FGSE02, a Novel Secreted Protein in Fusarium graminearum FG-12, Leads to Cell Death in Plant Tissues and Modulates Fungal Virulence
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strains, Plant Materials, and Growth Conditions
2.2. Infection Assays and Sample Collection
2.3. RNA Extraction, Library Construction, and Sequencing
2.4. Transcriptome Data Processing and Analysis
2.5. FGSE02 Bioinformatics Characterization
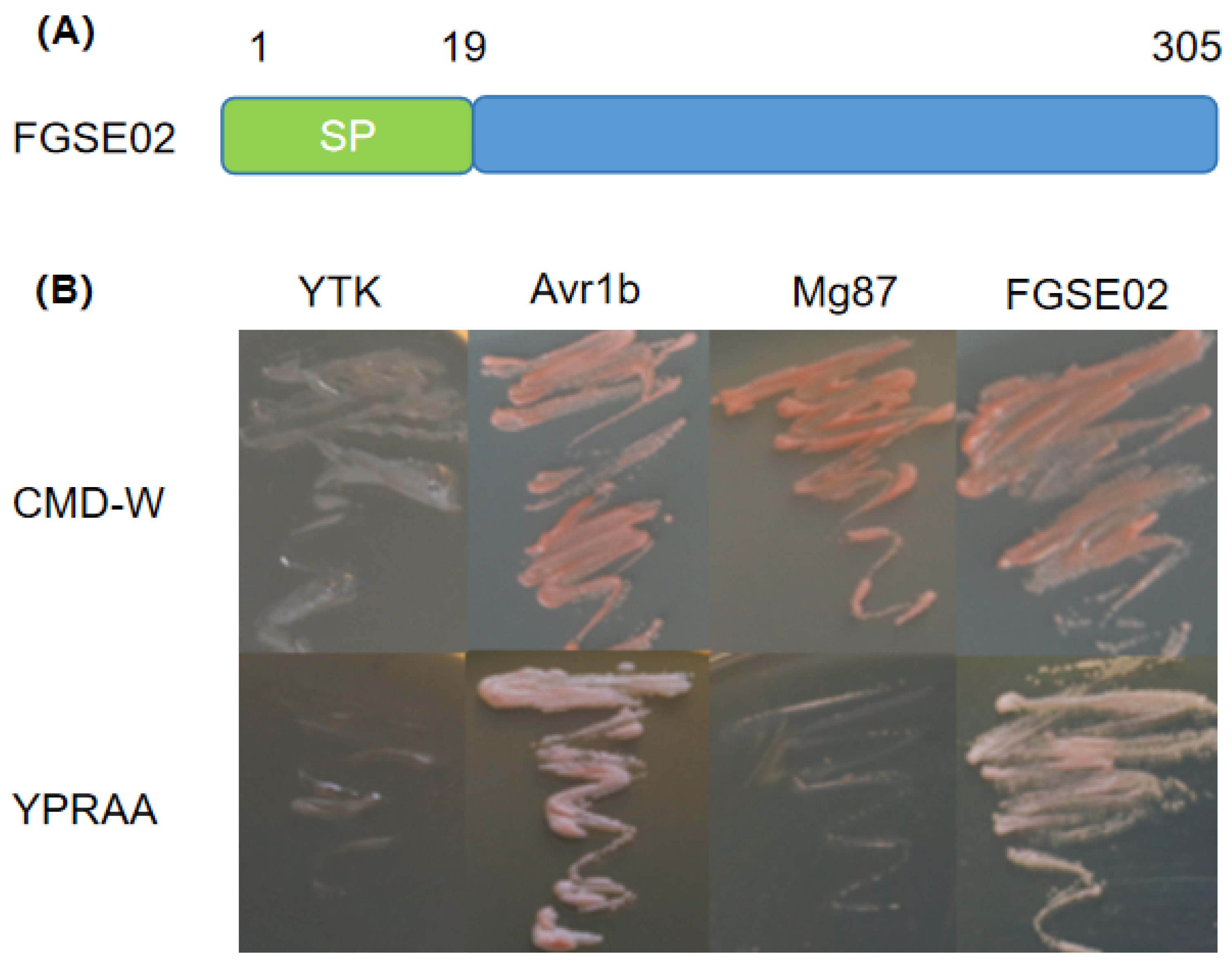
2.6. Yeast Secretion Assay
2.7. FGSE02 Knockout and Complementation
2.8. Transient Expression in Nicotiana benthamiana
2.9. Pathogenicity and Phenotypic Assays
2.10. Statistical Analysis
3. Results
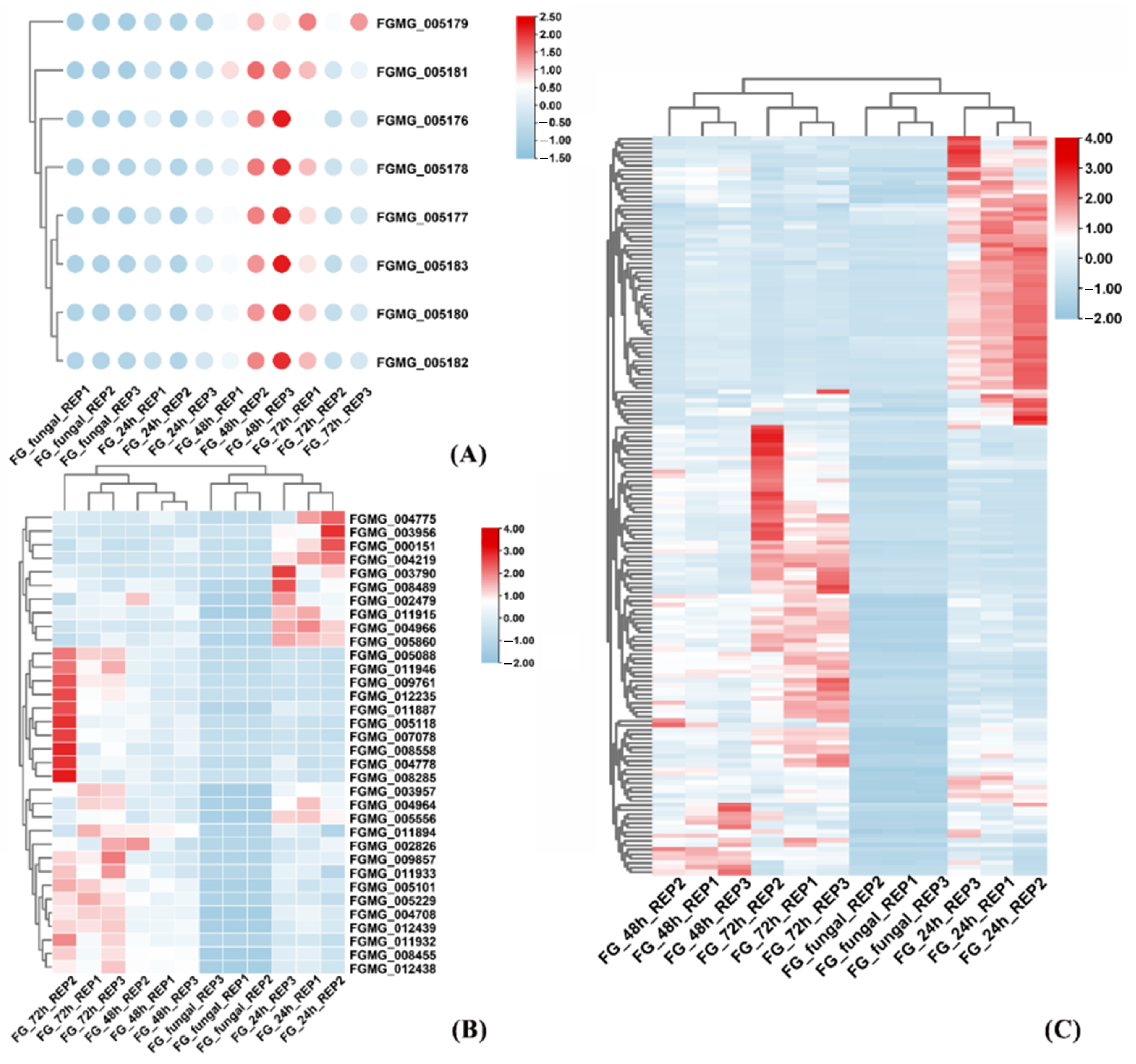
3.1. Dynamics of F. graminearum Gene Expression During Maize Infection
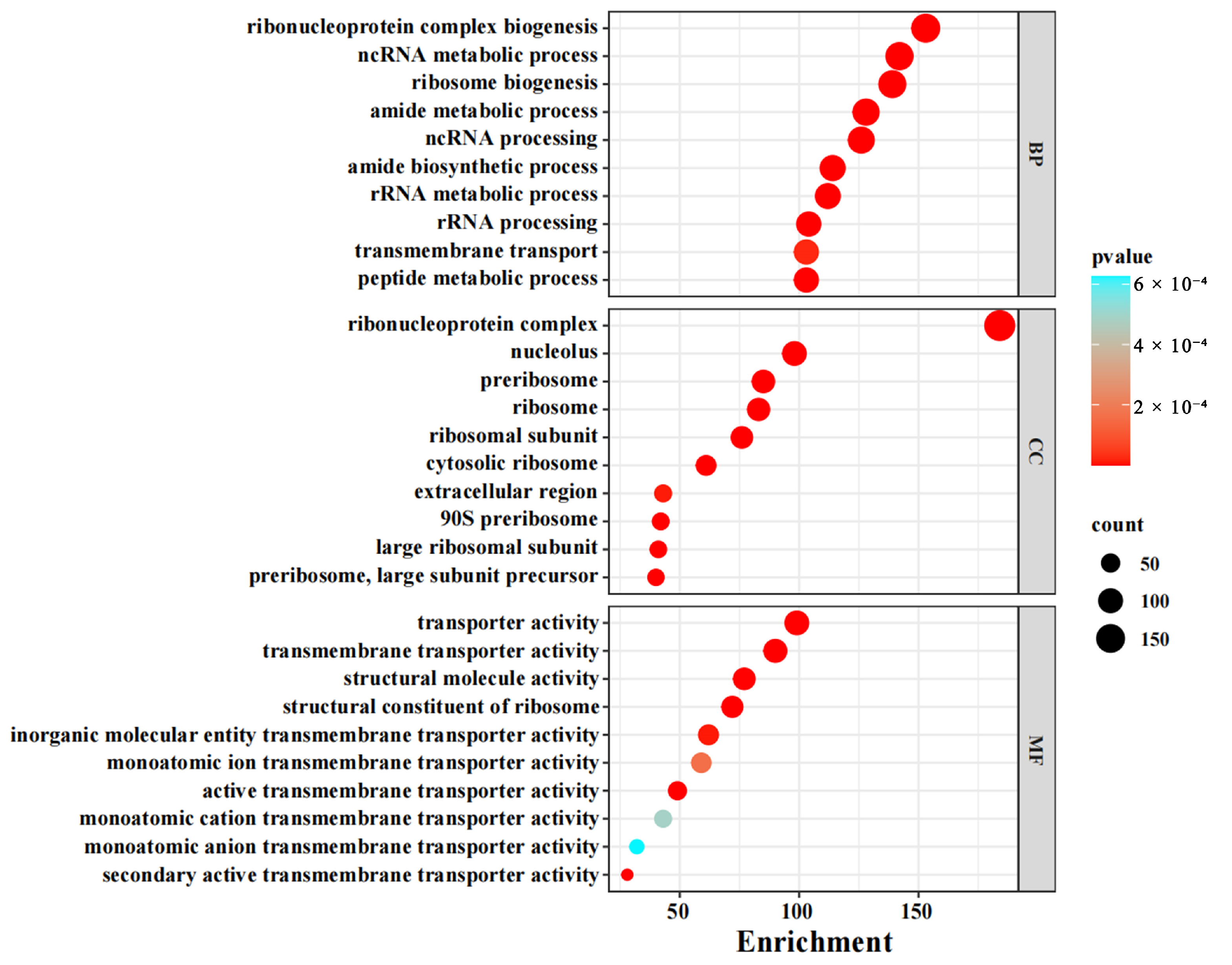
3.2. Functional Enrichment of Differentially Expressed Genes (DEGs)
3.3. Gene Analysis Related to Secondary Metabolite Biosynthesis
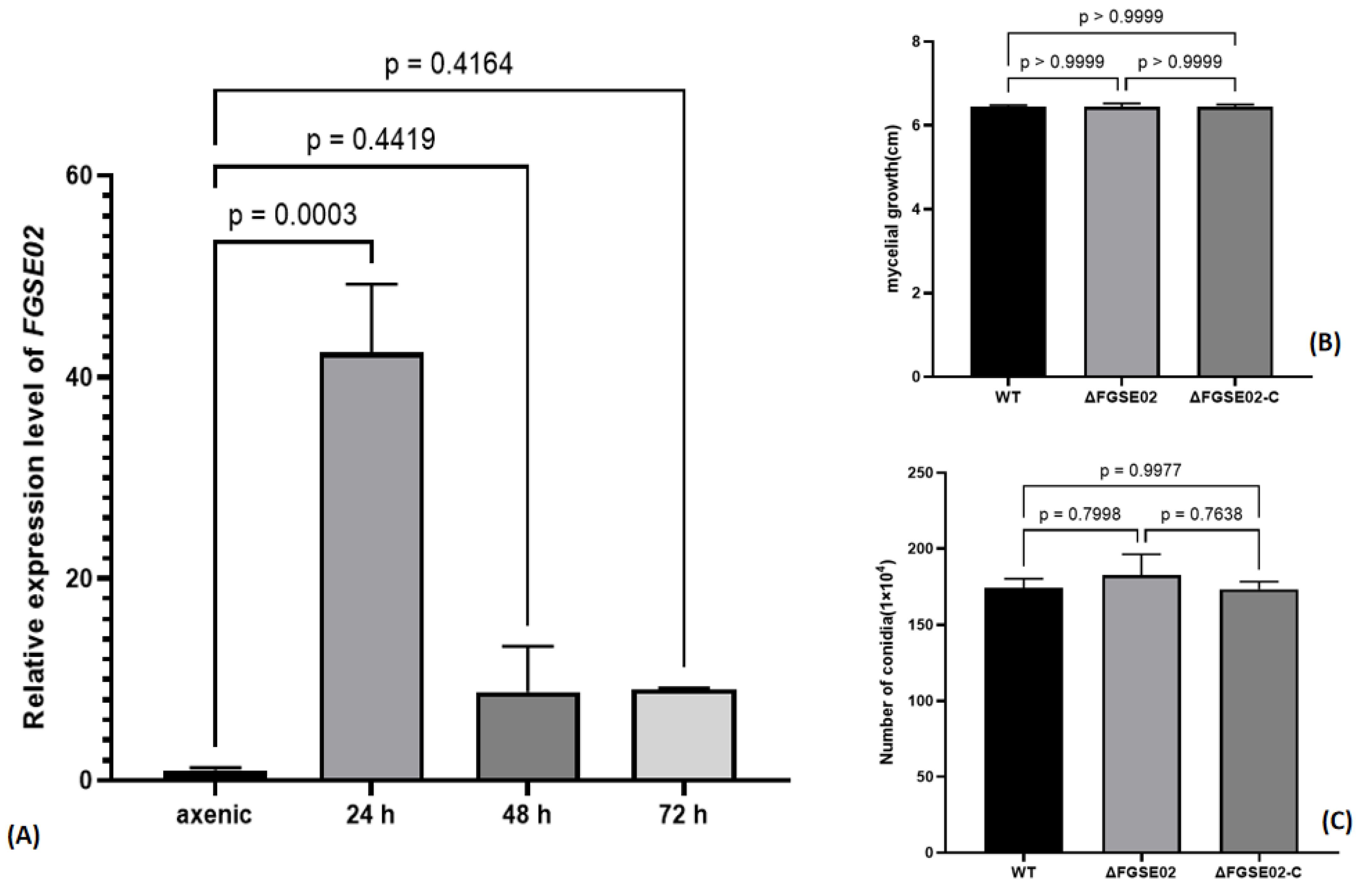
3.4. Identification and Characterization of FGSE02
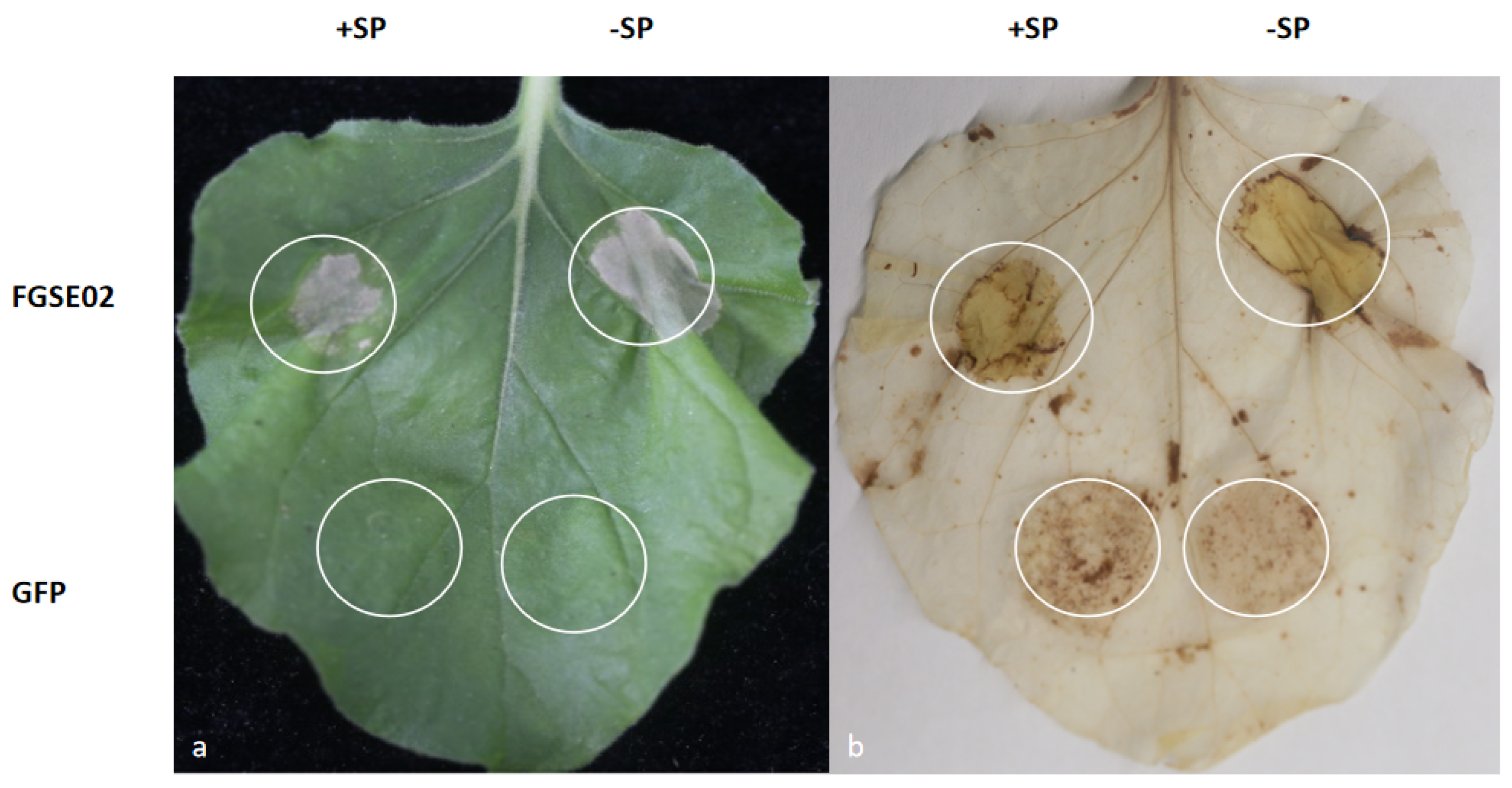
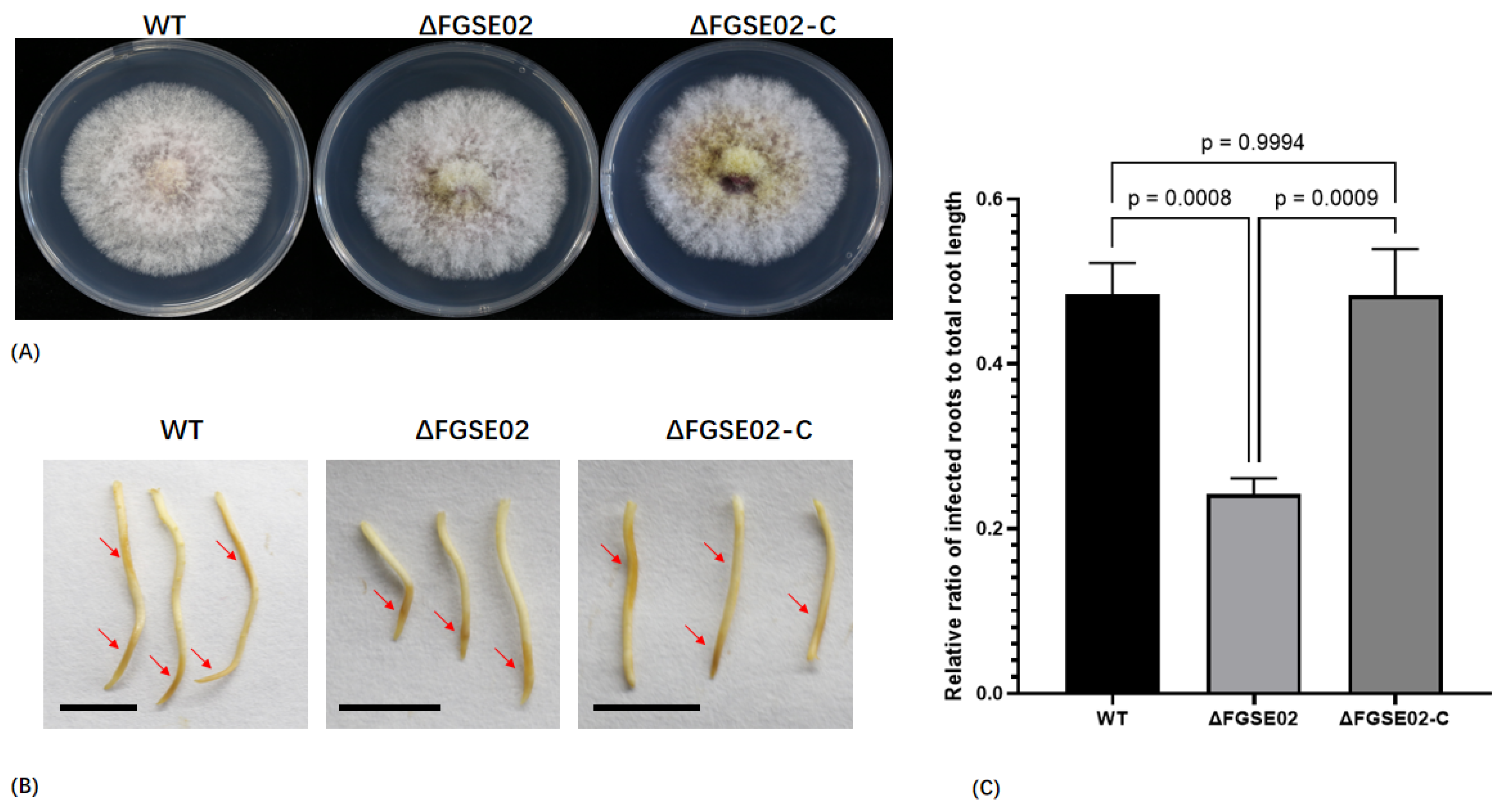
3.5. FGSE02 Induces Plant Cell Death and Modulates Pathogenicity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feng, G.; Gu, Y.; Wang, C.; Zhou, Y.; Huang, S.; Luo, B. Wheat Fusarium head blight automatic non-destructive detection based on multi-scale imaging: A technical perspective. Plants 2024, 13, 1722. [Google Scholar] [CrossRef] [PubMed]
