Autophagy-Related Proteins (ATGs) Are Differentially Required for Development and Virulence of Sclerotinia sclerotiorum
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strains of S. sclerotiorum and Culture Conditions
2.2. UV Mutagenesis
2.3. S. sclerotiorum Genomic DNA Extraction and NGS Analysis
2.4. Target Gene Knockout
2.5. Growth Rate Assessment and Colony Morphology Analysis of S. sclerotiorum
2.6. Plant Infection Assay
2.7. Observation of Compound Appressoria Formation
2.8. S. sclerotiorum Hyphal Allelism Test
2.9. Generation of Ssatg5-1::GFP-ATG8 Fusion Line
2.10. Fluorescence Microscopy
2.11. Fluorescence Intensity Quantification
2.12. Autophagosome Number Quantification
2.13. Statistical Analysis
3. Results
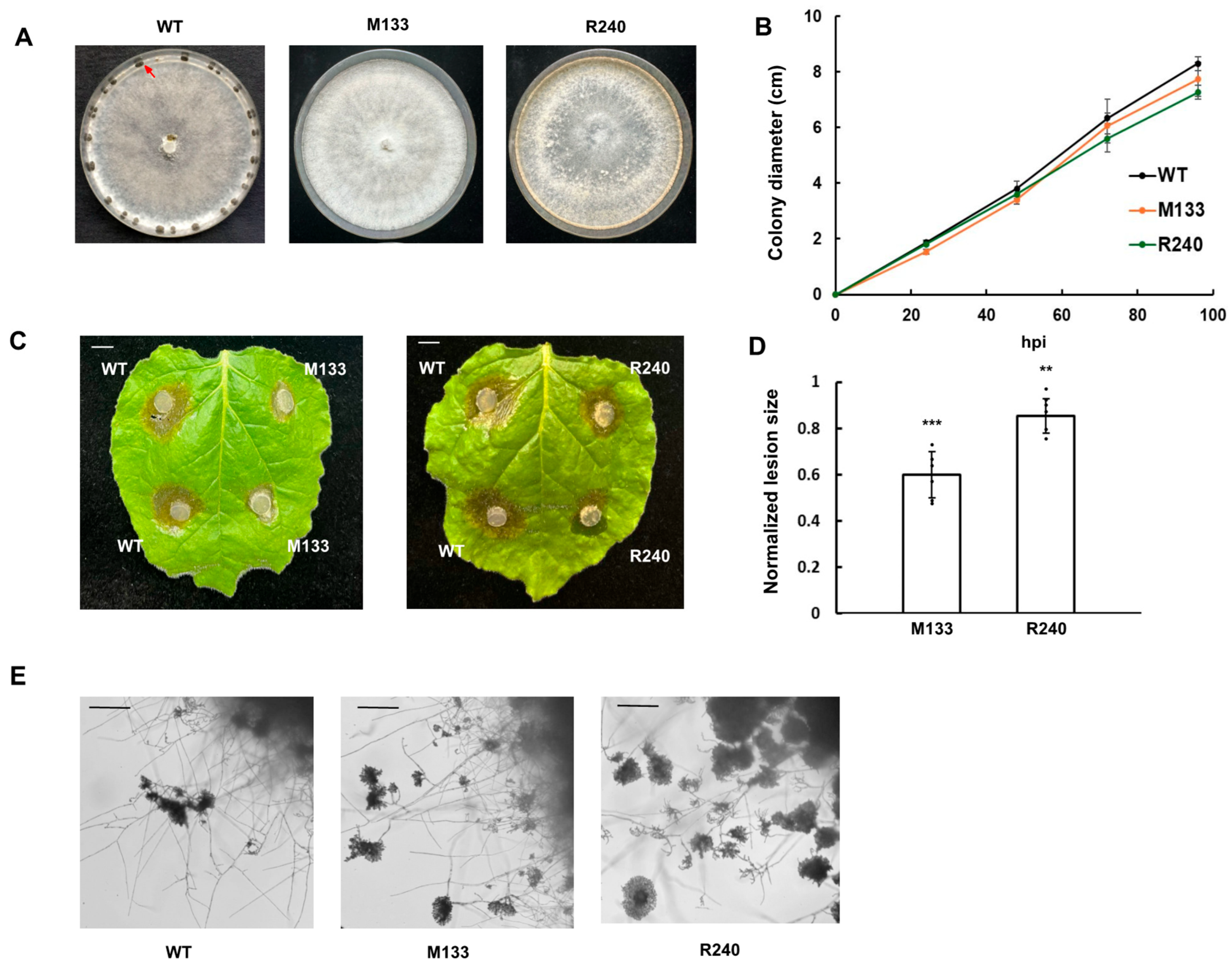
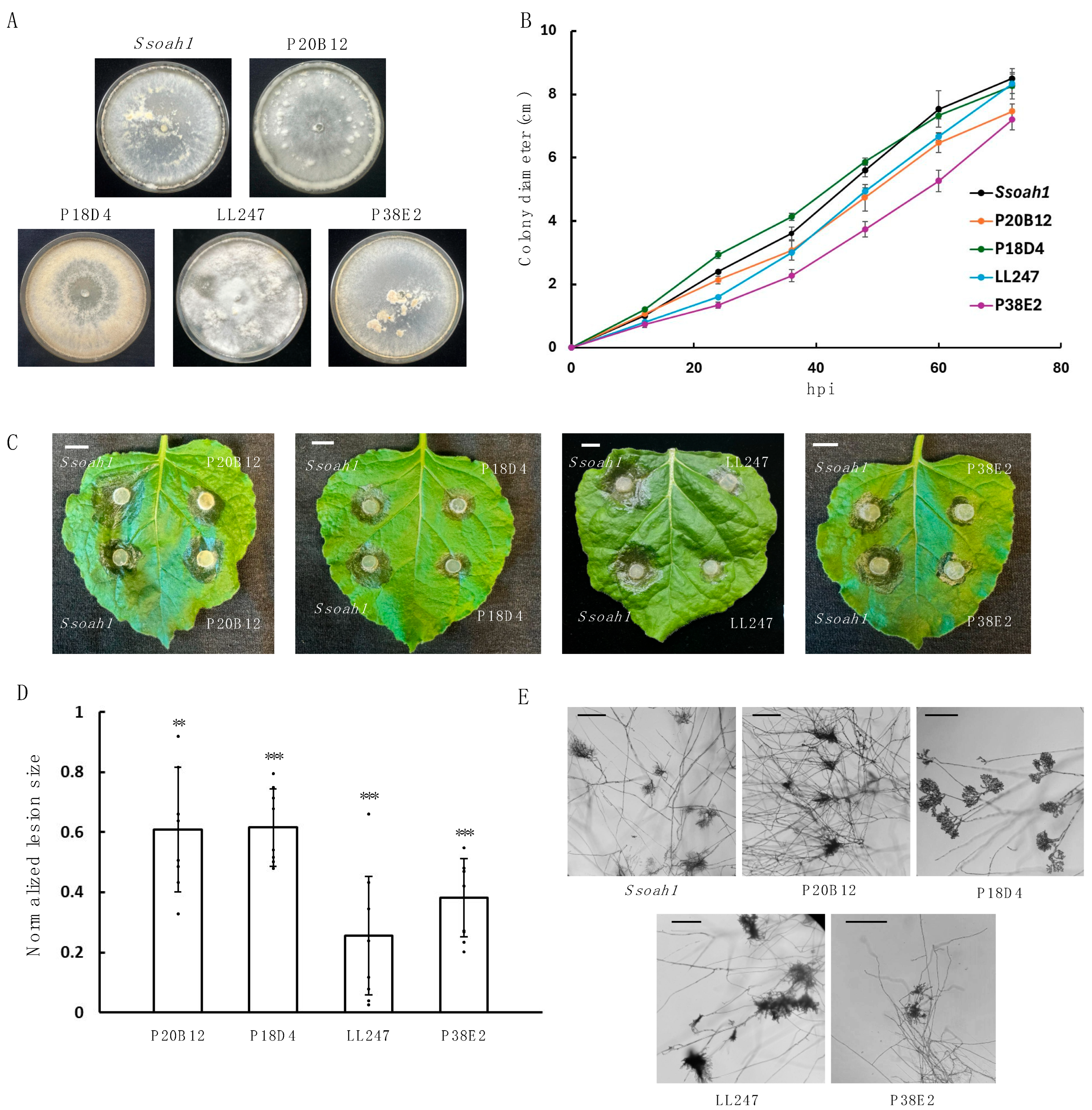
3.1. S. sclerotiorum Mutants That Cannot Form Sclerotia Were Obtained from UV Mutagenesis
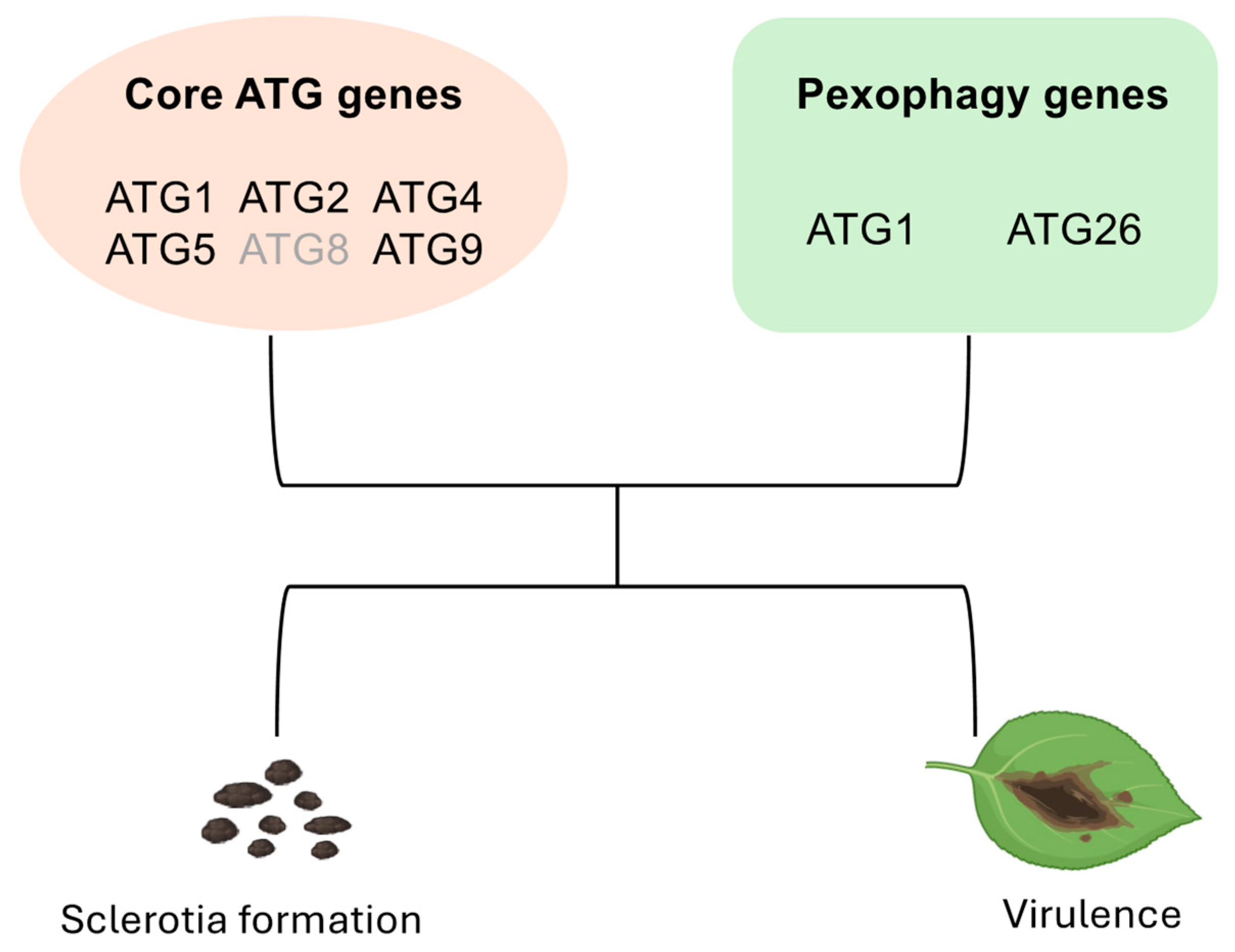
3.2. Distinct Autophagy-Related Genes Were Identified from Six UV Mutants
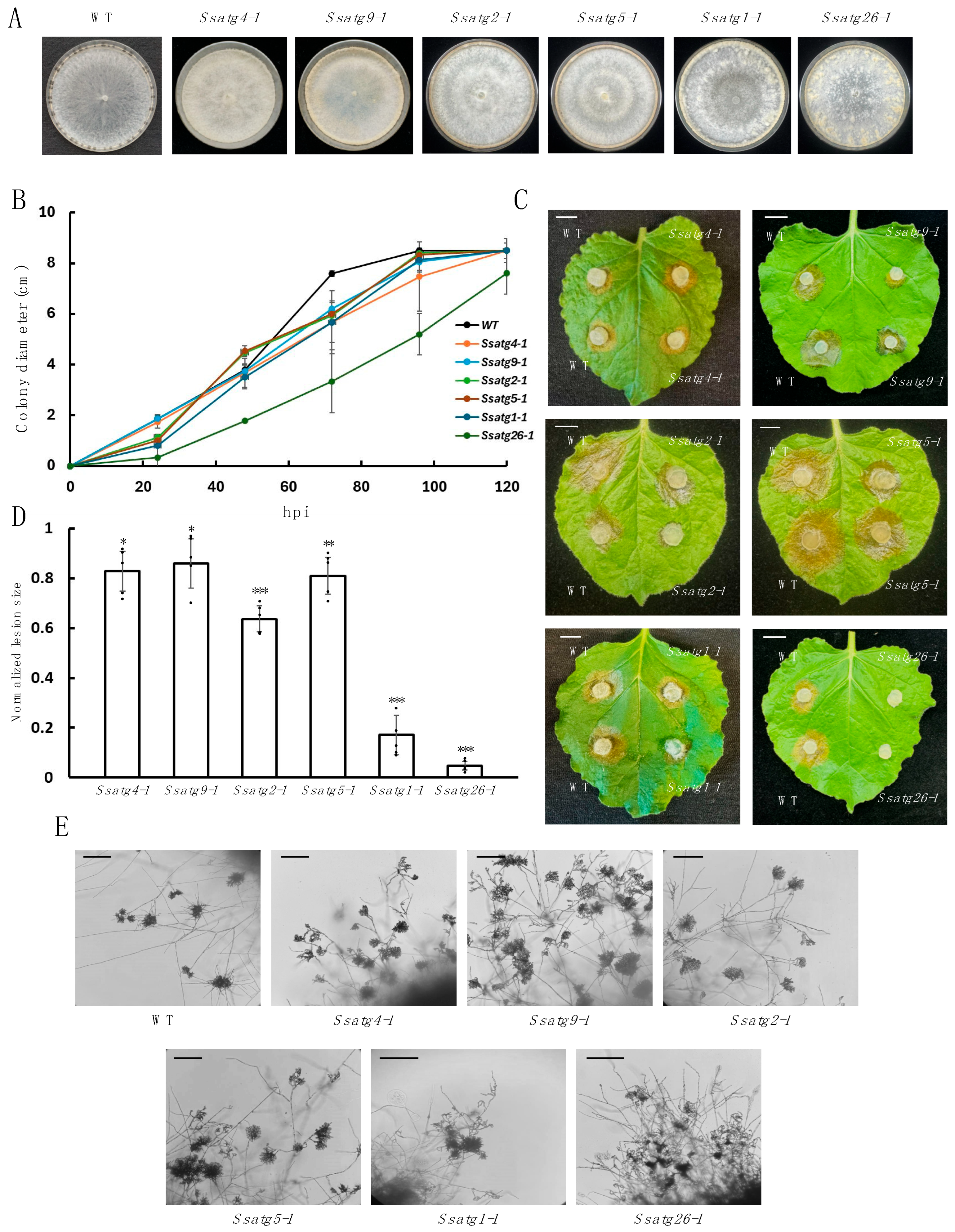
3.3. Knockouts of ATGs Exhibit Attenuated Sclerotia Formation and Virulence
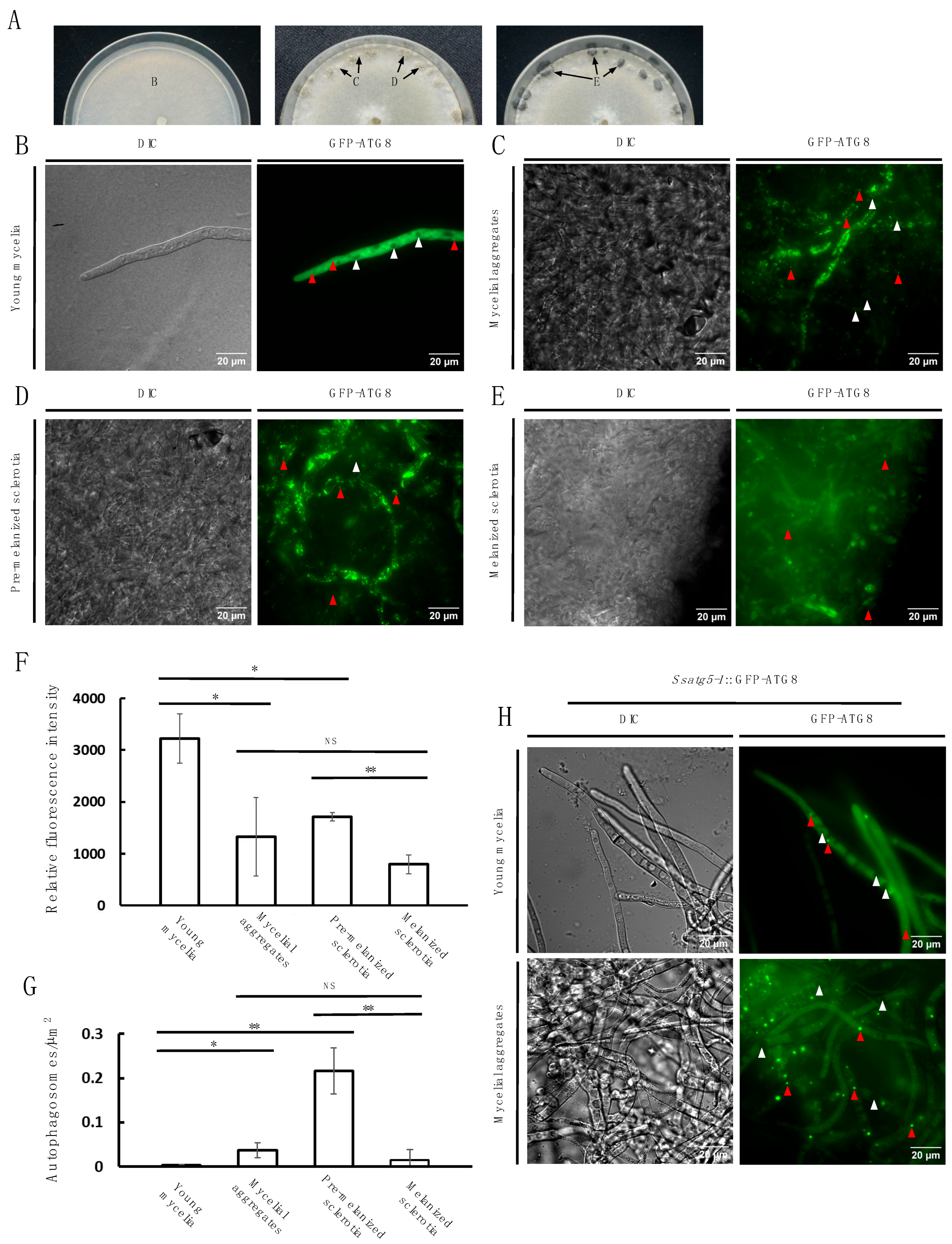
3.4. Autophagy Activity Is Dynamically Observed During Sclerotia Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xia, S.; Xu, Y.; Hoy, R.; Zhang, J.; Qin, L.; Li, X. The notorious soilborne pathogenic fungus Sclerotinia sclerotiorum: An update on genes studied with mutant analysis. Pathogens 2019, 9, 27. [Google Scholar] [CrossRef] [PubMed]
- Melzer, M.; Smith, E.; Boland, G. Index of plant hosts of Sclerotinia minor. Can. J. Plant Pathol. 1997, 19, 272–280. [Google Scholar] [CrossRef]
- Liang, X.; Rollins, J.A. Mechanisms of broad host range necrotrophic pathogenesis in Sclerotinia sclerotiorum. Phytopathology 2018, 108, 1128–1140. [Google Scholar] [CrossRef]
- Xu, Y.; Ao, K.; Tian, L.; Qiu, Y.; Huang, X.; Liu, X.; Hoy, R.; Zhang, Y.; Rashid, K.Y.; Xia, S.; et al. A forward genetic screen in Sclerotinia sclerotiorum revealed the transcriptional regulation of its sclerotial melanization pathway. Mol. Plant-Microbe Interact. 2022, 35, 244–256. [Google Scholar] [CrossRef]
