Abstract
Taxonomic and phylogenetic studies of Cotylidia (Hymenochaetales, Basidiomycota) were performed. A phylogenetic estimate based on six genetic markers revealed that Cotylidia in the current sense includes species belonging to three distantly related clades in the Hymenochaetales. Based on morphology, phylogeny, and ecological habitat, the name Cotylidia s.s. is proposed for the first clade, including the type species C. undulata and C. carpatica. Neocotylidia gen. nov. is proposed for the second clade, which includes N. diaphana, N. fibrae, the new species N. bambusicola, and two accessions recorded as Cotylidia aurantiaca var. alba and C. aurantiaca. Contrary to the findings in earlier studies, C. pannosa demonstrated a weak grouping affinity with Globulicium hiemale, Hastodontia hastata, Atheloderma mirabile, Tsugacorticium kenaicum, Lawrynomyces capitatus, and Lyoathelia laxa. The morphological characteristics of Cotylidia s.s. are restricted to species with hymenial cystidia, pileocystidia, and caulocystidia, as well as a muscicolous habitat. Neocotylidia species differ from Cotylidia s.s. in the lack of pileocystidia and caulocystidia and substrate preference for soil or wood. Illustrated descriptions of the new species and genus, as well as an identification key to the worldwide species of Cotylidia s.l. are provided.
1. Introduction
Stipitate stereoid fungi are species with spathulate, or infundibuliform basidiomata, a smooth hymenophore, and smooth, hyaline spores [1]. Previous studies have demonstrated that the stipitate stereoid basidiomata have evolved multiple times and are distributed across several orders of Agaricomycetes, at least including Agaricales Underw., Atheliales Jülich, Hymenochaetales Oberw., Polyporales Gäum., and Stereopsidales Sjökvist, E. Larss., B.E. Pfeil and K.H. Larss. [1,2,3,4,5,6,7,8,9].
As one of the important orders that include corticioid fungi, Hymenochaetales has been continually amended and by now consists of 15 families and 84 genera, among which 19 genera have no definite position at the family level [10,11]. Rickenellaceae Vizzini is a family in Hymenochaetales that includes about 20 genera [12]. Phylogenetic analyses have shown that the family is polyphyletic [1,3,7,8,13,14]. Wang et al. [10] recently amended the description of the family to be monotypic and restricted to the type genus Rickenella Raithelh.
Cotylidia P. Karst. was previously established under Rickenellaceae by P.A. Karsten [15] and typified through C. undulata (Fr.) P. Karst. However, Wang et al. [10] reported that the phylogenetic placement of Cotylidia at the family level remains undetermined in Hymenochaetales. Limited phylogenetic studies have grouped Cotylidia with species from Resiniciaceae L.W. Zhou and Xue W. Wang, Rickenellaceae, Peniophorellaceae L.W. Zhou, Xue W. Wang and S.L. Liu, and several allied genera that have no definite positions at the family level [8,9,10,11,16,17]. Only five species and one variety of Cotylidia have been sampled in previous studies and have never been found to constitute a monophyletic lineage in any monophyletic analysis [8,9]. Thus, further phylogenetic analyses and species collection are needed to clarify the relationships among Cotylidia and related genera.
Cotylidia species are characterized by infundibuliform or spathulate stipitate basidiomata; smooth or rugose hymenophore; a monomitic hyphal system without clamp connections; hyaline, smooth, inamyloid basidiospores; long protruding hymenial cystidia; and a muscicolous, terricolous, or lignicolous habitat [9,18,19,20,21,22]. Currently, approximately 11 taxa have been classified under Cotylidia [9,10,20,22,23,24,25,26,27,28], which includes seven species that were discovered in China [9,18,19,29,30].
During studies on macrofungi in Southwest China, a white, stipitate stereoid material/specimen was collected from the ground under bamboo in the Guizhou and Sichuan Provinces. Further studies showed that it belongs to Cotylidia s.l.; however, it could not be classified as any present species. The aim of the work has been to determine the identity of the material found, as well as to clarify the taxonomy and phylogeny of Cotylidia species based on morphological evaluation, phylogenetic analysis, and ecological data.
2. Materials and Methods
2.1. Site Description
The type specimen of the new species was collected from Qianling Park, Guiyang, Guizhou Province, Southwest China. This region has an annual rainfall of approximately 1129 mm, average temperature of 15.3 °C, and elevation of 1100–1396 m. In total, 128 families, 350 genera, and 476 species of vascular plants were discovered in Qianling Park. Its vegetation is tropical-subtropical forests biome dominated by Castanopsis eyrei (Champ. ex Benth.) Tutch., Quercus glauca Thunb., Q. fabri Hance, Q. acutissima Carr., Pinus massoniana Lamb., P. fenzeliana Hand.-Mzt., Cunninghamia lanceolata (Lamb.) Hook., Platycladus orientalis (L.) Franco, Liquidambar formosana Hance, Keteleeria davidiana (Bertr.) Beissn., Tsuga chinensis (Franch.) Pritz., Carpinus pubescens Burk., Taxus wallichiana var. chinensis (Pilger) Florin, Cornus controversa Hemsley, Ulmus pumila L., Celtis sinensis Pers., Betula luminifera H. Winkl., and bamboo, etc. [31,32].
2.2. Morphological Evaluation
All specimens were processed and deposited in the herbarium at the National Institute of Occupational Health and Poison Control, Chinese Center for Disease Control (NIOHP, China CDC). Macromorphological characteristics were investigated based on field notes and color photos of basidiomata. Color codes were verified as proposed by Petersen [33]. Microscopy studies were conducted as described by Kout and Zíbarová [22] and Yang et al. [9]. Sections were studied at up to 1000× magnification using a Nikon E 80i microscope and phase contrast illumination (Nikon, Japan). Detailed descriptions of microscopic structures were worded as in previous studies [34,35,36].