- Gargouri, S.; Masiello, M.; Somma, S.; Haidukowski, M.; Khaterchi, R.; Chekali, S.; Derouich, S.; Balmas, V.; Moretti, A. maize-fusarium interactions: Tunisian insights into mycotoxin ecology. Fungal Biol. 2024, 128, 2460–2470. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Bento, E.A.; de Moura, A.P.; Alves, T.; Da, S.I.; Fernandes, J.; Da, S.F.S.; Souza, V.; Da, S.W.; Ambrosio, M. First report of Fusarium falciforme and Fusarium pernambucanum causing root and stem rot on papaya plants in Brazil. Plant Dis. 2025. [Google Scholar] [CrossRef]
- Tang, Z.; Zhu, J.; Song, Q.; Daly, P.; Kong, L.; He, L.; Li, A.; Lou, J.; Wang, Z.; Zhang, L.; et al. Identification and pathogenicity of Fusarium spp. associated with tea wilt in Zhejiang Province, China. BMC Microbiol. 2024, 24, 38. [Google Scholar] [CrossRef]
- Vega, G.T.; Tirado, R.M.; Molina, C.L.; Lopez, U.G.; Lopez, O.C. Fusarium verticillioides causing root and stem rot in Papaya (Carica papaya) in Mexico. Plant Dis. 2023, 107, 2517. [Google Scholar] [CrossRef]
- Alisaac, E.; Mahlein, A.K. Fusarium head blight on wheat: Biology, modern detection and diagnosis and Integrated disease management. Toxins 2023, 15, 192. [Google Scholar] [CrossRef]
- Haile, J.K.; N’Diaye, A.; Walkowiak, S.; Nilsen, K.T.; Clarke, J.M.; Kutcher, H.R.; Steiner, B.; Buerstmayr, H.; Pozniak, C.J. Fusarium head blight in durum wheat: Recent status, breeding directions, and future research prospects. Phytopathology 2019, 109, 1664–1675. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.K.; Pandey, A.; Athar, T.; Choudhary, S.; Deval, R.; Gezgin, S.; Hamurcu, M.; Topal, A.; Atmaca, E.; Santos, P.A.; et al. Fusarium head blight in wheat: Contemporary status and molecular approaches. 3 Biotech. 2020, 10, 172. [Google Scholar] [CrossRef]
