Five New Species of Gibellula (Hypocreales, Cordycipitaceae) from China
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Collection and Fungus Isolation
2.2. Morphological Observations
2.3. DNA Extraction, Polymerase Chain Reaction (PCR), and Sequencing
2.4. Phylogenetic Analyses
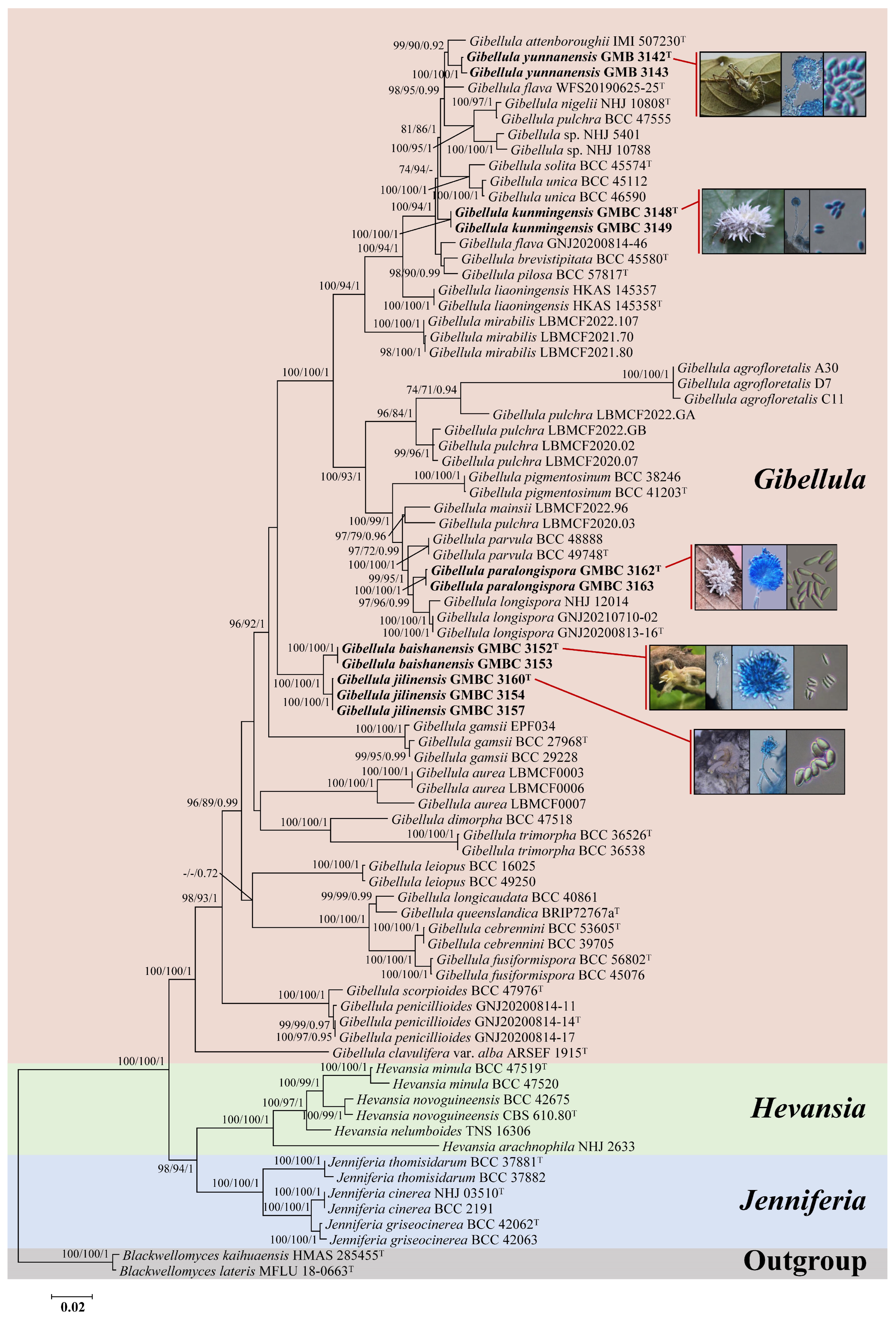
3. Results
3.1. Sequencing and Phylogenetic Analyses
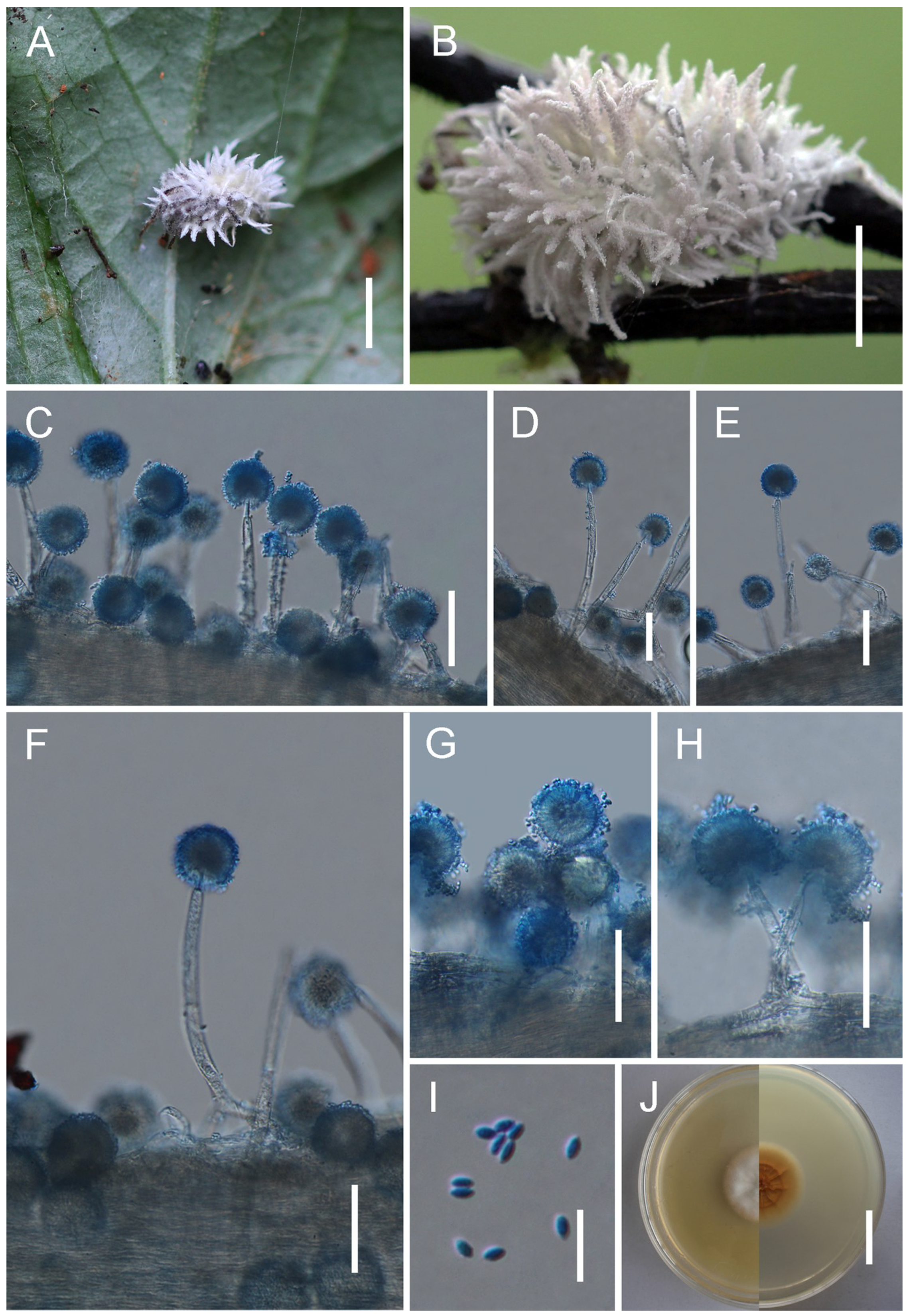
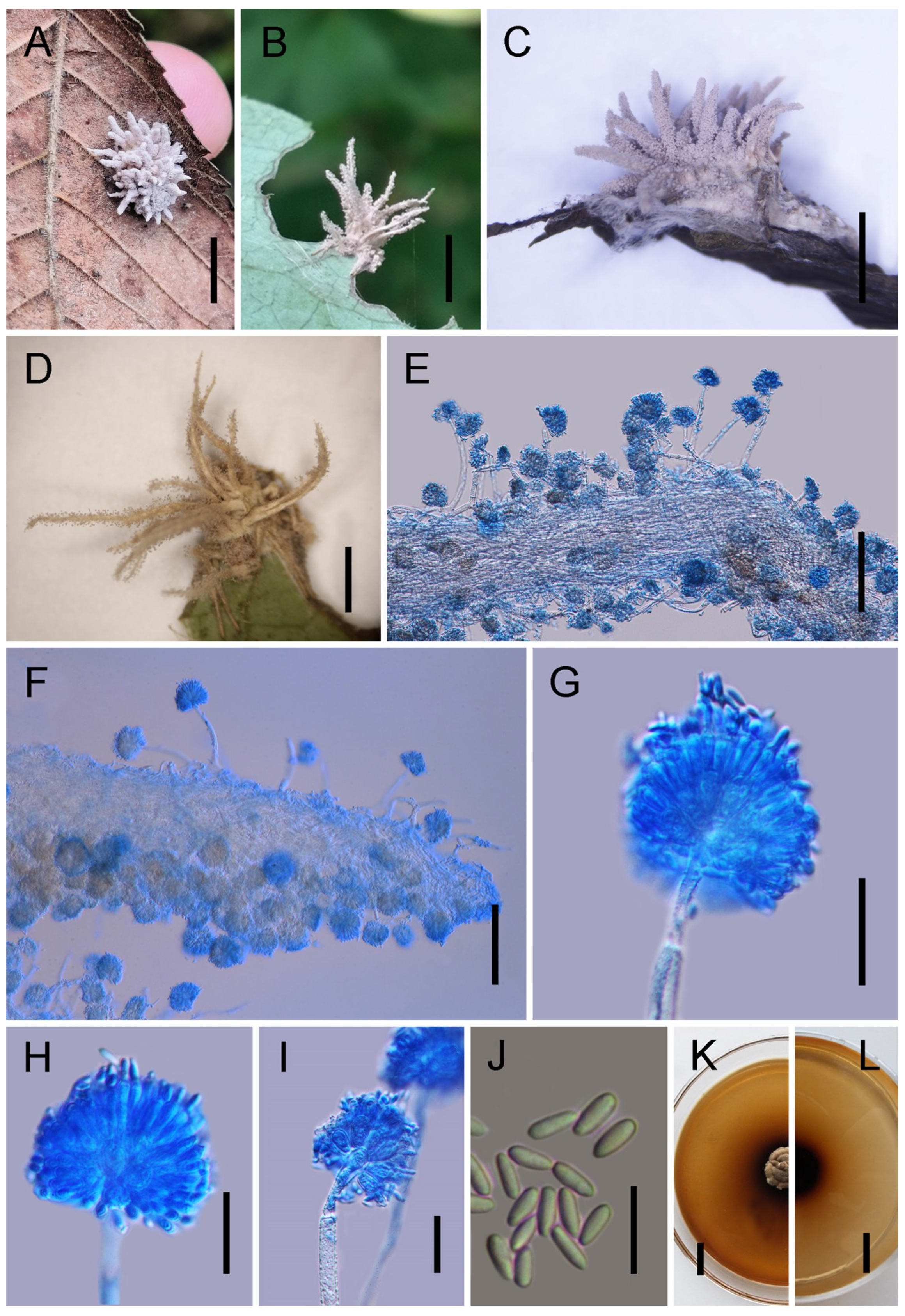
3.2. Taxonomy
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maharachchikumbura, S.S.; Hyde, K.D.; Jones, E.B.; McKenzie, E.H.; Huang, S.; Abdel-Wahab, M.A.; Daranagama, D.A.; Dayarathne, M.C.; D’souza, M.J.; Goonasekara, I.D.; et al. Towards a natural classification and backbone tree for Sordariomycetes. Fungal Divers. 2015, 72, 199–301. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Lumbsch, H.T.; Liu, J.; Maharachchikumbura, S.S.; Ekanayaka, A.H.; Tian, Q.; Phookamsak, R. Outline of Ascomycota: 2017. Fungal Divers. 2018, 88, 167–263. [Google Scholar] [CrossRef]