- Tariq, V.; Jeffries, P. Appressorium formation by Sclerotinia sclerotiorum: Scanning electron microscopy. Trans. Br. Mycol. Soc. 1984, 82, 645–651. [Google Scholar] [CrossRef]
- Yamamoto, H.; Zhang, S.; Mizushima, N. Autophagy genes in biology and disease. Nat. Rev. Genet. 2023, 24, 382–400. [Google Scholar] [CrossRef]
- Hollenstein, D.M.; Kraft, C. Autophagosomes are formed at a distinct cellular structure. Curr. Opin. Cell Biol. 2020, 65, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Marzella, L.; Ahlberg, J.; Glaumann, H. Autophagy, heterophagy, microautophagy and crinophagy as the means for intracellular degradation. Virchows Archiv. B Cell Pathol. Incl. Mol. Pathol. 1981, 36, 219–234. [Google Scholar]
- Ohsumi, Y. Historical landmarks of autophagy research. Cell Res. 2014, 24, 9–23. [Google Scholar] [CrossRef]
- Lynch-Day, M.A.; Klionsky, D.J. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010, 584, 1359–1366. [Google Scholar] [CrossRef]
- Oku, M.; Sakai, Y. Peroxisomes as dynamic organelles: Autophagic degradation. FEBS J. 2010, 277, 3289–3294. [Google Scholar] [CrossRef] [PubMed]
- Shintani, T.; Huang, W.P.; Stromhaug, P.E.; Klionsky, D.J. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell 2002, 3, 825–837. [Google Scholar] [CrossRef] [PubMed]
- van Zutphen, T.; Veenhuis, M.; van der Klei, I.J. Pex14 is the sole component of the peroxisomal translocon that is required for pexophagy. Autophagy 2008, 4, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Noda, N.N.; Inagaki, F. Mechanisms of autophagy. Annu. Rev. Biophys. 2015, 44, 101–122. [Google Scholar] [CrossRef]
- Stjepanovic, G.; Davies, C.W.; Stanley, R.E.; Ragusa, M.J.; Kim, D.J.; Hurley, J.H. Assembly and dynamics of the autophagy-initiating Atg1 complex. Proc. Natl. Acad. Sci. USA 2014, 111, 12793–12798. [Google Scholar] [CrossRef]
- Matsuura, A.; Tsukada, M.; Wada, Y.; Ohsumi, Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 1997, 192, 245–250. [Google Scholar] [CrossRef]