2.3. DNA Extraction and Sequencing
To obtain polymerase chain reaction (PCR) products from six dried specimens of the new species and Cotylidia fibrae, the Phire® Plant Direct PCR Kit (Finnzymes Oy, Espoo, Finland) was used according to the manufacturer’s instructions with some modifications [36]. The ITS1-5.8S-ITS2 region (nuclear ribosomal internal transcribed spacer, ITS), nuclear ribosomal large subunit (nrLSU), nuclear ribosomal small subunit (nrSSU), mitochondrial small subunit (mtSSU), RNA polymerase II second largest subunit (rpb2), and translation elongation factor 1α (tef1α) regions were amplified using the selected primer pairs ITS5/ITS4 [37], LR0R/LR5 [38], PNS1/NS41 [39], MS1/MS2 [37], bRPB2-6F/bRPB2-7.1R [40], and EF1-983F/EF1-1953R [41]. PCR was performed as described by Tang et al. [36]. All newly generated sequences in this study were deposited in GenBank and are listed in bold in Table S1.
2.4. Phylogenetic Analyses
In addition to the newly generated sequences for this study, additional related sequences, mainly based on the work of Yang et al. [9], Wang et al. [10], and Wang and Zhou [11], were also integrated in phylogenetic analyses (Table S1).
All sequences were aligned using ClustalX v.1.83 [42], manually optimized in BioEdit v.7.0.5.3 [43], and compiled in one concatenated dataset for ITS1-5.8S-ITS2, nrLSU, nrSSU, mtSSU, rpb2, and tef1α markers to explore the phylogenetic position of the newly sequenced specimens in Hymenochaetales. Following the work of Wang and Zhou [11], two species from Polyporales, viz. Fomitopsis pinicola and Grifola frondosa, were also included, and two species from Thelephorales, viz. Boletopsis leucomelaena and Thelephora ganbajun, were selected as outgroups. The final alignments and the topologies were deposited in TreeBase (http://treebase.org/treebase-web/home.html, submission ID: 31700, accessed on 14 September 2024).
Maximum likelihood (ML) analyses and Bayesian inference (BI) were carried out using RAxML v.8.2.10 [44] and MrBayes v.3.2.6 [45], respectively. In ML analyses, statistical support values were obtained using rapid bootstrapping with 1000 replicates, with default settings used for the other parameters. For BI analyses, the best-fit models for nucleotide substitution were estimated using jModeltest v.2.17 [46]. Four Markov chains were run for 2,000,000 generations until the split deviation frequency values were lower than 0.01. Trees were sampled every 100th generation. The first quarter of the sampled trees, which represented the burn-in phase of the analyses, was discarded, whereas the remaining ones were used to calculate the Bayesian posterior probabilities (BPPs) in the majority rule consensus.
Phylogenetic trees were visualized using TreeView [47]. Branches that received bootstrap supports for ML (≥50%) and BPPs (≥0.95) were considered significantly supported.
Genetic distances of nrLSU sequences of species belonging to different genera within Hymenochaetales were calculated with the Kimura 2-parameter (K2P) model using MEGA v.11 software [48,49].
3. Results
3.1. Phylogeny
The concatenated ITS1-5.8S-ITS2-nrLSU-nrSSU-mtSSU-rpb2-tef1α dataset contained 107 ITS, 111 nrLSU, 66 nrSSU, 62 mtSSU, 71 rpb2, and 45 tef1α sequences from 114 samples, representing 101 ingroup taxa and the outgroup (Table S1), and had an aligned length of 5579 characters. jModelTest suggested SYM+I+G, SYM+G, GTR+G, GTR+I+G, GTR+I+G, GTR+G, GTR+I+G, and GTR+I+G to be the best-fit models of nucleotide evolution for ITS1, 5.8S, ITS2, nrLSU, nrSSU, mtSSU, rpb2, and tef1α markers, respectively, for the Bayesian analysis. The average standard deviation of split frequencies of BI was 0.009318 at the end of the run. BI analyses yielded nearly identical tree topologies with the ML analyses. Only the ML tree is provided in Figure 1 with the likelihood bootstrap values and BPPs labelled along the branches.
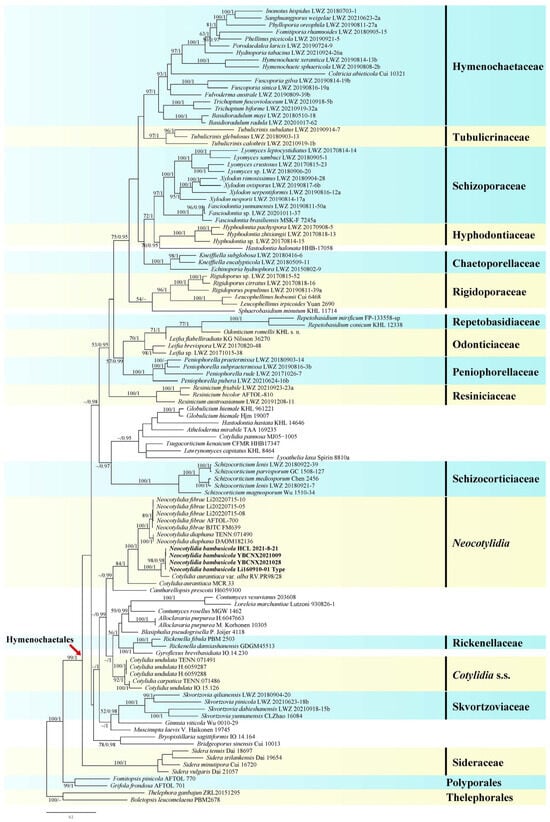
Figure 1.
Phylogeny of Cotylidia s.l. as determined using maximum likelihood (ML) analyses based on internal transcribed spacer (ITS), nuclear ribosomal large subunit (nrLSU), nuclear ribosomal small subunit (nrSSU), mitochondrial small subunit (mtSSU), RNA polymerase II second largest subunit (rpb2), and translation elongation factor 1α (tef1α) sequences. Branches are labeled with an ML bootstrap value of >50% and Bayesian posterior probability (BPP) of >0.90. New species are indicated in bold.
