An Optimized In-House Protocol for Cryptococcus neoformans DNA Extraction from Whole Blood: “Comparison of Lysis Buffer and Ox-Bile Methods”
Abstract
1. Introduction
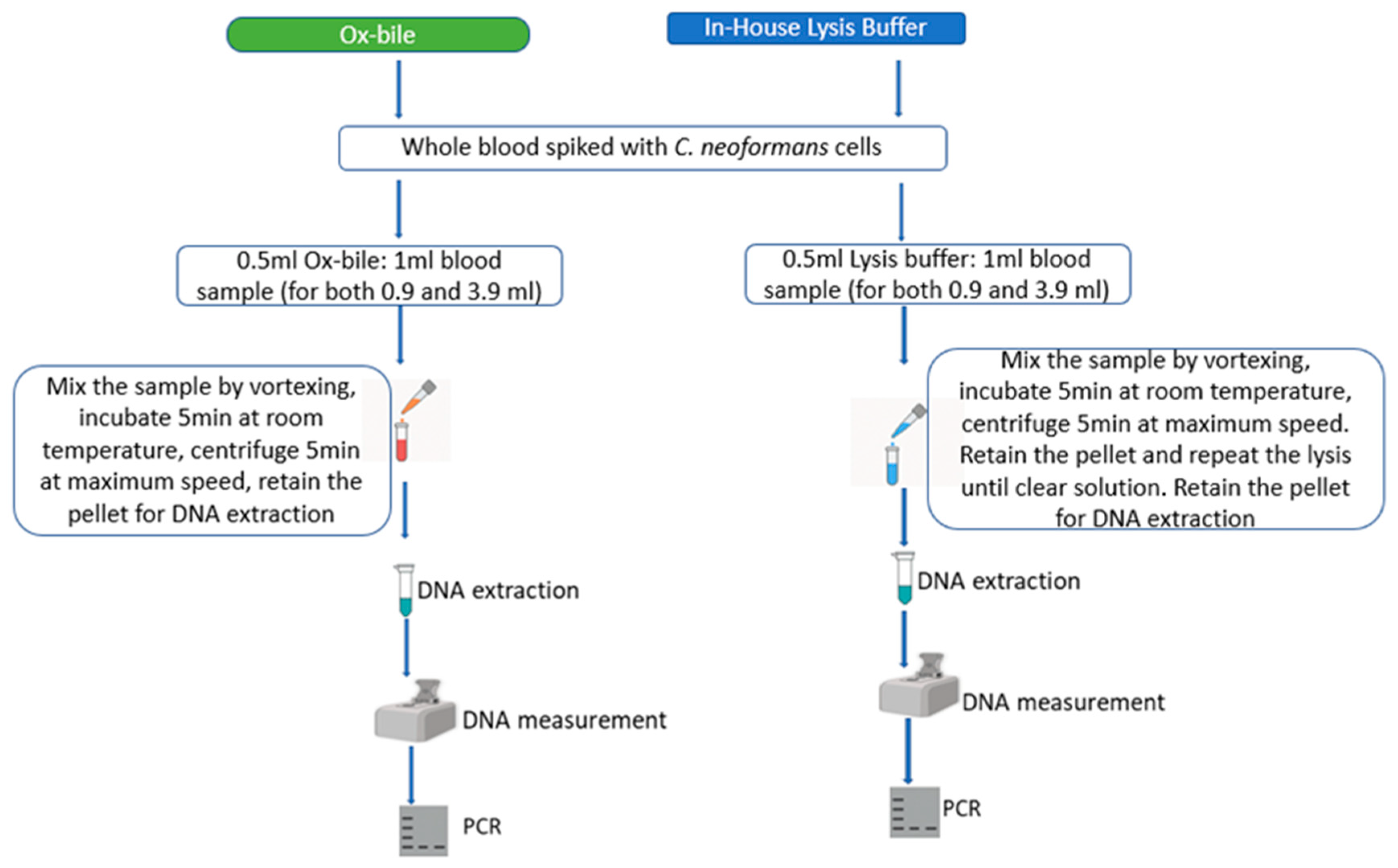
2. Materials and Methods
- 1
- Lysis Buffer: Prepared in-house (5 mL of 2 M Tris pH 7.6, 5 mL of 1 M MgCl2, 3.3 mL of 3 M NaCl, and up to 1000 mL of distilled water).