- Kabeya, Y.; Noda, N.N.; Fujioka, Y.; Suzuki, K.; Inagaki, F.; Ohsumi, Y. Characterization of the ATG17–ATG29–ATG31 com-plex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2009, 389, 612–615. [Google Scholar] [CrossRef]
- Suzuki, S.W.; Yamamoto, H.; Oikawa, Y.; Kondo-Kakuta, C.; Kimura, Y.; Hirano, H.; Ohsumi, Y. Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc. Natl. Acad. Sci. USA 2015, 112, 3350–3355. [Google Scholar] [CrossRef]
- Sekito, T.; Kawamata, T.; Ichikawa, R.; Suzuki, K.; Ohsumi, Y. Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells 2009, 14, 525–538. [Google Scholar] [CrossRef]
- Mailler, E.; Guardia, C.M.; Bai, X.; Jarnik, M.; Williamson, C.D.; Li, Y.; Maio, N.; Golden, A.; Bonifacino, J.S. The autophagy protein ATG9A enables lipid mobilization from lipid droplets. Nat. Commun. 2021, 12, 6750. [Google Scholar] [CrossRef]
- Baskaran, S.; Ragusa, M.J.; Boura, E.; Hurley, J.H. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol. Cell 2012, 47, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Osawa, T.; Noda, N.N. ATG2: A novel phospholipid transfer protein that mediates de novo autophagosome biogenesis. Protein Sci. 2019, 28, 1005–1012. [Google Scholar] [CrossRef]
- Rieter, E.; Vinke, F.; Bakula, D.; Cebollero, E.; Ungermann, C.; Proikas-Cezanne, T.; Reggiori, F. Atg18 function in autophagy is regulated by specific sites within its β-propeller. J. Cell Sci. 2013, 126, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Kotani, T.; Kirisako, H.; Koizumi, M.; Ohsumi, Y.; Nakatogawa, H. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. USA 2018, 115, 10363–10368. [Google Scholar] [CrossRef] [PubMed]
- Le Bars, R.; Marion, J.; Le Borgne, R.; Satiat-Jeunemaitre, B.; Bianchi, M.W. ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat. Commun. 2014, 5, 4121. [Google Scholar] [CrossRef]
- Hanada, T.; Noda, N.N.; Satomi, Y.; Ichimura, Y.; Fujioka, Y.; Takao, T.; Inagaki, F.; Ohsumi, Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007, 282, 37298–37302. [Google Scholar] [CrossRef]