Cotylidia species were recovered in three distinct clades (Figure 1), namely the Cotylidia s.s. clade including the type species C. undulata and C. carpatica (ML = 92%, BI = 1); the new genus Neocotylidia clade including N. diaphana, N. fibrae, the new species N. bambusicola, and two accessions recorded as Cotylidia aurantiaca var. alba and C. aurantiaca. (ML = 84%, BI = 1); and C. pannosa (Sowerby) D.A. Reid weakly grouped with Globulicium hiemale (Laurila) Hjortstam, Hastodontia hastata (Litsch.) Hjortstam and Ryvarden, Atheloderma mirabile Parmasto, Tsugacorticium kenaicum Nakasone and Burds., Lawrynomyces capitatus (J. Erikss. and Å. Strid) Karasiński, and Lyoathelia laxa (Burt) Hjortstam and Ryvarden.
The pairwise genetic distances between the clade of Neocotylidia and other known genera in the order Hymenochaetales ranged from 0.0422 to 0.1551, which were not lower than the pairwise distance (from 0.0207 to 0.1994) of other genera in Hymenochaetales (Table S2). Hence, we feel that the clade of Neocotylidia can be phylogenetically treated as an independent genus in Hymenochaetales.
3.2. Taxonomy
- Neocotylidia Jing Si and Hai J. Li, gen. nov.
MycoBank No. 850556.
Etymology—Neocotylidia: refers to the resemblance to Cotylidia.
Type species—Neocotylidia bambusicola Jing Si and Hai J. Li.
Description—Basidiomata annual, solitary or gregarious, sometimes confluent, infundibuliform, flabelliform or spathulate, stipitate, coriaceous, usually papery-thin. Pileal surface from white or cream to alutaceous brown, zonate or not, sometimes darker when dry. Hymenophore smooth or rugose, similar to pileal surface color. Stipe central, eccentric to lateral, usually tomentose, floccose or pubescent. Hyphal system monomitic, hyphae simple septate. Hymenial cystidia of tramal origin, hyaline, thin- to thick-walled, aseptate or septate, the cystidia are long and projecting beyond the basidia. Pileocystidia and caulocystidia absent. Basidiospores hyaline, thin-walled, smooth, mostly more or less ellipsoid, rarely cylindrical, clavate to ovate, with or without guttules, IKI−, CB−. Terricolous or lignicolous habitat.
Remarks—Compared with Cotylidia, Neocotylidia species have no pileocystidia and caulocystidia and grow on soil or wood. The two species subsumed under Cotylidia s.s. (C. undulata and C. carpatica) grow on moss [22,50]. In particular, some fungal anatomical structures in Mnium thalli indicate biotrophic parasitism in connection with C. carpatica [50].

Figure 2.
Basidiomata of Neocotylidia bambusicola (holotype; bar = 1 cm).
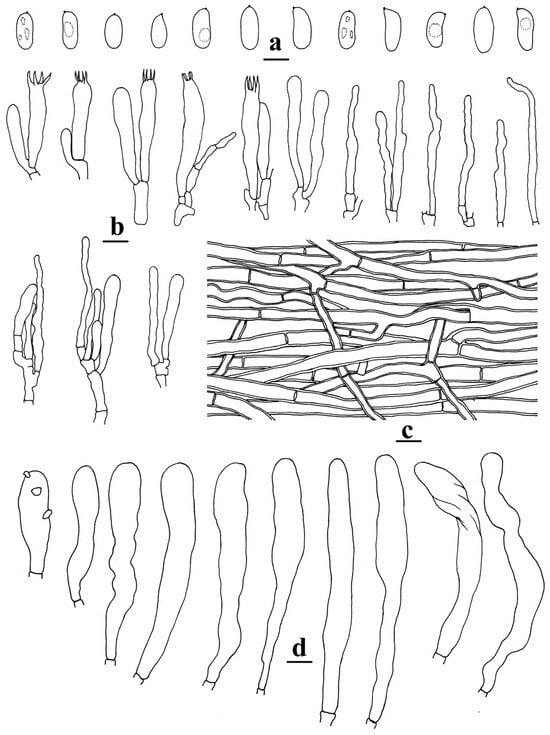
Figure 3.
Microscopic structures of Neocotylidia bambusicola (160910-01, holotype). (a) Basidiospores; (b) basidia, basidioles, and hyphal ends; (c) hyphae from context; (d) hymenial cystidia. (bars: (a) = 5 µm; (b–d) = 10 µm). Illustrated by Hai-Jiao Li.
MycoBank No. 850559.
Etymology—bambusicola (Lat.): refers to this species that grows under bamboo.
Diagnosis—Neocotylidia bambusicola is characterized by stipitate basidiomata; infundibuliform, flabelliform to spathulate pilei; glabrous, radially furrowed, white, cream to pale flesh pink pileal surface; tomentose stipe; thick-walled, cylindrical to clavate hymenial cystidia; lack of pileocystidia and caulocystidia; hyaline, cylindrical, clavate to oblong ellipsoid, thin-walled basidiospores, 7–9.3 × 3.2–4.4 µm.
Habitat and distribution—Terricolous under bamboo, at present only reported in Southwest China, summer to autumn.
Type—CHINA. Guizhou Province, Guiyang, Qianling Park, altitude: 1145 m, 26°36′22.36″ N, 106°41′52.33″ E, on the ground under bamboo, 10 September 2016, Li160910-01 (holotype, deposited at the NIOHP). GenBank accession numbers: ITS OQ376551, nrLSU OQ372914, rpb2 PP764668, and tef1α PP764673.
Description—Basidiomata gregarious, annual, fasciculate, stipitate, coriaceous when fresh and hard, corky upon drying, without odor or taste. Pilei infundibuliform, flabelliform to spathulate, very thin, up to 3.5-cm high and 3-cm wide. Pileal surface glabrous, concentrically and radially zonate, white, cream, and pale flesh pink toward the center. Stipe surface tomentose, white to cream, up to 2 cm in length and 5 mm in diameter. Hymenophore glabrous, white, cream, and pale flesh pink toward the stipe. Flesh under hymenophore very thin, usually less than 1-mm thick, coriaceous.
Hyphal structure—Hyphal system monomitic; all hyphae without clamps, hyphae IKI−, CB−, tissue unchanged in KOH. Generative hyphae in the pileal context hyaline, thick-walled with a wide lumen, branched, more or less parallel along the pileal surface, 4–7 μm in diameter; generative hyphae in the stipe trama similar to those in the pileal context, 3–7.5 μm in diameter.