- 2
- Ox-Bile Solution: 10 g ox-bile powder (made from cow by Solarbio Beijing China), known for lysing red blood cells [41] was dissolved in 100 mL of distilled water.
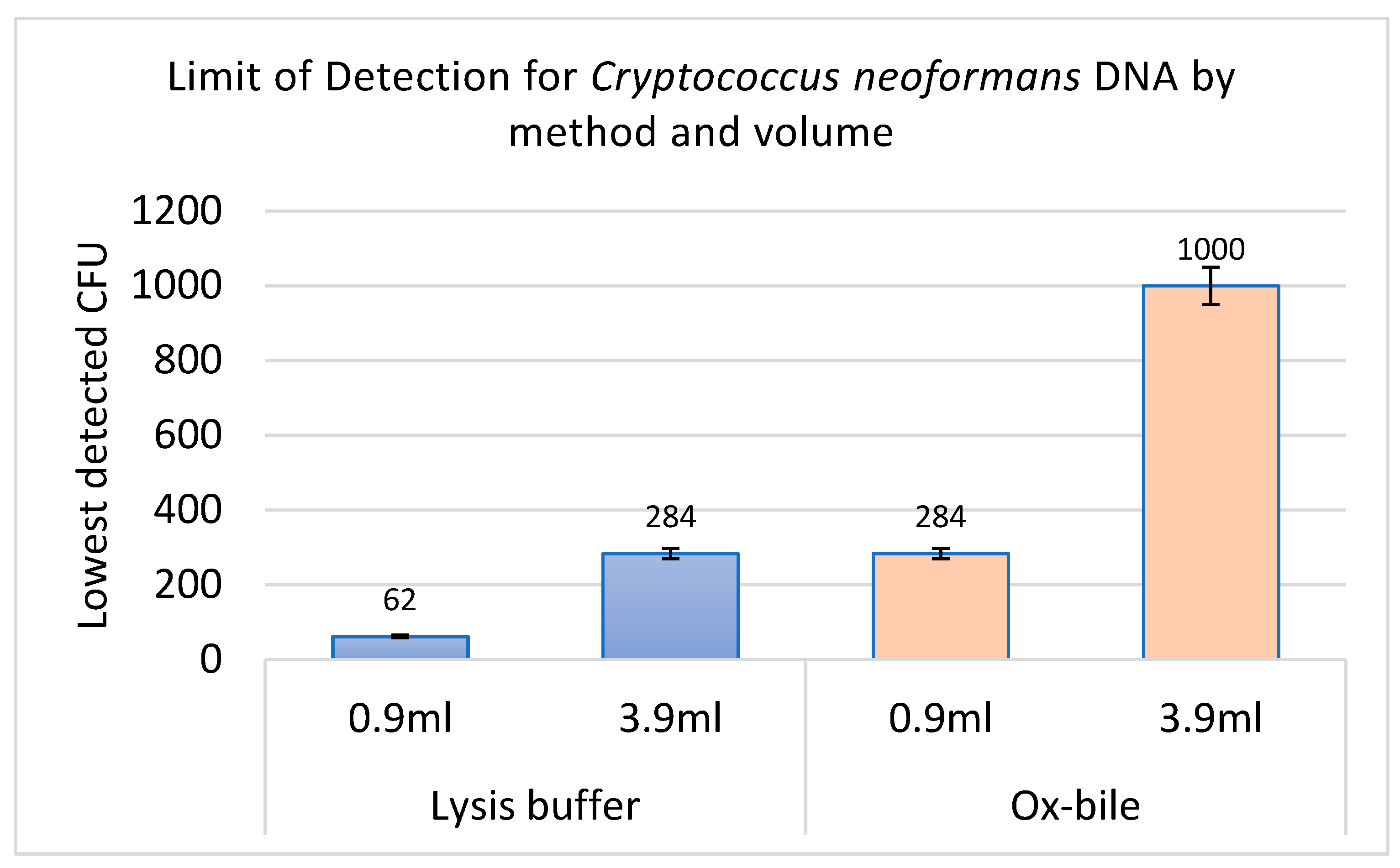
3. Results and Discussion
4. Conclusions
5. Recommendation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CSF | Cerebrospinal fluid |
| CFU | Colony-forming units |
| CNS | Central nervous system |
| CrAg | Cryptococcal antigen |
| LFA | Lateral flow assay |
| LOD | Limit of Detection |
| GTRL | Genomic Translation and Research Laboratory |
| MUST | Mbarara University of Science and Technology |
| C. neoformans | Cryptococcus neoformans |
| YPD | Yeast extract peptone dextrose |
References
- Botts, M.R.; Hull, C.M. Dueling in the lung: How Cryptococcus spores race the host for survival. Curr. Opin. Microbiol. 2010, 13, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, K.L.; Murphy, J.W. What makes Cryptococcus neoformans a pathogen? Emerg. Infect. Dis. 1998, 4, 71. [Google Scholar] [CrossRef] [PubMed]
- Bulmer, G.; Sans, M.D.; Gunn, M.C. Cryptococcus neoformans I. Nonencapsulated mutants. J. Bacteriol. 1967, 94, 1475–1479. [Google Scholar] [CrossRef]
- Vecchiarelli, A. Immunoregulation by capsular components of Cryptococcus neoformans. Med. Mycol. 2000, 38, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Goldman, D.; Lee, S.C.; Casadevall, A. Pathogenesis of pulmonary Cryptococcus neoformans infection in the rat. Infect. Immun. 1994, 62, 4755–4761. [Google Scholar] [CrossRef]
- Jongwutiwes, U.; Sungkanuparph, S.; Kiertiburanakul, S. Comparison of Clinical Features and Survival between Cryptococcosis in Human Immunodeficiency Virus (HIV)-Positive and HIV-Negative Patients. Jpn. J. Infect. Dis. 2008, 61, 111–115. [Google Scholar] [CrossRef]
- Yoon, H.A.; Felsen, U.; Wang, T.; Pirofski, L.-A. Cryptococcus neoformans infection in Human Immunodeficiency Virus (HIV)-infected and HIV-uninfected patients at an inner-city tertiary care hospital in the Bronx. Med. Mycol. 2020, 58, 434–443. [Google Scholar] [CrossRef]
- Anjum, S.; Williamson, P.R. Clinical aspects of immune damage in cryptococcosis. Curr. Fungal. Infect. Rep. 2019, 13, 99–108. [Google Scholar] [CrossRef]