- Maruyama, T.; Noda, N.N. Autophagy-regulating protease Atg4: Structure, function, regulation and inhibition. J. Antibiot. 2018, 71, 72–78. [Google Scholar] [CrossRef]
- Martens, S.; Fracchiolla, D. Activation and targeting of ATG8 protein lipidation. Cell Discov. 2020, 6, 23. [Google Scholar] [CrossRef]
- Xie, Z.; Nair, U.; Klionsky, D.J. ATG8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 2008, 19, 3290–3298. [Google Scholar] [CrossRef]
- Yin, Z.; Chen, C.; Yang, J.; Feng, W.; Liu, X.; Zuo, R.; Wang, J.; Yang, L.; Zhong, K.; Gao, C.; et al. Histone acetyltransferase MoHat1 acetylates autophagy-related proteins MoATG3 and MoATG9 to orchestrate functional appressorium formation and pathogenicity in Magnaporthe oryzae. Autophagy 2019, 15, 1234–1257. [Google Scholar] [CrossRef]
- Liu, X.H.; Zhao, Y.H.; Zhu, X.M.; Zeng, X.Q.; Huang, L.Y.; Dong, B.; Su, Z.Z.; Wang, Y.; Lu, J.P.; Lin, F.C. Autophagy-related protein MoATG14 is involved in differentiation, development and pathogenicity in the rice blast fungus Magnaporthe oryzae. Sci. Rep. 2017, 7, 40018. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, L.; Xiang, Y.; Du, L.; Huang, X.; Liu, Y. Comparative transcriptome analysis of Sclerotinia sclerotiorum revealed its response mechanisms to the biological control agent, Bacillus amyloliquefaciens. Sci. Rep. 2020, 10, 12576. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Lai, W.; Huang, K.; Li, Y.; Wang, Z.; Chen, X.; Wang, A. SsATG8 and SsNBR1 mediated-autophagy is required for fungal development, proteasomal stress response and virulence in Sclerotinia sclerotiorum. Fungal Genet. Biol. 2021, 157, 103632. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Liberti, D.; Li, M.; Kim, Y.T.; Hutchens, A.; Wilson, R.; Rollins, J.A. Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants. Mol. Plant Pathol. 2015, 16, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xu, Y.; Li, L.; Wu, M.; Fang, Z.; Tan, J.; Rollins, J.A.; Lin, H.; Huang, X.; Mansfield, S.D.; et al. A GDP-mannose-1-phosphate guanylyltransferase as a potential HIGS target against Sclerotinia sclerotiorum. PLoS Pathog. 2025, 21, e1013129. [Google Scholar] [CrossRef]
