D.; Fraser, J.A.; Clarke, D.F.; Hynes, M.J.; Davis, M.A.; Todd, R.B. Multiple nuclear localization signals mediate nuclear localization of the GATA transcription factor AreA. Eukaryot. Cell 2014, 13, 527–538. [Google Scholar] [CrossRef]
- Todd, R.B.; Fraser, J.A.; Wong, K.H.; Davis, M.A.; Hynes, M.J. Nuclear accumulation of the GATA factor AreA in response to complete nitrogen starvation by regulation of nuclear export. Eukaryot. Cell 2005, 4, 1646–1653. [Google Scholar] [CrossRef] [PubMed]
- Hortschansky, P.; Haas, H.; Huber, E.M.; Groll, M.; Brakhage, A.A. The CCAAT-binding complex (CBC) in Aspergillus species. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Scheven, M.T.; Misslinger, M.; Zhao, C.; Hoefgen, S.; Gsaller, F.; Lau, J.; Jöchl, C.; Donaldson, I.; Valiante, V.; et al. The fungal CCAAT-binding complex and HapX display highly variable but evolutionary conserved synergetic promoter-specific DNA recognition. Nucleic Acids Res. 2020, 48, 3567–3590. [Google Scholar] [CrossRef]
- Steidl, S.; Tüncher, A.; Goda, H.; Guder, C.; Papadopoulou, N.; Kobayashi, T.; Tsukagoshi, N.; Kato, M.; Brakhage, A.A. A single subunit of a heterotrimeric CCAAT-binding complex carries a nuclear localization signal: Piggy back transport of the pre-assembled complex to the nucleus. J. Mol. Biol. 2004, 342, 515–524. [Google Scholar] [CrossRef]
- Goda, H.; Nagase, T.; Tanoue, S.; Sugiyama, J.; Steidl, S.; Tüncher, A.; Kobayashi, T.; Tsukagoshi, N.; Brakhage, A.A.; Kato, M. Nuclear translocation of the heterotrimeric CCAAT binding factor of Aspergillus oryzae is dependent on two redundant localising signals in a single subunit. Arch. Microbiol. 2005, 184, 93–100. [Google Scholar] [CrossRef]
- Ren, Y.; Zhang, C.; Chen, Z.; Lu, L. The heterotrimeric transcription factor CCAAT-binding complex and Ca2+-CrzA signaling reversely regulate the transition between fungal hyphal growth and asexual reproduction. mBio 2021, 12, e0300721. [Google Scholar] [CrossRef]
- Oakley, C.E.; Ahuja, M.; Sun, W.W.; Entwistle, R.; Akashi, T.; Yaegashi, J.; Guo, C.J.; Cerqueira, G.C.; Russo Wortman, J.; Wang, C.C.C.; et al. Discovery of McrA, a master regulator of Aspergillus secondary metabolism. Mol. Microbiol. 2017, 103, 347–365. [Google Scholar] [CrossRef]
- Grau, M.F.; Entwistle, R.; Oakley, C.E.; Wang, C.C.C.; Oakley, B.R. Overexpression of an LaeA-like methyltransferase upregulates secondary metabolite production in Aspergillus nidulans. ACS Chem. Biol. 2019, 14, 1643–1651. [Google Scholar] [CrossRef]
- Dao, T.T.; de Mattos-Shipley, K.M.J.; Prosser, I.M.; Williams, K.; Zacharova, M.K.; Lazarus, C.M.; Willis, C.L.; Bailey, A.M. Cleaning the cellular factory-deletion of McrA in Aspergillus oryzae NSAR1 and the generation of a novel kojic acid deficient strain for cleaner heterologous production of secondary metabolites. Front. Fungal Biol. 2021, 2, 632542. [Google Scholar] [CrossRef]