- Chae, H.S.; Jang, G.E.; Kim, N.H.; Son, H.R.; Lee, J.H.; Kim, S.H.; Park, G.N.; Jo, H.J.; Kim, J.T.; Chang, K.S. Classification of Cryptococcus neoformans and Yeast-Like Fungus Isolates from Pigeon Droppings by Colony Phenotyping and ITS Genotyping and Their Seasonal Variations in Korea. Avian Dis. 2012, 56, 58–64. [Google Scholar] [CrossRef]
- Archibald, L.K.; Quisling, R.G. Central nervous system infections. In Textbook of Neurointensive Care; Springer: London, UK, 2013; pp. 427–517. [Google Scholar]
- Dromer, F.; Levitz, S.M. Invasion of Cryptococcus into the central nervous system. In Cryptococcus: From Human Pathogen to Model Yeast; ASM Press: Washington, DC, USA, 2010; pp. 465–471. [Google Scholar]
- Giovane, R.A.; Lavender, P.D. Central nervous system infections. Prim Care 2018, 45, 505–518. [Google Scholar] [CrossRef]
- Le Govic, Y.; Demey, B.; Cassereau, J.; Bahn, Y.-S.; Papon, N. Pathogens infecting the central nervous system. In Advances in Surgical and Medical Specialties; Jenny Stanford Publishing: Singapore, 2023; pp. 1129–1142. [Google Scholar]
- Liu, T.-B.; Perlin, D.S.; Xue, C. Molecular mechanisms of cryptococcal meningitis. Virulence 2012, 3, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Tugume, L.; Ssebambulidde, K.; Kasibante, J.; Ellis, J.; Wake, R.M.; Gakuru, J.; Lawrence, D.S.; Abassi, M.; Rajasingham, R.; Meya, D.B. Cryptococcal meningitis. Nat. Rev. Dis. Primers 2023, 9, 62. [Google Scholar] [CrossRef]
- Bava, A.J.; Solari, R.; Isla, G.; Troncoso, A. Atypical forms of Cryptococcus neoformans in CSF of an AIDS patient. J. Infect. Dev. Ctries. 2008, 2, 403–405. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dantas, K.C.; de Freitas—Xavier, R.S.; Spina Lombardi, S.C.F.; Júnior, A.M.; da Silva, M.V.; Criado, P.R.; de Freitas, V.L.T.; de Almeida, T.M.B. Comparative analysis of diagnostic methods for the detection of Cryptococcus neoformans meningitis. PLoS Negl. Trop. Dis. 2023, 17, e0011140. [Google Scholar] [CrossRef]
- Mahajan, K.R.; Roberts, A.L.; Curtis, M.T.; Fortuna, D.; Dharia, R.; Sheehan, L. Diagnostic Challenges of Cryptococcus neoformans in an Immunocompetent Individual Masquerading as Chronic Hydrocephalus. Case Rep. Neurol. Med. 2016, 2016, 7381943. [Google Scholar] [CrossRef] [PubMed]
- Chikusu, C.M.P. How to perform a lumbar puncture. South Sudan Med. J. 2016, 9, 85–88. [Google Scholar]