- Athukorala, S.N.; Fernando, W.D.; Rashid, K.Y.; de Kievit, T. The role of volatile and non-volatile antibiotics produced by Pseudomonas chlororaphis strain PA23 in its root colonization and control of Sclerotinia sclerotiorum. Biocontrol. Sci. Technol. 2010, 20, 875–890. [Google Scholar] [CrossRef]
- Cubero, O.; Crespo, A.; Fatehi, J.; Bridge, P. DNA extraction and PCR amplification method suitable for fresh, herbarium-stored, lichenized, and other fungi. Plant Syst. Evol. 1999, 216, 243–249. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Li, X.; Tian, L.; Xu, Y.; Tan, J.; Li, J.; O’Neil, N.; Hirst, M.; Hieter, P.; Zhang, Y. Distribution of haploid chromosomes into separate nuclei in two pathogenic fungi. Science 2025, 380, 123–127. [Google Scholar]
- Liang, X.; Moomaw, E.W.; Rollins, J.A. Fungal oxalate decarboxylase activity contributes to Sclerotinia sclerotiorum early infection by affecting both compound appressoria development and function. Mol. Plant Pathol. 2015, 16, 825–836. [Google Scholar] [CrossRef] [PubMed]
- Jiao, W.; Yu, H.; Chen, X.; Xiao, K.; Jia, D.; Wang, F.; Zhang, Y.; Pan, H. The SsATG1 activating autophagy is required for sclerotia formation and pathogenicity in Sclerotinia sclerotiorum. J. Fungi 2022, 8, 1314. [Google Scholar] [CrossRef]
- Xu, Y.; Qiu, Y.; Zhang, Y.; Li, X. A cAMP phosphodiesterase is essential for sclerotia formation and virulence in Sclerotinia sclerotiorum. Front. Plant Sci. 2023, 14, 1175552. [Google Scholar] [CrossRef]
- Ford, E.; Miller, R.; Gray, H.; Sherwood, J. Heterokaryon formation and vegetative compatibility in Sclerotinia sclerotiorum. Mycol. Res. 1995, 99, 241–247. [Google Scholar] [CrossRef]
- Li, Y.; Shu, P.; Xiang, L.; Sheng, J.; Shen, L. CRISPR/Cas9-Mediated SlATG5 mutagenesis reduces the resistance of tomato fruit to Botrytis cinerea. Foods 2023, 12, 2750. [Google Scholar] [CrossRef]