Microstructure—Hymenial cystidia of tramal origin, hyaline, thick-walled, aseptate, more or less cylindrical to clavate, sometimes encrusted with irregular crystals, remarkably projecting above the hymenium up to 55 μm, 77–104 × 8–11 μm. Pileocystidia and caulocystidia absent. Basidia clavate, with four sterigmata, 30–47 × 6–8 µm; basidioles shaped similarly to basidia but slightly smaller; hyphal ends in the hymenium usually with narrow tips, sometimes with one or two secondary septa. Basidiospores hyaline, cylindrical, clavate to oblong ellipsoid, thin-walled, smooth, sometimes with one to several guttules, IKI−, CB−, [80/4/2] (6.9–)7–9.3(–10.1) × (3–)3.2–4.4(–4.6) µm; L = 8.02 µm; W = 3.7 µm; Q = (1.6–)1.85–2.66(–3); Qm = 2.19 ± 0.25.
Additional specimens examined (paratypes)—CHINA. Guizhou Province, Guiyang, Qianling Park, altitude: 1145 m, 26°36′22.36″ N, 106°41′52.33″ E, on the ground under bamboo, 21 September 2014, Li 140921-03; Sichuan Province, Yibin, Changning County, Huatan Town, Shiliang Village, altitude: 378 m, 28°28′18″ N, 104°48′16″ E, on the ground under bamboo, 20 July 2021, YBCNX2021009; Meitong Town, Yihong Village, altitude: 528 m, 28°18′25″ N, 105°0′42″ E, on the ground under bamboo, 21 July 2021, YBCNX2021028.
Remarks—Neocotylidia fibrae (comb. nov. in the present paper), a new species first discovered in China [9] is similar to N. bambusicola by its more or less white basidiomata. However, N. fibrae has a hymenophore covered with distinct white fibres, wider hymenial cystidia (70–115 × 12.5–20 µm), and smaller basidiospores (5–5.5 × 3–3.5 µm), grows on the ground under Pinus tabuliformis Carr., and is distributed in temperate regions in China [9].
Cotylidia aurantiaca var. alba also forms white, papyraceous basidiomata and forms a clade with the new species N. bambusicola (Figure 1). However, C. aurantiaca var. alba has septate hymenial cystidia with 1–3 transverse septa and ellipsoid basidiospores, usually grows on trunks or woody debris, and was first described in Argentina [18].
Cotylidia diaphana, which is combined with N. diaphana in a forthcoming manuscript, has more or less white basidiomata similar to those of N. bambusicola; however, N. diaphana has hymenial cystidia with one septum and smaller, ellipsoid basidiospores (4–6 × 2.75–3.5 µm), and grows on the ground in damp coniferous or frondose woods, or on lawns under trees [24].
Cotylidia harmandii was discovered in Japan and is similar to N. bambusicola in its creamy to beige pileus, protruding hymenial cystidia, lack of pileocystidia and caulocystidia, and growth on the ground. However, C. harmandii differs from N. bambusicola by its pinkish or purplish-red pileus when dry, shorter hymenial cystidia (up to 78 µm long), and smaller basidiospores (6 × 3 µm) [18].
Morphologically, Cotylidia komabensis is in good accordance with the definition of Neocotylidia and has characteristics similar to those of N. bambusicola, such as stipitate, white basidiomata, hymenial cystidia, lack of pileocystidia and caulocystidia, and growth on the ground. The type material of Stereum albidum Lloyd (1916), a synonym of C. komabensis, was found in association with bamboo roots [18]. However, C. komabensis differs from N. bambusicola by its thin-walled hymenial cystidia and smaller basidiospores (6–8.5 × 2.5–3.5 µm) [18].
Cotylidia marsicana is similar to N. bambusicola in terms of stipitate, white basidiomata, but it differs from N. bambusicola by its distinctly smaller hymenial cystidia (40–60 × 4–6 μm), basidia (15–20 × 4–5 µm), basidiospores (4–5 × 3–3.5 µm), and growth on burnt wood [19].
Cotylidia pannosa differs from N. bambusicola by its bright red-orange pileal surface margin (all basidiomata when young), larger hymenial cystidia (70–160 × 10–14 μm), wider ellipsoid basidiospores (8–10 × 4–4.6 μm), and growth on bare soil or litter from deciduous trees [22].
- Neocotylidia diaphana (Cooke) Jing Si and Hai J. Li, comb. nov.
MycoBank No. 850561.
Basionym—Stereum diaphanum Cooke, Syll. fung. (Abellini) 6: 558. 1888.
=Cotylidia diaphana (Cooke) Lentz, Agric. Monogr. U.S.D.A. 24: 12. 1955.
Remarks—This species is characterized by stipitate basidiomata; thin coriaceous, normally discrete, and frequently infundibuliform but sometimes pseudo-infundibuliform or even spathulate pileus, which is white or pallid when fresh, becoming creamy-buff, pale ochraceous or straw-colored with a slight sheen when dry; protruding thin- to slightly thick-walled hymenial cystidia that sometimes has 1–3 transverse septa; only hymenial cystidia appearing on the hymenium; ellipsoid, thin-walled basidiospores; and growth on the ground or on the wood of hardwood trees [20]. Phylogenetically, this species is grouped in the clade we have defined as Neocotylidia (Figure 1).
- Neocotylidia fibrae (L. Fan and C. Yang) Jing Si and Hai J. Li, comb. nov.
MycoBank No. 850562.
Basionym—Cotylidia fibrae L. Fan and C. Yang, Phytotaxa. 487(2): 7. 2021.
Remarks—This species was discovered in a temperate region in China in 2021 [9]. It is characterized by more or less white basidiomata with a hymenophore covered with distinct white fibres; protruding hymenial cystidia; lack of pileocystidia and caulocystidia; ellipsoid to oblong, thin-walled basidiospores; and growth on the ground under Pinus tabuliformis [9]. Based on the morphological characteristics and phylogenetic analyses (Figure 1) [9], we propose the classification of this species as Neocotylidia fibrae.
- Cotylidia aurantiaca (Pat.) A.L. Welden, Lloydia 21: 40. 1958.