- Karlsson, I.; Persson, P.; Friberg, H. Fusarium head blight from a microbiome perspective. Front. Microbiol. 2021, 12, 628373. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, J.; Mu, P.; Lin, R.; Wen, J.; Deng, Y. Toxicokinetics and metabolism of deoxynivalenol in animals and humans. Arch. Toxicol. 2022, 96, 2639–2654. [Google Scholar] [CrossRef]
- Vatzia, E.; Pierron, A.; Saalmuller, A.; Mayer, E.; Gerner, W. Deoxynivalenol affects proliferation and expression of activation-related molecules in major porcine T-Cell subsets. Toxins 2019, 11, 644. [Google Scholar] [CrossRef]
- Alkadri, D.; Nipoti, P.; Doll, K.; Karlovsky, P.; Prodi, A.; Pisi, A. Study of fungal colonization of wheat kernels in syria with a focus on fusarium species. Int. J. Mol. Sci. 2013, 14, 5938–5951. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminearum trichothecene mycotoxins: Biosynthesis, regulation, and management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef]
- Miltenburg, M.G.; Bonner, C.; Hepworth, S.; Huang, M.; Rampitsch, C.; Subramaniam, R. Proximity-dependent biotinylation identifies a suite of candidate effector proteins from Fusarium graminearum. Plant J. 2022, 112, 369–382. [Google Scholar] [CrossRef] [PubMed]
- Rauwane, M.E.; Ogugua, U.V.; Kalu, C.M.; Ledwaba, L.K.; Woldesemayat, A.A.; Ntushelo, K. Pathogenicity and Virulence Factors of Fusarium graminearum including factors discovered using next generation sequencing technologies and proteomics. Microorganisms 2020, 8, 305. [Google Scholar] [CrossRef]