- Tumani, H.; Petereit, H.F.; Gerritzen, A.; Gross, C.C.; Huss, A.; Isenmann, S.; Jesse, S.; Khalil, M.; Lewczuk, P.; Lewerenz, J.; et al. S1 guidelines “lumbar puncture and cerebrospinal fluid analysis” (abridged and translated version). Neurol. Res. Pract. 2020, 2, 1–28. [Google Scholar] [CrossRef]
- Kwizera, R.; Omali, D.; Tadeo, K.; Kasibante, J.; Rutakingirwa, M.K.; Kagimu, E.; Ssebambulidde, K.; Williams, D.A.; Rhein, J.; Boulware, D.; et al. Evaluation of the Dynamiker cryptococcal antigen lateral flow assay for the diagnosis of HIV-associated cryptococcosis. J. Clin. Microbiol. 2021, 59. [Google Scholar] [CrossRef]
- Matsuo, T.; Wurster, S.; Hoenigl, M.; Kontoyiannis, D.P. Current and emerging technologies to develop Point-of-Care Diagnostics in medical mycology. Expert Rev. Mol. Diagn. 2024, 24, 841–858. [Google Scholar] [CrossRef]
- Rajasingham, R.; Wake, R.M.; Beyene, T.; Katende, A.; Letang, E.; Boulware, D.R. Cryptococcal meningitis diagnostics and screening in the era of point-of-care laboratory testing. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef]
- Vianna, C.M.d.M.; Mosegui, G.B.G. Cost-effectiveness analysis and budgetary impact of the Cryptococcal Antigen Lateral Flow Assay (CRAG-LFA) implementation for the screening and diagnosis of cryptococcosis in asymptomatic people living with HIV in Brazil. Rev. Inst. Med. Trop. Sao Paulo 2021, 63, e57. [Google Scholar] [CrossRef]
- Boyd, K.; Kouamou, V.; Hlupeni, A.; Tangwena, Z.; Ndhlovu, C.E.; Makadzange, A.T.; CryptoART Study Team. Diagnostic Accuracy of Point of Care Cryptococcal Antigen Lateral Flow Assay in Fingerprick Whole Blood and Urine Samples for the Detection of Asymptomatic Cryptococcal Disease in Patients with Advanced HIV Disease. Microbiol. Spectr. 2022, 10, e0107522. [Google Scholar] [CrossRef]
- Abassi, M.; Boulware, D.R.; Rhein, J. Cryptococcal Meningitis: Diagnosis and Management Update. Curr. Trop. Med. Rep. 2015, 2, 90–99. [Google Scholar] [CrossRef]
- Malavika, K.; Premamalini, T.; Kindo, A. Evaluation of Lateral Flow Assay for the Diagnosis of Cryptococcal Meningitis and its Comparison with the Gold Standard and Other Laboratory Tests. J. Clin. Diagn. Res. 2022, 16, DC25–DC30. [Google Scholar] [CrossRef]
- Maziarz, E.K.; Perfect, J.R. Cryptococcosis. In Diagnosis and Treatment of Fungal Infections; Springer International Publishing: Cham, Switzerland, 2023; pp. 245–265. [Google Scholar]