- Onodera, J.; Ohsumi, Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J. Biol. Chem. 2005, 280, 31582–31586. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.Z.; Ramos-Pamplona, M.; Naqvi, N.I. Autophagy-assisted glycogen catabolism regulates asexual differentiation in Magnaporthe oryzae. Autophagy 2009, 5, 33–43. [Google Scholar] [CrossRef]
- Richie, D.L.; Askew, D.S. Autophagy in the filamentous fungus Aspergillus fumigatus. Methods Enzymol. 2008, 451, 241–250. [Google Scholar]
- Liu, N.; Zhu, M.; Zhang, Y.; Wang, Z.; Li, B.; Ren, W. Involvement of the autophagy protein Atg1 in development and virulence in Botryosphaeria dothidea. J. Fungi 2022, 8, 904. [Google Scholar] [CrossRef]
- Corral-Ramos, C.; Barrios, R.; Ayté, J.; Hidalgo, E. TOR and MAP kinase pathways synergistically regulate autophagy in response to nutrient depletion in fission yeast. Autophagy 2022, 18, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, T.; Utsumi, R.; Shimada, E.; Bamba, A.; Hagihara, K.; Satoh, R.; Sugiura, R. Atg1, a key regulator of autophagy, functions to promote MAPK activation and cell death upon calcium overload in fission yeast. Microb. Cell 2023, 10, 133. [Google Scholar] [CrossRef] [PubMed]
- Madrid, M.; Vázquez-Marín, B.; Franco, A.; Soto, T.; Vicente-Soler, J.; Gacto, M.; Cansado, J. Multiple crosstalk between TOR and the cell integrity MAPK signaling pathway in fission yeast. Sci. Rep. 2016, 6, 37515. [Google Scholar] [CrossRef]
- Cansado, J.; Soto, T.; Franco, A.; Vicente-Soler, J.; Madrid, M. The fission yeast cell integrity pathway: A functional hub for cell survival upon stress and beyond. J. Fungi 2021, 8, 32. [Google Scholar] [CrossRef]
- Tian, L.; Li, J.; Xu, Y.; Qiu, Y.; Zhang, Y.; Li, X. A MAP kinase cascade broadly regulates the lifestyle of Sclerotinia sclerotiorum and can be targeted by HIGS for disease control. Plant J. 2024, 118, 324–344. [Google Scholar] [CrossRef]
- Liu, X.H.; Lu, J.P.; Zhang, L.; Dong, B.; Min, H.; Lin, F.C. Involvement of a Magnaporthe grisea Serine/Threonine kinase gene, Mg ATG1, in appressorium turgor and pathogenesis. Eukaryot. Cell 2007, 6, 997–1005. [Google Scholar] [CrossRef]