- Voigt, C.A.; Schafer, W.; Salomon, S. A secreted lipase of Fusarium graminearum is a virulence factor required for infection of cereals. Plant J. 2005, 42, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Hao, G.; McCormick, S.; Vaughan, M.M.; Naumann, T.A.; Kim, H.S.; Proctor, R.; Kelly, A.; Ward, T.J. Fusarium graminearum arabinanase (Arb93B) enhances wheat head blight susceptibility by suppressing plant immunity. Mol. Plant Microbe Interact. 2019, 32, 888–898. [Google Scholar] [CrossRef]
- Hao, G.; Naumann, T.A.; Chen, H.; Bai, G.; McCormick, S.; Kim, H.S.; Tian, B.; Trick, H.N.; Naldrett, M.J.; Proctor, R. Fusarium graminearum effector FgNls1 targets plant nuclei to induce wheat head blight. Mol. Plant Microbe Interact. 2023, 36, 478–488. [Google Scholar] [CrossRef]
- Jin, M.; Hu, S.; Wu, Q.; Feng, X.; Zhang, Y.; Jiang, Q.; Ma, J.; Qi, P.; Chen, G.; Jiang, Y.; et al. An effector protein of Fusarium graminearum targets chloroplasts and suppresses cyclic photosynthetic electron flow. Plant Physiol. 2024, 196, 2422–2436. [Google Scholar] [CrossRef]
- Darino, M.; Jaiswal, N.; Darma, R.; Kroll, E.; Urban, M.; Xiang, Y.; Srivastava, M.; Kim, H.S.; Myers, A.; Scofield, S.R.; et al. The Fusarium graminearum effector protease FgTPP1 suppresses immune responses and facilitates Fusarium head blight disease. Mol. Plant Microbe Interact. 2025, 38, I8240103F. [Google Scholar] [CrossRef]
- Shang, S.; He, Y.; Hu, Q.; Fang, Y.; Cheng, S.; Zhang, C.J. Fusarium graminearum effector FgEC1 targets wheat TaGF14b protein to suppress TaRBOHD-mediated ROS production and promote infection. J. Integr. Plant Biol. 2024, 66, 2288–2303. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Hei, R.; Yang, Y.; Zhang, S.; Wang, Q.; Wang, W.; Zhang, Q.; Yan, M.; Zhu, G.; Huang, P.; et al. An orphan protein of Fusarium graminearum modulates host immunity by mediating proteasomal degradation of TaSnRK1alpha. Nat. Commun. 