- Xiao, Y.P.; Wen, T.C.; Hongsanan, S.; Jeewon, R.; Luangsa-Ard, J.J.; Brooks, S.; Wanasinghe, D.N.; Long, F.Y.; Hyde, K.D. Multigene phylogenetics of Polycephalomyces (Ophiocordycipitaceae, Hypocreales), with two new species from Thailand. Sci. Rep. 2018, 8, 18087. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.P.; Wang, Y.B.; Hyde, K.D.; Eleni, G.; Sun, J.Z.; Yang, Y.; Meng, J.; Yu, H.; Wen, T.C. Polycephalomycetaceae, a new family of clavicipitoid fungi segregates from Ophiocordycipitaceae. Fungal Divers. 2023, 120, 1–76. [Google Scholar] [CrossRef]
- Wei, D.P.; Wanasinghe, D.N.; Xu, J.C.; To-Anun, C.; Mortimer, P.E.; Hyde, K.D.; Elgorban, A.M.; Madawala, S.; Suwannarach, N.; Karunarathna, S.C.; et al. Three novel entomopathogenic fungi from China and Thailand. Front. Microbiol. 2021, 11, 608991. [Google Scholar] [CrossRef]
- Wei, D.P.; Gentekaki, E.; Wanasinghe, D.; Tang, S.; Hyde, K.D. Diversity, molecular dating and ancestral character state reconstruction of entomopathogenic fungi in Hypocreales. Mycosphere 2023, 13, 281–351. [Google Scholar] [CrossRef]