- Tan, J.; Zhao, H.; Li, J.; Gong, Y.; Li, X. The devastating rice blast airborne pathogen Magnaporthe oryzae—A review on genes studied with mutant analysis. Pathogens 2023, 12, 379. [Google Scholar] [CrossRef] [PubMed]
- Bolton, M.D.; Thomma, B.P.; Nelson, B.D. Sclerotinia sclerotiorum (Lib.) de Bary: Biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 2006, 7, 1–16. [Google Scholar] [CrossRef]
- Sakai, Y.; Oku, M.; van der Klei, I.J.; Kiel, J.A. Pexophagy: Autophagic degradation of peroxisomes. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2006, 1763, 1767–1775. [Google Scholar] [CrossRef]
- Asakura, M.; Ninomiya, S.; Sugimoto, M.; Oku, M.; Yamashita, S.I.; Okuno, T.; Sakai, Y.; Takano, Y. Atg26-mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbiculare. Plant Cell 2009, 21, 1291–1304. [Google Scholar] [CrossRef]
- Komduur, J.A.; Veenhuis, M.; Kiel, J.A. The Hansenula polymorpha PDD7 gene is essential for macropexophagy and microautophagy. FEMS Yeast Res. 2003, 3, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Pereira de Sa, N.; Del Poeta, M. Sterylglucosides in Fungi. J. Fungi 2022, 8, 1130. [Google Scholar] [CrossRef] [PubMed]

| Mutant | Nucleotide Change | Amino Acid Change | Reference Gene | Predicted Function |
|---|---|---|---|---|
| M133 | T407C T356C A574T | F136S L119S M192L | sscle_01g000110 sscle_01g000110 sscle_05g044500 | Mitochondrial substrate/solute carrier Mitochondrial substrate/solute carrier SIT4 phosphatase-associated protein |
| G299A | G100E | sscle_09g072660 | Nucleic acid-binding protein | |
| C376T | R126STOP | sscle_11g082070 | Autophagy-related protein ATG9 | |
| G766A | E256K | sscle_13g092650 | Histidine phosphatase | |
| G843A | M281I | sscle_13g092670 | Serine/threonine protein kinase | |
| R240 | A360T G1037A | Q120H G346E | sscle_03g027360 sscle_05g040590 | None Acyl-coenzyme A diphosphatase NUDT19 |
| C1120T | Q374STOP | sscle_09g070220 | Fructose-2,6bisphosphatase | |
| C485T | S162F | sscle_12g089570 | Autophagy-related protein ATG4 | |