Basionym—Podoscypha aurantiaca Pat., Enum. Champ. Guadeloupe (Lons-le-Saunier): 20. 1903.
Remarks—This species is characterized by stipitate, paper-thin, basidiomata; bright yellow pileus when fresh with fimbriate margin; protruding cylindrical, clavate or slightly capitate to subglobose or pyriform hymenial cystidia; lack of pileocystidia and caulocystidia; thin-walled, ellipsoid basidiospores, and growth primarily on woody substrates, but it may also be terrestrial [18,20], These characteristics indicate that this species may be a Neocotylidia species. The type locality of C. aurantiaca is in Rio de Janeiro, Brazil. No sequences from the type specimen or the type locality were obtained. Only one sequence (AF261460) from the USA was available at present (Figure 1). However, the sequences (AF261460) from the USA may not be the real C. aurantiaca, and further studies are needed to obtain sequences from the type specimen of C. aurantiaca or at least from the type locality before a final decision on C. aurantiaca.
- Cotylidia aurantiaca var. alba D.A. Reid, Beih. Nova Hedwig. 18: 67. 1965.
Remarks—One sequences (AF261458) recorded as Cotylidia aurantiaca var. alba from Puerto Rico was used in our phylogenetic analyses, which strongly grouped with the new species Neocotylidia bambusicola (ML = 100%, BI = 1), indicating that this specimen (RV.PR98/28) was undoubtedly a Neocotylidia species. Phylogenetic results (Figure 1) also showed that the two sequences recorded as C. aurantiaca and C. aurantiaca var. alba represent two independent species. The type locality of C. aurantiaca var. alba is from Argentina, and thus the sequences (AF261458) are questionable. Further efforts are needed to acquire sequences from the type specimen or at least other specimens from the type locality to determine its final classification status.
- Notes on other species of Cotylidia recorded without molecular data
- Cotylidia guttulata L. Rémy, Bull. trimest. Soc. mycol. Fr. 80(4): 579. 1965.
Remarks—Morphologically, this species is very similar in features to C. muscigena except for its biguttulate spores. However, the number of guttules in each spore may vary. Jülich [51] synonymized C. guttulata with C. muscigena, whereas Moreau et al. [52] consider C. guttulata as a form of C. muscigena f. guttulata (Rémy) P.-A. Moreau, Wuilb. and Courtec. The muscicolous habitat preference of C. guttulata and the presence of hymenial cystidia, pileocystidia, and caulocystidia [18] indicate that C. guttulata is a Cotylidia s.s. species.
- Cotylidia harmandii (Lloyd) D.A. Reid, Beih. Nova Hedwigia. 18: 76. 1965.
Basionym—Stereum harmandii Lloyd, Mycol. Writ. (Cincinnati) 4(46): 22. 1913.
Remarks—This Japanese species produces stipitate basidiomata; thin, translucent, flabelliform pileus creamy to beige in color, without zonation, and split into several segments; protruding abundant hymenial cystidia (up to 78 × 7–9 μm) which are long, thin-walled, and non-septate; lacks pileocystidia and caulocystidia; and grows on the ground [18]. These distinct features suggest this species belongs to Neocotylidia.
- Cotylidia komabensis (Henn.) D.A. Reid, Beih. Nova Hedwigia. 18: 77. 1965.
Basionym—Thelephora komabensis Henn., Bot. Jb. 31: 736. 1902.
Remarks—This species is characterized by stipitate, white basidiomata; thin, mostly translucent, spathulate, or reniform pileus with a crenulate, incised, or entire margin; long cylindrical, thin-walled, non-septate hymenial cystidia (up to 104 × 9–11 μm); lack of pileocystidia and caulocystidia; elliptical basidiospores (6–8.5 × 2.5–3.5 μm); and growth on the ground in large groups [18], which indicate that it may be a Neocotylidia species.
- Cotylidia marsicana Lonati, Micol. Veg. Medit. 15(1): 3. 2000.
Remarks—This Mediterranean species is characterized by small, stipitate, white basidiomata; infundibuliform pileus (5–15 × 3–8 mm); white, pruinose stipe (3–5 × 1–2 mm); small, thin-walled, ellipsoid to broadly ellipsoid basidiospores (4–5 × 3–3.5 μm); cylindrical hymenial cystidia (40–60 × 4–6 μm); lack of pileocystidia and caulocystidia; and growth on burnt wood [19], most of which indicate that it may be a Neocotylidia species.
- Cotylidia muscigena L. Rémy, Bull. trimest. Soc. mycol. Fr. 80(4): 579. 1965.
Remarks—This species is characterized by small, stipitate basidiomata; very thin, semi-translucent, flabelliform to semi-infundibuliform pileus, which is often longitudinally incised; pale yellowish to brownish, sometimes zonate pileal surface, which is finely pruinose; ellipsoid, hyaline, smooth, inamyloid basidiospores (6–8 × 2.5–2.8 μm), protruding aseptate, cylindrical hymenial cystidia (43–70 × 6–10 μm); pileocystidia (40 × 7–9 μm) and caulocystidia (40–70 × 6–10 μm), and growth on living mosses (Brachythecium, Bryum, and Mnium) [18,22]. These features indicate that this species is a Cotylidia s.s. species.
- Cotylidia pannosa (Sowerby) D.A. Reid, Beih. Nova Hedwigia 18: 81. 1965.
Basionym—Helvella pannosa Sowerby, Col. fig. Engl. Fung. Mushr. (London) 2(14): 155. 1799.
Remarks—This species is characterized by stipitate basidiomata, which in groups often become confluent; bright red-orange pileal surface at the margin, ochre toward the center, and changes from rose to cream; large, aseptate hymenial cystidia (70–160 × 10–14 μm); lack of pileocystidia and caulocystidia; large ellipsoid basidiospores (8–10 × 4–4.6 μm), and growth on bare soil or litter from deciduous trees [22]. These characteristics indicate that this species may be a Neocotylidia species. A previous phylogenetic analysis based on ITS sequences showed that C. pannosa is grouped with N. fibrae but is weakly supported by the analyses (ML = 56%, BI < 0.90) [9]. However, C. pannosa is weakly grouped with Globulicium hiemale, Hastodontia hastata, Atheloderma mirabile, Tsugacorticium kenaicum, Lawrynomyces capitatu, and Lyoathelia laxa (Figure 1) in the present study.