- Peng, X.C.; Wen, T.C.; Wei, D.P.; Liao, Y.H.; Wang, Y.; Zhang, X.; Wang, G.Y.; Zhou, Y.; Tangtrakulwanich, K.; Liang, J.D. Two new species and one new combination of Ophiocordyceps (Hypocreales, Ophiocordycipitaceae) in Guizhou. MycoKeys 2024, 102, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Kuephadungphan, W.; Petcharad, B.; Tasanathai, K.; Thanakitpipattana, D.; Kobmoo, N.; Khonsanit, A.; Samson, R.A.; Luangsa-Ard, J.J. Multilocus phylogeny unmasks hidden species within the specialised spider-parasitic fungus, Gibellula (Hypocreales, Cordycipitaceae) in Thailand. Stud. Mycol. 2022, 101, 245–286. [Google Scholar] [CrossRef]
- Shrestha, B.; Kubátová, A.; Tanaka, E.; Oh, J.; Yoon, D.H.; Sung, J.M.; Sung, G.H. Spider-pathogenic fungi within Hypocreales (Ascomycota): Their current nomenclature, diversity, and distribution. Mycol. Prog. 2019, 18, 983–1003. [Google Scholar] [CrossRef]
- Mongkolsamrit, S.; Noisripoom, W.; Tasanathai, K.; Kobmoo, N.; Thanakitpipattana, D.; Khonsanit, A.; Petcharad, B.; Himaman, W. Comprehensive treatise of Hevansia and three new genera Jenniferia, Parahevansia and Polystromomyces on spiders in Cordycipitaceae from Thailand. MycoKeys 2022, 91, 113–149. [Google Scholar] [CrossRef]