- Nalintya, E.; Kiggundu, R.; Meya, D. Evolution of Cryptococcal Antigen Testing: What Is New? Curr. Fungal Infect. Rep. 2016, 10, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Saha, D.C.; Xess, I.; Jain, N. Evaluation of conventional & serological methods for rapid diagnosis of cryptococcosis. Indian J. Med. Res. 2008, 127, 483–488. [Google Scholar]
- Tang, M.W.; Clemons, K.V.; Katzenstein, D.A.; Stevens, D.A. The cryptococcal antigen lateral flow assay: A point-of-care diagnostic at an opportune time. Crit. Rev. Microbiol. 2015, 42, 634–642. [Google Scholar] [CrossRef]
- Saha, D.C.; Xess, I.; Biswas, A.; Bhowmik, D.M.; Padma, M.V. Detection of Cryptococcus by conventional, serological and molecular methods. J. Med. Microbiol. 2009, 58, 1098–1105. [Google Scholar] [CrossRef] [PubMed]
- Sidrim, J.J.C.; Costa, A.K.F.; Cordeiro, R.A.; Brilhante, R.S.m.N.; Moura, F.E.A.; Castelo-Branco, D.S.C.M.; Neto, M.P.d.A.; Rocha, M.F.G. Molecular methods for the diagnosis and characterization of Cryptococcus: A review. Can. J. Microbiol. 2010, 56, 445–458. [Google Scholar] [CrossRef]
- Tay, E.; Chen, S.C.-A.; Green, W.; Lopez, R.; Halliday, C.L. Development of a Real-Time PCR Assay to Identify and Distinguish between Cryptococcus neoformans and Cryptococcus gattii Species Complexes. J. Fungi 2022, 8, 462. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, M.; Xia, Y.; Yan, J.; Zou, J.; Zhang, D. Application and evaluation of nucleic acid sequence-based amplification, PCR and cryptococcal antigen test for diagnosis of cryptococcosis. BMC Infect. Dis. 2021, 21, 1020. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Wickes, B.L.; Ilkit, M.; Pincus, D.H.; Daneshnia, F.; Pan, W.; Fang, W.; Boekhout, T. Identification of Mycoses in Developing Countries. J. Fungi 2019, 5, 90. [Google Scholar] [CrossRef] [PubMed]
- Zolotareva, M.; Cascalheira, F.; Caneiras, C.; Bárbara, C.; Caetano, D.M.; Teixeira, M.C. In the flow of molecular miniaturized fungal diagnosis. Trends Biotechnol. 2024, 42, 1628–1643. [Google Scholar] [CrossRef]
- Lee, J.S.; Kim-Shapiro, D.B. Stored blood: How old is too old? J. Clin. Investig. 2017, 127, 100–102. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Estimation of Cell Number by Hemocytometry Counting. Cold Spring Harb. Protoc. 2019, 2019, pdb-rot097980. [Google Scholar] [CrossRef]
- PowerPoint. Available online: https://powerpoint.cloud.microsoft/en-us/ (accessed on 10 March 2025).