- Lee, M.K.; Son, Y.E.; Park, H.S.; Alshannaq, A.; Han, K.H.; Yu, J.H. Velvet activated McrA plays a key role in cellular and metabolic development in Aspergillus nidulans. Sci. Rep. 2020, 10, 15075. [Google Scholar] [CrossRef]
- Pfannenstiel, B.T.; Zhao, X.; Wortman, J.; Wiemann, P.; Throckmorton, K.; Spraker, J.E.; Soukup, A.A.; Luo, X.; Lindner, D.L.; Lim, F.Y.; et al. Revitalization of a forward genetic screen identifies three new regulators of fungal secondary metabolism in the genus Aspergillus. mBio 2017, 8, e01246-17. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Lyu, H.; Zhou, S.; Yu, J.; Keller, N.P.; Chen, L.; Yin, W.B. Deletion of a global regulator LaeB leads to the discovery of novel polyketides in Aspergillus nidulans. Org. Biomol. Chem. 2018, 16, 4973–4976. [Google Scholar] [CrossRef] [PubMed]
- Cary, J.W.; Harris-Coward, P.; Scharfenstein, L.; Mack, B.M.; Chang, P.K.; Wei, Q.; Lebar, M.; Carter-Wientjes, C.; Majumdar, R.; Mitra, C.; et al. The Aspergillus flavus homeobox gene, hbx1, is required for development and aflatoxin production. Toxins 2017, 9, 315. [Google Scholar] [CrossRef] [PubMed]
- Son, S.H.; Son, Y.E.; Cho, H.J.; Chen, W.; Lee, M.K.; Kim, L.H.; Han, D.M.; Park, H.S. Homeobox proteins are essential for fungal differentiation and secondary metabolism in Aspergillus nidulans. Sci. Rep. 2020, 10, 6094. [Google Scholar] [CrossRef]
- Yin, W.B.; Reinke, A.W.; Szilágyi, M.; Emri, T.; Chiang, Y.M.; Keating, A.E.; Pócsi, I.; Wang, C.C.C.; Keller, N.P. bZIP transcription factors affecting secondary metabolism, sexual development and stress responses in Aspergillus nidulans. Microbiology 2013, 159, 77–88. [Google Scholar] [CrossRef]
- Sekonyela, R.; Palmer, J.M.; Bok, J.W.; Jain, S.; Berthier, E.; Forseth, R.; Schroeder, F.; Keller, N.P. RsmA regulates Aspergillus fumigatus gliotoxin cluster metabolites including cyclo(L-Phe-L-Ser), a potential new diagnostic marker for invasive aspergillosis. PLoS ONE 2013, 8, e62591. [Google Scholar] [CrossRef]
- Wang, X.; Zha, W.; Liang, L.; Fasoyin, O.E.; Wu, L.; Wang, S. The bZIP transcription factor AflRsmA regulates aflatoxin B1 biosynthesis, oxidative stress response and sclerotium formation in Aspergillus flavus. Toxins 2020, 12, 271. [Google Scholar] [CrossRef]
- Yao, G.; Bai, X.; Zhang, B.; Wang, L.; Chen, S.; Wang, Z. Enhanced production of terrein in marine-derived Aspergillus terreus by refactoring both global and pathway-specific transcription factors. Microb. Cell Fact. 2022, 21, 136. [Google Scholar] [CrossRef]
- Itoh, E.; Odakura, R.; Oinuma, K.I.; Shimizu, M.; Masuo, S.; Takaya, N. Sirtuin E is a fungal global transcriptional regulator that determines the transition from the primary growth to the stationary phase. J. Biol. Chem. 2017, 292, 11043–11054. [Google Scholar] [CrossRef]
- Wen, M.; Lan, H.; Sun, R.; Chen, X.; Zhang, X.; Zhu, Z.; Tan, C.; Yuan, J.; Wang, S. Histone deacetylase SirE regulates development, DNA damage response and aflatoxin production in Aspergillus flavus. Environ. Microbiol. 2022, 24, 5596–5610. [Google Scholar] [CrossRef]