| P20B12 | A938G G4417A | K313R A1473T | sscle_02g020610 sscle_03g023750 | RNA-dependent RNA polymerase Autophagy-related protein ATG2 |
| C167T | S56F | sscle_04g034370 | Glycosyltransferase | |
| 648delT | Y216STOP | sscle_08g062260 | Sterol reductase | |
| P18D4 | A5309G | K1770R | sscle_07g055370 | Class II myosin |
| T656G | L219STOP | sscle_08g066910 | Autophagy-related protein ATG5 | |
| LL247 | A2776G | N92D6 | sscle_04g032930 | E3 ubiquitin-protein ligase |
| C1066T | R356STOP | sscle_12g087380 | Autophagy-related protein ATG1 | |
| P38E2 | C940T T271G | P314S Y91D | sscle_01g004320 sscle_01g010390 | Not known Mitochondrial chaperone BCS1 |
| T547G | L183V | sscle_09g069660 | Not known | |
| A3875G | Q1292R | sscle_11g081870 | Autophagy-related protein ATG26 | |
| G515A | G172E | sscle_11g086620 | Nucleoporin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weerasinghe, T.; Li, J.; Chen, X.; Gao, J.; Tian, L.; Xu, Y.; Gong, Y.; Huang, W.; Zhang, Y.; Jiang, L.; et al. Autophagy-Related Proteins (ATGs) Are Differentially Required for Development and Virulence of Sclerotinia sclerotiorum. J. Fungi 2025, 11, 391. https://doi.org/10.3390/jof11050391
Weerasinghe T, Li J, Chen X, Gao J, Tian L, Xu Y, Gong Y, Huang W, Zhang Y, Jiang L, et al. Autophagy-Related Proteins (ATGs) Are Differentially Required for Development and Virulence of Sclerotinia sclerotiorum. Journal of Fungi. 2025; 11(5):391. https://doi.org/10.3390/jof11050391
Chicago/Turabian StyleWeerasinghe, Thilini, Josh Li, Xuanye Chen, Jiayang Gao, Lei Tian, Yan Xu, Yihan Gong, Weijie Huang, Yuelin Zhang, Liwen Jiang, and et al. 2025. "Autophagy-Related Proteins (ATGs) Are Differentially Required for Development and Virulence of Sclerotinia sclerotiorum" Journal of Fungi 11, no. 5: 391. https://doi.org/10.3390/jof11050391
APA StyleWeerasinghe, T., Li, J., Chen, X., Gao, J., Tian, L., Xu, Y., Gong, Y., Huang, W., Zhang, Y., Jiang, L., & Li, X. (2025). Autophagy-Related Proteins (ATGs) Are Differentially Required for Development and Virulence of Sclerotinia sclerotiorum. Journal of Fungi, 11(5), 391. https://doi.org/10.3390/jof11050391