2020, 11, 4382. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Wang, Y.; Tian, M.; Dai, K.; Zheng, W.; Liu, Z.; Yang, S.; Liu, X.; Shi, D.; Zhang, H.; et al. Fg12 ribonuclease secretion contributes to Fusarium graminearum virulence and induces plant cell death. J. Integr. Plant Biol. 2021, 63, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yang, S.; Dai, K.; Zheng, W.; Zhang, X.; Yang, B.; Ye, W.; Zheng, X.; Wang, Y. The effector Fg62 contributes to Fusarium graminearum virulence and induces plant cell death. Phytopathol. Res. 2023, 5, 12. [Google Scholar] [CrossRef]
- Xiong, J.; Luo, M.; Chen, Y.; Hu, Q.; Fang, Y.; Sun, T.; Hu, G.; Zhang, C.J. Subtilisin-like proteases from Fusarium graminearum induce plant cell death and contribute to virulence. Plant Physiol. 2024, 195, 1681–1693. [Google Scholar] [CrossRef]
- Fatima, M.; Anjum, B.H.; Rebekah, N.; Murugasamy, S.; Makandar, R. Genome-wide search and gene expression studies reveal candidate effectors with a role in pathogenicity and virulence in Fusarium graminearum. Mycologia 2024, 116, 708–728. [Google Scholar] [CrossRef]
- Hao, Z.; Li, Y.; Jiang, Y.; Xu, J.; Li, J.; Luo, L. Genome sequence analysis of the fungal pathogen Fusarium graminearum using oxford nanopore technology. J. Fungi 2021, 7, 699. [Google Scholar] [CrossRef]
- Chen, L.; Wu, Q.; He, T.; Lan, J.; Ding, L.; Liu, T.; Wu, Q.; Pan, Y.; Chen, T. Transcriptomic and metabolomic changes triggered by Fusarium solani in Common Bean (Phaseolus vulgaris L.). Genes 2020, 11, 177. [Google Scholar] [CrossRef]