- Zehetbauer, F.; Seidl, A.; Berger, H.; Sulyok, M.; Kastner, F.; Strauss, J. RimO (SrrB) is required for carbon starvation signaling and production of secondary metabolites in Aspergillus nidulans. Fungal Genet. Biol. 2022, 162, 103726. [Google Scholar] [CrossRef] [PubMed]
- Pandit, S.S.; Zheng, J.; Yin, Y.; Lorber, S.; Puel, O.; Dhingra, S.; Espeso, E.A.; Calvo, A.M. Homeobox transcription factor HbxA influences expression of over one thousand genes in the model fungus Aspergillus nidulans. PLoS ONE 2023, 18, e0286271. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, D.C.; Doedt, T.; Chiang, L.Y.; Kim, H.S.; Chen, D.; Nierman, W.C.; Filler, S.G. The Aspergillus fumigatus StuA protein governs the up-regulation of a discrete transcriptional program during the acquisition of developmental competence. Mol. Biol. Cell 2005, 16, 5866–5879. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.Y.; Wu, J.; Miller, B.L. StuA is required for cell pattern formation in Aspergillus. Genes Dev. 1992, 6, 1770–1782. [Google Scholar] [CrossRef]
- Yu, Z.; Hübner, J.; Herrero, S.; Gourain, V.; Fischer, R. On the role of the global regulator RlcA in red-light sensing in Aspergillus nidulans. Fungal Biol. 2020, 124, 447–457. [Google Scholar] [CrossRef]
- Collemare, J.; Seidl, M.F. Chromatin-dependent regulation of secondary metabolite biosynthesis in fungi: Is the picture complete? FEMS Microbiol. Rev. 2019, 43, 591–607. [Google Scholar] [CrossRef]
- Moon, H.; Lee, M.K.; Bok, I.; Bok, J.W.; Keller, N.P.; Yu, J.H. Unraveling the gene regulatory networks of the global regulators VeA and LaeA in Aspergillus nidulans. Microbiol. Spectr. 2023, 11, e00166-23. [Google Scholar] [CrossRef]

| Aspergillus Strain | Deletion or Overexpression | Upregulated SMs | Downregulated SMs | Ref. |
|---|---|---|---|---|
| A. nidulans FGSC 26 | Deletion | - | Sterigmatocystin (ST, 1), monacolin J (MONJ, 2), and penicillin G (3) | [46] |
| Overexpression | Penicillin G (3) and MONJ (2) | - | ||
| A. fumigatus AF293 | Deletion | - | Gliotoxin (4) | |
| A. terreus ATCC 20542 | Deletion | - | Lovastatin (5) | |
| Overexpression | Lovastatin (5) | - | ||
| A. fumigatus AF293 | Deletion | - | ST (1) | [55] |
| A. flavus CA14 | Deletion | - | Aflatoxin B1 (6) and aflatoxin B2 (7) | [56] |
| A. fumigatus CEA17 | Deletion | - | Endocrocin (8) | [57] |
| A. oryzae RIB40 | Deletion | - | Kojic acid (9) | [58] |
| A. niger ATCC9029 | Deletion | BMS-192548 (13) and aspernigrin A (14) | Asperrubrol (10), atromentin (11) and JBIR-86 (12) | [59] |
| A. carbonarius UdL-TA 3.83 | Deletion | - | Ochratoxin A (OTA, 15) | [60] |
| A. ochraceus fc-1 | Deletion | - | OTA (15) | [61] |
| A. carbonarius Ac ITEM 5010 | Deletion | - | OTA (15) | [62] |
| A. pseudoterreus ATCC 32359 | Deletion | - | Itaconic acid (16) | [63] |