4. Discussion
Our phylogenetic analyses revealed that Cotylidia species in the current sense are classified into three distantly related clades, confirming that Cotylidia is not monophyletic, and the three clades form genus-level lineages in Hymenochaetales [8,9,10,11,16]. One clade included the type species C. undulata and C. carpatica (Figure 1), which produce stipitate, coriaceous basidiomata with hymenial cystidia, pileocystidia, and caulocystidia. These basidiomata grow on mosses [9,18,22,52]. The second clade Neocotylidia is composed of N. diaphana, N. fibrae, and the new species N. bambusicola, and two samples/specimens recorded as Cotylidia aurantiaca var. alba and C. aurantiaca (Figure 1). All of them grow on soil or wood, only produce hymenial cystidia, and lack pileocystidia and caulocystidia [9,18,22,52]. Unlike the classification reported in a previous study of Yang et al. [9], Cotylidia pannosa is weakly grouped with Globulicium hiemale, Hastodontia hastata, Atheloderma mirabile, Tsugacorticium kenaicum, Lawrynomyces capitatus, and Lyoathelia laxa (Figure 1).
Cotylidia was previously established under Rickenellaceae [12,16]. Wang et al. [10] and Wang and Zhou [11] reported that the phylogenetic placement of Cotylidia (the specimen AFTOL-700 labelled as Cotylidia sp.) was also used in the present study, which found it to belong to Neocotylidia, at the family level remains undetermined in Hymenochaetales. Phylogenetic analyses have shown that Rickenellaceae is polyphyletic [1,3,7,8,13,14], and it was recently amended to be monotypic and restricted for the type genus Rickenella [10].
Limited phylogenetic studies have grouped Cotylidia with species from Resiniciaceae, Rickenellaceae, Peniophorellaceae, and several allied genera, Alloclavaria, Atheloderma, Blasiphalia, Bryopistillaria, Cantharellopsis, Contumyces, Globulicium, Gyroflexus, Hastodontia, Lawrynomyces, Loreleia, Lyoathelia, Muscinupta, Sphaerobasidium, Tsugacorticium, and the new genus Neocotylidia that have no definite positions at the family level [8,9,10,11,17].
Phylogenetically, N. bambusicola is clustered with one specimen Cotylidia sp. (HCL 2021-8-21) from China and strongly supported by the phylogenetic analyses (ML = 98%, BI = 0.98; Figure 1); thus, it could be classified as N. bambusicola. Current and previous phylogenetic analyses support that the two sequences recorded as C. aurantiaca and C. aurantiaca var. alba (AF261460 and AF261458 for nrLSU) represent two independent Neocotylidia species, and these two specimens need further identification [9,18,22,52]. The two sequences were not obtained from the type specimens or specimens from the type localities, which made us doubt whether the identification was accurate. Therefore, before sequences from the type specimens of C. aurantiaca and C. aurantiaca var. alba, or at least sequences from the type locality are available, we would likely retain C. aurantiaca and the variety in Cotylidia.
No molecular sequences are currently available for C. guttulata, C. harmandii, C. komabensis, C. marsicana, and C. muscigena; however, based on their morphological characteristics, C. harmandii, C. komabensis, C. marsicana, and C. pannosa may be Neocotylidia species, whereas C. muscigena may belong to Cotylidia, and C. guttulata may be a form of C. muscigena f. guttulata belonging to Cotylidia [9,18,22,24,52]. Cotylidia decolorans was synonymized with C. aurantiaca by Ryvarden [20]. Further molecular studies are urgently needed to resolve the taxonomic and phylogenetic locations of these species.
| Key to the worldwide species of Cotylidia s.l. |
| 1. Producing hymenial cystidia, pileocystidia, and caulocystidia, growth on moss············2 |
| 1. Only producing hymenial cystidia, growth on soil or wood··············································5 |
| 2. Basidiospores with two guttules································································Cotylidia guttulata |
| 2. Basidiospores without guttules······························································································3 |
| 3. Basidiospores > 5 µm in length·································································Cotylidia muscigena |
| 3. Basidiospores < 5 µm in length·······························································································4 |
| 4. Pileus pale alutaceous brown; stipe lateral; basidiospores 3–3.75 × 1.2–2.5 µm················· |
| ····························································································································Cotylidia carpatica |
| 4. Pileus buff to brownish, darker in the center; stipe central to eccentric; basidiospores |
| 4–5 × 2–2.5 µm···································································································Cotylidia undulata |
| 5. Basidiomata growing on wood·······························································································6 |
| 5. Basidiomata growing on soil···································································································8 |
| 6. Basidiomata growing on burnt wood, with white basidiomata, small hymenial cystidia |
| (40–60 × 4–6 μm) and basidiospores (4–5 × 3–3.5 µm)································Cotylidia marsicana |
| 6. Basidiomata most frequently growing on woody substrates, with bright yellow or white |
| basidiomata, large hymenial cystidia (up to 122 × 6–26 μm) and basidiospores (6.8–7.5 × |
| 3–3.75 µm)·····································································································································7 |
| 7. Pileus bright yellow··························································Cotylidia aurantiaca var. aurantiaca |
| 7. Pileus white to cream··································································Cotylidia aurantiaca var. alba |
| 8. Hymenial cystidia septate······································································Neocotylidia diaphana |
| 8. Hymenial cystidia nonseptate·································································································9 |
| 9. Basidiomata more or less white to cream·············································································10 |
| 9. Basidiomata pinkish, purplish-red, or at least bright red-orange at the margin·············12 |
| 10. Growing on the ground under Pinus tabuliformis, hymenophore covered with distinct |
| white fibers, hymenial cystidia (70–115 × 12.5–20 µm), basidiospores (5–5.5 × 3–3.5 µm)···· |
| ····························································································································Neocotylidia fibrae |
| 10. Not growing on the ground under Pinus tabuliformis, hymenophore without white |
| fibers··············································································································································11 |
| 11. Hymenial cystidia thick-walled, basidiospores large (7–9.3 × 3.2–4.4 µm)························· |
| ···················································································································Neocotylidia bambusicola |
| 11. Hymenial cystidia thin-walled, basidiospores small (6–8.5 × 2.5–3.5 µm)························· |
| ··························································································································Cotylidia komabensis |
| 12. Pileal surface bright red-orange at the margin (all basidiomata when young), large |
| hymenial cystidia (70–160 × 10–14 μm) and basidiospores (8–10 × 4–4.6 µm)························· |
| ······························································································································Cotylidia pannosa |
| 12. Pileus of dried material pinkish or purplish-red, small hymenial cystidia (up to 78 × |
| 7–9 μm) and basidiospores (6 × 3 µm)··························································Cotylidia harmandii |
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof11050390/s1, Table S1: Sources of specimens and GenBank accession numbers for the sequences used in this study. Newly generated sequences are in bold. Table S2: The genetic distances between different genera in the order Hymenochaetales based on nrLSU sequences using the software MEGA v.11. The pairwise genetic distances between the clade of Neocotylidia and other known genera are in bold, with the minimum and maximum values on a yellow background. The minimum value and maximum values of pairwise distance of other genera in Hymenochaetales are also in bold with a yellow background.