- Zhou, L.; Pollard, A.J. A novel method of selective removal of human DNA improves PCR sensitivity for detection of Salmonella Typhi in blood samples. BMC Infect. Dis. 2012, 12, 164. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard Wayne, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008; pp. 6–12. [Google Scholar]
- Sidstedt, M.; Hedman, J.; Romsos, E.L.; Waitara, L.; Wadsö, L.; Steffen, C.R.; Vallone, P.M.; Rådström, P. Inhibition mechanisms of hemoglobin, immunoglobulin G, and whole blood in digital and real-time PCR. Anal. Bioanal. Chem. 2018, 410, 2569–2583. [Google Scholar] [CrossRef]
- Schmidt, J.; Blessing, F.; Fimpler, L.; Wenzel, F. Nanopore Sequencing in a Clinical Routine Laboratory: Challenges and Opportunities. Clin. Lab. 2020, 66, 1097–1104. [Google Scholar] [CrossRef]
- Houghton, S.G.; Cockerill, F.R. Real-time PCR: Overview and applications. Surgery 2006, 139, 1–5. [Google Scholar] [CrossRef]

| DNA Quality (A260/A280) | DNA Quantity (ng/µL) | |||||||
|---|---|---|---|---|---|---|---|---|
| Method | C. neoformans Cells | Blood Volume (mL) | Mean (95% CI) | SD (95% CI) | p-Value | Mean (95% CI) | SD (95% CI) | p-Value |
| Lysis buffer | 1 × 101–10 × 104 | 0.9 | 1.88 (1.87–1.89) | 0.014 (0.008–0.018) | 0.341 | 290.17 (267.17–310.08) | 32.188 (17.07–38.13) | 0.003 |
| 1–5 McFarland | 3.9 | 1.873 (1.87–1.88) | 0.013 (0–0.018) | 853.47 (624.86–999.7) | 285.95 (38.6–406.5) | |||
| Ox-bile | 1 × 101–10 × 104 | 0.9 | 2.36 (1.69–3.57) | 1.676 (0.015–2.46) | 0.366 | 199.28 (110.79–268.19) | 122.19 (19.99–143.07) | 0.656 |
| 1–5 McFarland | 3.9 | 1.84 (1.833–1.85) | 0.01 (0.004–0.015) | 266.73 (173.85–359.09) | 137.75 (64.132–179.49) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wasswa, F.B.; Kassaza, K.; Nielsen, K.; Bazira, J. An Optimized In-House Protocol for Cryptococcus neoformans DNA Extraction from Whole Blood: “Comparison of Lysis Buffer and Ox-Bile Methods”. J. Fungi 2025, 11, 430. https://doi.org/10.3390/jof11060430
Wasswa FB, Kassaza K, Nielsen K, Bazira J. An Optimized In-House Protocol for Cryptococcus neoformans DNA Extraction from Whole Blood: “Comparison of Lysis Buffer and Ox-Bile Methods”. Journal of Fungi. 2025; 11(6):430. https://doi.org/10.3390/jof11060430
Chicago/Turabian StyleWasswa, Fredrickson B, Kennedy Kassaza, Kirsten Nielsen, and Joel Bazira. 2025. "An Optimized In-House Protocol for Cryptococcus neoformans DNA Extraction from Whole Blood: “Comparison of Lysis Buffer and Ox-Bile Methods”" Journal of Fungi 11, no. 6: 430. https://doi.org/10.3390/jof11060430
APA StyleWasswa, F. B., Kassaza, K., Nielsen, K., & Bazira, J. (2025). An Optimized In-House Protocol for Cryptococcus neoformans DNA Extraction from Whole Blood: “Comparison of Lysis Buffer and Ox-Bile Methods”. Journal of Fungi, 11(6), 430. https://doi.org/10.3390/jof11060430