| A. flavipes (507) | Deletion | Flavipamide A and B (17,18), N-benzoylphenylalaniny-N-benzoylphenyl-alaninate (19), 4′-OMe-asperphenamate (20), cyclic Pro-Gly-Val-Gly-Try (/8-OH, 3-prenyl)-Gly-Trp (21) | - | [64] |
| Aspergillus sp. Z5 | Overexpression | Diorcinol (22) and quinolactacin A (23) | - | [45] |
| A. fumisynnematus F746 | Overexpression | Cyclopiazonic acid (24) | - | [65] |
| A. versicolor 0312 | Overexpression | Versicolor A (25), acetylaranotin (26), acetylapoaranotin (27), ergosterol (28) and diisobutyl phthalate (29) | - | [66] |
| A. niger FGSC A1279 | Overexpression | Flaviolin (30), orlandin (31), and kotanin (32) | - | [67] |
| A. terreus RA2905 | Overexpression | Dihydroisoflavipucines 1 and 2 (33, 34) | - | [68] |
| A. niger L14 | Overexpression | Aspochracin (35), JBIR-15 (36), sclerotiotide C (37), kojic acid (9), and penicillic acid (38) | - | [69] |
| Aspergillus Strain | Deletion or Overexpression | Upregulated SMs | Downregulated SMs | Ref. |
|---|---|---|---|---|
| A. nidulans FGSC4 | Deletion | - | ST (1) and penicillin G (3) | [76] |
| A. nidulans AXB4A2 | Deletion | - | Penicillin G (3) | [77] |
| Overexpression | - | Penicillin G (3) | ||
| A. oryzae RIB40 | Deletion | - | Penicillin G (3) | [78] |
| A. flavus ATCC MYA384 | Deletion | - | Aflatoxin, aflatrem (39) and CPA (24) | [79] |
| A. fumigatus CEA10 | Deletion | - | Gliotoxin (4) | [81] |
| A. carbonarius UdL-TA 3.83 | Deletion | - | OTA (15) | [60] |
| A. niger CICC 41702 | Deletion | - | OTA (15) | [82] |
| A. fumigatus CEA10 | Deletion | - | Fumagillin (40), fumitremorgin G (41), fumigaclavine C (42), and glionitrin A (43) | [83] |
| Overexpression | - | fumagillin (40), fumitremorgin G (41), fumigaclavine C (42), and glionitrin A (43) | ||
| A. pachycristatus NRRL 11440 | Deletion | - | Echinocandin B (ECB,44) and ST (1) | [84] |
| A. nidulans RDIT9.32 | Deletion | F9775A (45), F9775B (46), and orsellinic acid (47) | - | [85] |
| GTRs | Aspergillus Strain | Deletion or Overexpression | Upregulated SMs | Downregulated SMs | Ref. |
|---|---|---|---|---|---|
| PacC | A. nidulans | Overexpression | Penicillin G (3) | - | [99] |
| A. parasiticus RHN1 | Deletion of the PacC binding site | - | O-methylsterigmatocystin (48) | [100] | |
| A. ochraceus fc-1 | Deletion | - | OTA (15) | [101] | |
| A. carbonarius NRRL 368 | Deletion | - | OTA (15) | [102] | |
| CreA | A. flavus CA14PTS | Overexpression | Aflatoxin B1 (6) | - | [103] |
| Deletion | - | Aflatoxin B1 (6) | |||
| A. ochraceus fc-1 | Deletion | - | OTA (15) | [104] | |
| AreA | A. terreus SBUG844 | Deletion | - | Terrein (49) | [105] |
| A. flavus SRRC1709 | Overexpression | Aflatoxin B1 (6) | - | [106] | |
| Deletion | Aflatoxin B1 (6) | ||||
| CBC | A. nidulans MH8193 | Deletion of hapC | - | Penicillin (3) | [107] |
| A. fumigatus A1160P+ | Deletion of hapE | Ergosta-5,7,24(28)-trien-3β-ol (50) and episterol (51) | - | [108] |