- Satterlee, T.R.; Williams, F.N.; Nadal, M.; Glenn, A.E.; Lofton, L.W.; Duke, M.V.; Scheffler, B.E.; Gold, S.E. Transcriptomic response of Fusarium verticillioides to variably inhibitory environmental isolates of streptomyces. Front. Fungal Biol. 2022, 3, 894590. [Google Scholar] [CrossRef]
- Yang, T.; Liu, J.; Li, X.; Amanullah, S.; Lu, X.; Zhang, M.; Zhang, Y.; Luan, F.; Liu, H.; Wang, X. Transcriptomic Analysis of Fusarium oxysporum Stress-Induced Pathosystem and Screening of Fom-2 Interaction Factors in Contrasted Melon Plants. Front. Plant Sci. 2022, 13, 961586. [Google Scholar] [CrossRef]
- Guo, L.; Han, L.; Yang, L.; Zeng, H.; Fan, D.; Zhu, Y.; Feng, Y.; Wang, G.; Peng, C.; Jiang, X.; et al. Genome and transcriptome analysis of the fungal pathogen Fusarium oxysporum f. sp. cubense causing banana vascular wilt disease. PLoS ONE 2014, 9, e95543. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Z.; Sun, C.H.; Liu, Y.; Feng, H.Q.; Chang, H.W.; Cao, S.N.; Li, G.H.; Yang, S.; Hou, J.; Zhu-Salzman, K.; et al. Transcriptome analysis and functional validation reveal a novel gene, BcCGF1, that enhances fungal virulence by promoting infection-related development and host penetration. Mol. Plant Pathol. 2020, 21, 834–853. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, gix120. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Gueguen, L.; Coppee, J.Y.; Dillies, M.A. SARTools: A DESeq2- and edgeR-based R pipeline for comprehensive differential analysis of RNA-Seq Data. PLoS ONE 2016, 11, e157022. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Lu, Y.Y.; Noble, W.S.; Keich, U. A BLAST from the past: Revisiting blastp’s E-value. Bioinformatics 2024, 40, btae729. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Teufel, F.; Almagro, A.J.; Johansen, A.R.; Gislason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, P.; Luo, J.; Jiang, Y. Secreted protein prediction system combining CJ-SPHMM, TMHMM, and PSORT. Mamm. Genome 2003, 14, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Pierleoni, A.; Martelli, P.L.; Casadio, R. PredGPI: A GPI-anchor predictor. BMC Bioinform. 2008, 9, 392. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Xu, C.; Li, L.; Fan, L.; Li, R.; He, J.; Li, H.; Deng, W.; Kang, Z.; Li, Z.; et al. A conserved fungal effector disturbs Ca(2+) sensing and ROS homeostasis to induce plant cell death. Nat. Commun. 2025, 16, 3523. [Google Scholar] [CrossRef]
- Ribeiro, S.; Tran, D.M.; Deon, M.; Clement-Demange, A.; Garcia, D.; Soumahoro, M.; Masson, A.; Pujade-Renaud, V. Gene deletion of Corynespora cassiicola cassiicolin Cas1 suppresses virulence in the rubber tree. Fungal Genet. Biol. 2019, 129, 101–114. [Google Scholar] [CrossRef]
- Audenaert, K.; Vanheule, A.; Hofte, M.; Haesaert, G. Deoxynivalenol: A major player in the multifaceted response of Fusarium to its environment. Toxins 2013, 6, 1–19. [Google Scholar] [CrossRef]

| Sample | Clean Reads | Clean Bases | Read Length | Q20 (%) | GC (%) | Mapping to Fungi |
| FG_fungal_1 | 82,073,226 | 12,310,983,900 | 150 | 96.91 | 51.85 | 97.91% |
| FG_fungal_2 | 81,677,390 | 12,251,608,500 | 150 | 97.15 | 51.90 | 98.05% |
| FG_fungal_3 | 81,157,202 | 12,173,580,300 | 150 | 97.13 | 51.84 | 97.56% |
| FG_24h_1 | 80,689,404 | 12,103,410,600 | 150 | 96.58 | 53.26 | 5.77% |
| FG_24h_2 | 81,429,004 | 12,214,350,600 | 150 | 97.12 | 53.66 | 5.82% |