Author Contributions
Conceptualization, J.S. and H.L.; methodology, J.S. and H.L.; software, Y.L., H.Z. and H.L.; validation, J.S. and H.L.; formal analysis, J.M., Y.Z., J.L., Y.L., H.Z. and H.L.; investigation, J.M., Y.Z., J.L., Y.L., H.Z., Z.S., J.S. and H.L.; resources, H.L.; data curation, J.M., Y.Z., J.S. and H.L.; writing-original draft preparation, J.S. and H.L.; writing-review and editing, J.S. and H.L.; visualization, J.M., Y.Z., J.L., J.S. and H.L.; supervision, J.S. and H.L.; project administration, J.S. and H.L.; funding acquisition, J.S. and H.L. All authors have read and agreed to the published version of the manuscript.
Funding
The research was financed by the National Natural Science Foundation of China (32270016, 32070016, and 32270021) and Beijing Forestry University Undergraduate Training Programs for Innovation and Entrepreneurship (X202410022297).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequence data generated for this study can be accessed via GenBank: https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 15 May 2025).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sjökvist, E.; Larsson, E.; Eberhardt, U.; Ryvarden, L.; Larsson, K.H. Stipitate stereoid basidiocarps have evolved multiple times. Mycologia 2012, 104, 1046–1055. [Google Scholar] [CrossRef] [PubMed]
- Pine, E.M.; Hibbett, D.S.; Donoghue, M.J. Phylogenetic relationships of cantharelloid and clavarioid homobasidiomycetes based on mitochondrial and nuclear rDNA sequences. Mycologia 1999, 91, 944–963. [Google Scholar] [CrossRef][Green Version]
- Moncalvo, J.M.; Vilgalys, R.; Redhead, S.A.; Johnson, J.E.; James, T.Y.; Aime, M.C.; Hofstetter, V.; Verduin, S.J.W.; Larsson, E.; Baroni, T.J.; et al. One hundred and seventeen clades of euagarics. Mol. Phylogenet. Evol. 2002, 23, 357–400. [Google Scholar] [CrossRef] [PubMed]
- Larsson, K.H.; Larsson, E.; Kõljalg, U. High phylogenetic diversity among corticioid homobasidiomycetes. Mycol. Res. 2004, 108, 983–1002. [Google Scholar] [CrossRef]
- Larsson, K.H. Re-thinking the classification of corticioid fungi. Mycol. Res. 2007, 111, 1040–1063. [Google Scholar] [CrossRef]
- Binder, M.; Larsson, K.H.; Matheny, P.B.; Hibbett, D.S. Amylocorticiales ord. nov. and Jaapiales ord. nov.: Early diverging clades of Agaricomycetidae dominated by corticioid forms. Mycologia 2010, 102, 865–880. [Google Scholar] [CrossRef]
- Sjökvist, E.; Pfeil, B.E.; Larsson, E.; Larsson, K.H. Stereopsidales—A new order of mushroom-forming fungi. PLoS ONE 2014, 9, e106204. [Google Scholar] [CrossRef]
- Wu, S.H.; Wei, C.L.; Chen, Y.P.; Chen, C.C.; Chen, S.Z. Schizocorticium gen. nov. (Hymenochaetales, Basidiomycota) with three new species. Mycol. Prog. 2021, 20, 769–779. [Google Scholar] [CrossRef]
- Yang, C.; Xu, Y.Y.; Fan, L. Cotylidia fibrae (Rickenellaceae, Hymenochaetales), a new species from China. Phytotaxa 2021, 487, 139–148. [Google Scholar] [CrossRef]
- Wang, X.W.; Liu, S.L.; Zhou, L.W. An updated taxonomic framework of Hymenochaetales (Agaricomycetes, Basidiomycota). Mycosphere 2023, 14, 452–496. [Google Scholar] [CrossRef]
- Wang, X.W.; Zhou, L.W. Umbellaceae fam. nov. (Hymenochaetales, Basidiomycota) for Umbellus sinensis gen. et sp. nov. and three new combinations. J. Fungi 2024, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Vizzini, A. Segnalazioni di muscinupta laevis (Basidiomycota, Agaricomycetes) per il Nord Italia. Micol. Veg. Mediterr. 2010, 25, 141–148. [Google Scholar]
- Liu, S.L.; Gafforov, Y.; Zhang, X.Y.; Wang, H.L.; Wang, X.W.; Zhou, L.W. Reinstatement of the corticioid genus Leifia (Hymenochaetales, Basidiomycota) with a new species L. brevispora from Hubei, Central China. MycoKeys 2019, 51, 85–96. [Google Scholar] [CrossRef]
- Yu, J.; Wang, X.W.; Liu, S.L.; Shen, S.; Zhou, L.W. Taxonomy and phylogeny of Resinicium sensu lato from Asia-Pacific revealing a new genus and five new species (Hymenochaetales, Basidiomycota). IMA Fungus 2021, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Karsten, P.A. Enumeratio Thelephorearum Fr. et Clavariearum Fr. Fennicarum, systemate novo dispositarum. Rev. Mycol. Toulouse 1881, 3, 21–23. [Google Scholar]
- Liu, Z.B.; Zhou, M.; Yuan, Y.; Dai, Y.C. Global diversity and taxonomy of Sidera (Hymenochaetales, Basidiomycota): Four new species and keys to species of the genus. J. Fungi 2021, 7, 251. [Google Scholar] [CrossRef]
- Korotkin, H.B.; Swenie, R.A.; Miettinen, O.; Budke, J.M.; Chen, K.H.; Lutzoni, F.; Smith, M.E.; Matheny, P.B. Stable isotope analyses reveal previously unknown trophic mode diversity in the Hymenochaetales. Am. J. Bot. 2018, 105, 1869–1887. [Google Scholar] [CrossRef]