| GTRs | Aspergillus Strain | Deletion or Overexpression | Upregulated SMs | Downregulated SMs | Ref. |
|---|---|---|---|---|---|
| McrA | A. Nidulans FGSC A4 | Deletion | 1,3-Dihydro-6-hydroxy-4-methoxy-5-methyl-1-oxo-2H-isoindole-2-pentanoic acid (52), 4-[hydroxy(4-hydroxy-2-methoxy-3,6-dimethylphenyl)methoxy]-2-methoxy-3,6-dimethylbenzaldehyde (53) | - | [139] |
| A. nidulans FGSCA442 | Deletion | Cichorine intermediate (54) | - | [140] | |
| A. wentii IMI 49129 | Deletion | Emodin (55), physcion (56), sulochrin (57), 14-O-demethylsulochrin (58), physcion bianthrone (59), (trans)-emodin bianthrone (60), (cis)-emodin bianthrone (61), (trans)-emodin physcion bianthrone (62), (cis)-emodin physcion bianthrone (63), aspergillus acid B (64), and aspergillus acid E (65) | - | [35] | |
| A. melleus IMV 01140 | Deletion | Neoaspergillic acid (66), and neohydroxyaspergillic acid (67) | - | [36] | |
| McrA | A. oryzae NSAR1 | Deletion | Kojic acid (9) | - | [141] |
| A. nidulans FGSC4 | Overexpression | - | ST (1) | [142] | |
| LaeB | A. Nidulans BTP69 | Deletion | - | ST (1) | [143] |
| A. flvus NRRL3357 | Deletion | - | Aflatoxin | ||
| LaeB | A. nidulans RJMP1.49 | Deletion | Gibellulin C (68), gibellulin D (69), 3-methoxyporriolide (70), 7-methoxyporriolide (71), porriolide (72), cichorine (73), asperthecin (74), microperfuranone (75) | ST (1) | [144] |
| HbxA/Hbx1 | A. flavus CA14 | Deletion of hbx1 | - | CPA (24), aflatrem (39) and aflatoxin | [145] |
| A. nidulans FGSC4 | Deletion of hbxA | ST (1) | - | [146] | |
| RsmA | A. nidulans RJMP1.49 | Overexpression | ST (1) | - | [147] |
| A. fumigatus AF293 | Overexpression | Gliotoxin (4) and cyclo(L-Phe-L-Ser) (76) | - | [148] | |
| A. flavus CA14 | Overexpression | Aflatoxin B1 (6) | - | [149] | |
| StuA | A. terreus RA2905 | Deletion | Terrein(49) | Butyrolactones | [150] |
| SirE | A. nidulans A26 | Deletion | - | ST (1) | [151] |
| A. flavus CA14 PTS | Deletion | Aflatoxin | - | [152] | |
| RimO | A. nidulans FGSC4 | Overexpression | ST (1) and penicillin G (3) | - | [153] |
| Deletion | - | ST (1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Gong, Q.; Zhang, H. Engineering of Global Transcriptional Regulators (GTRs) in Aspergillus for Natural Product Discovery. J. Fungi 2025, 11, 449. https://doi.org/10.3390/jof11060449
Zhao Y, Gong Q, Zhang H. Engineering of Global Transcriptional Regulators (GTRs) in Aspergillus for Natural Product Discovery. Journal of Fungi. 2025; 11(6):449. https://doi.org/10.3390/jof11060449
Chicago/Turabian StyleZhao, Yujie, Qing Gong, and Huawei Zhang. 2025. "Engineering of Global Transcriptional Regulators (GTRs) in Aspergillus for Natural Product Discovery" Journal of Fungi 11, no. 6: 449. https://doi.org/10.3390/jof11060449
APA StyleZhao, Y., Gong, Q., & Zhang, H. (2025). Engineering of Global Transcriptional Regulators (GTRs) in Aspergillus for Natural Product Discovery. Journal of Fungi, 11(6), 449. https://doi.org/10.3390/jof11060449