| FG_24h_3 | 82,074,668 | 12,311,200,200 | 150 | 97.02 | 53.71 | 8.06% |
| FG_48h_1 | 80,281,800 | 12,042,270,000 | 150 | 96.49 | 53.62 | 22.06% |
| FG_48h_2 | 81,366,832 | 12,205,024,800 | 150 | 96.48 | 54.17 | 36.49% |
| FG_48h_3 | 79,535,168 | 11,930,275,200 | 150 | 96.41 | 53.44 | 17.25% |
| FG_72h_1 | 81,867,320 | 12,280,098,000 | 150 | 96.38 | 53.45 | 42.29% |
| FG_72h_2 | 77,635,306 | 11,645,295,900 | 150 | 96.42 | 53.80 | 37.64% |
| FG_72h_3 | 81,744,194 | 12,261,629,100 | 150 | 96.51 | 54.03 | 32.88% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, Z.; Pan, L.; Xu, J.; Yu, C.; Li, J.; Luo, L. FGSE02, a Novel Secreted Protein in Fusarium graminearum FG-12, Leads to Cell Death in Plant Tissues and Modulates Fungal Virulence. J. Fungi 2025, 11, 397. https://doi.org/10.3390/jof11050397
Hao Z, Pan L, Xu J, Yu C, Li J, Luo L. FGSE02, a Novel Secreted Protein in Fusarium graminearum FG-12, Leads to Cell Death in Plant Tissues and Modulates Fungal Virulence. Journal of Fungi. 2025; 11(5):397. https://doi.org/10.3390/jof11050397
Chicago/Turabian StyleHao, Zhigang, Lei Pan, Jiaqing Xu, Chengxuan Yu, Jianqiang Li, and Laixin Luo. 2025. "FGSE02, a Novel Secreted Protein in Fusarium graminearum FG-12, Leads to Cell Death in Plant Tissues and Modulates Fungal Virulence" Journal of Fungi 11, no. 5: 397. https://doi.org/10.3390/jof11050397
APA StyleHao, Z., Pan, L., Xu, J., Yu, C., Li, J., & Luo, L. (2025). FGSE02, a Novel Secreted Protein in Fusarium graminearum FG-12, Leads to Cell Death in Plant Tissues and Modulates Fungal Virulence. Journal of Fungi, 11(5), 397. https://doi.org/10.3390/jof11050397