- Reid, D.A. A Monograph of the Stipitate Stereoid Fungi; Beihefte zur Nova Hedwigia; Cramer: Weinheim, Germany, 1965; Volume 18, pp. 1–382. [Google Scholar]
- Lonati, G. Una nuova specie: Cotylidia marsicana. Micol. Veg. Mediterranea 2000, 15, 3–5. [Google Scholar]
- Ryvarden, L. Stereoid fungi of America. Fungiflora 2009, 28, 1–235. [Google Scholar]
- Bernicchia, A.; Gorjón, S.P. Corticiaceae s. l., Fungi Europaei; Candusso: Alassio, Italy, 2010; Volume 12, pp. 1–1008. [Google Scholar]
- Kout, J.; Zíbarová, L. Revision of the genus Cotylidia (Basidiomycota, Hymenochaetales) in the Czech Republic. Czech Mycol. 2013, 65, 1–13. [Google Scholar] [CrossRef]
- Huijsman, H.S.C. Cotylidia carpatica (Pilát) comb. nov. Bull. Société Mycol. France 1954, 70, 57–62. [Google Scholar]
- Lentz, P.L. Stereum and Allied Genera of Fungi in the Upper Mississippi Valley; Agriculture Monograph; US Department of Agriculture: Washington, DC, USA, 1955; Volume 24, pp. 1–74.
- Welden, A.L. A contribution toward a monograph of Colytidia. Lloydia 1958, 21, 38–44. [Google Scholar]
- Boidin, J. Hétérobasidiomycèstes saprophytes et Homobasidiomycètes résupinés: VI. Essai sur le genre Stereum sensu lato. Rev. Mycol. 1959, 24, 197–225. [Google Scholar]
- Reid, D.A. Notes on fungi which have been referred to the Thelephoraceae senso lato. Persoonia 1962, 2, 109–170. [Google Scholar]
- Rémy, L. Contribution à ľétude de la flore mycologique briançonnaise (Basidiomycètes et Discomycètes). Bull. Société Mycol. France 1965, 80, 459–585. [Google Scholar]
- Shang, B.; He, W.; Yu, C.J. Cotylidia harmandii−a new corticioid fungus in China. For. Res. 2006, 19, 114–116. [Google Scholar]
- Dai, Y.C. A revised checklist of corticioid and hydnoid fungi in China for 2010. Mycoscience 2011, 52, 69–79. [Google Scholar] [CrossRef]
- Yang, L. Discussion upon the vegetational types and the regularities of their distribution of the Qingling Mountain in Guiyang. J. Guizhou Norm. Univ. 1987, 1, 25–31. [Google Scholar]
- Chen, X.J.; Ren, X.B.; Huang, Y.; Chen, X. Investigation on ecological characteristics of Cornus controversa Hensl in Qianling Mountain. Guizhou Sci. 2006, 24, 75–80. [Google Scholar]
- Petersen, J.H. The Danish Mycological Society’s Colour-Chart; Foreningen til Svampekundskabens Fremme: Greve, Denmark, 1996. [Google Scholar]
- Li, Y.; He, S.H.; Zhang, Y.Z.; Liang, J.Q.; Sheng, W.Y.; Si, J.; Li, H.J. Crinipellis deutziae, Marasmius pinicola spp. nov., and C. rhizomaticola (Agaricales, Basidiomycota) new to China from Beijing. Forests 2023, 14, 1480. [Google Scholar] [CrossRef]
- Si, J.; Zhang, Y.Z.; Liang, J.Q.; Li, H.J. Morphology and phylogeny identify two new species and one new subspecies of Podoscypha from Yunnan Province, Southwest China. Front. Microbiol. 2023, 14, 1151365. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.X.; Zhang, Y.Z.; Liang, J.Q.; Jiang, S.F.; Si, J.; Li, H.J. Pyrofomes lagerstroemiae (Polyporaceae, Basidiomycota), a new species from Hubei Province, Central China. Phytotaxa 2023, 583, 191–198. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Hibbett, D.S. Phylogenetic evidence for horizontal transmission of group I introns in the nuclear ribosomal DNA of mushroomforming fungi. Mol. Biol. Evol. 1996, 13, 903–917. [Google Scholar] [CrossRef]
- Matheny, P.B. Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Mol. Phylogenet. Evol. 2005, 35, 1–20. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Chenna, R.; Sugawara, H.; Koike, T.; Lopez, R.; Gibson, T.J.; Higgins, D.G.; Thompson, J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003, 31, 3497–3500. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Stamatakis, A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hőhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Page, R.D.M. TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996, 12, 357–358. [Google Scholar] [PubMed]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Moreau, P.A.; Audet, S. Une récolte du champignon Cotylidia carpatica au Québec. Le Nat. Can. 2008, 132, 5–9. [Google Scholar]
- Jülich, W. Die Nichtblätterpilze, Gallertpilze und Bauchpilze. Aphyllophorales, Heterobasidiomycetes, Gastromycetes. In Kleine Kryptogamenflora Band; Gams, H., Ed.; G. Fischer: Stuttgart, Germany, 1984; Volume IIb/1, pp. 1–626. [Google Scholar]
- Moreau, P.A.; Wuilbaut, J.J.; Courtecuisse, R. Cyphellostereum, Cotylidia et autres Podoscyphaceae stipitées d’Europe. Doc. Mycol. 2008, 34, 53–75. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).