- Joseph, R.A.; Masoudi, A.; Valdiviezo, M.J.; Keyhani, N.O. Discovery of Gibellula floridensis from infected spiders and analysis of the surrounding fungal entomopathogen community. J. Fungi 2024, 10, 694. [Google Scholar] [CrossRef]
- Khonsanit, A.; Thanakitpipattana, D.; Mongkolsamrit, S.; Kobmoo, N.; Phosrithong, N.; Samson, R.A.; Crous, P.W.; Luangsa-Ard, J.J. A phylogenetic assessment of Akanthomyces sensu lato in Cordycipitaceae (Hypocreales, Sordariomycetes): Introduction of new genera, and the resurrection of Lecanicillium. Fungal Syst. Evol. 2024, 14, 271–305. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.A.; Evans, H.C. Notes on entomogenous fungi from Ghana: I. The genera Gibellula and Pseudogibellula. Acta Bot. Neerl. 1973, 22, 522–528. [Google Scholar] [CrossRef]
- Huang, B.; Ding, D.G.; Fan, M.Z.; Li, Z.Z. A new entomopathogenic fungus on spiders. Mycosystema 1998, 17, 109–113. [Google Scholar]
- Han, Y.F.; Chen, W.H.; Zou, X.; Liang, Z.Q. Gibellula curcispora, a new species of Gibellula. Mycosystema 2013, 32, 777–780. [Google Scholar]
- Chen, W.H.; Han, Y.F.; Liang, Z.Q.; Wang, Y.R.; Zou, X. Classification of Gibellula spp. by DELTA system. Microbiol. China 2014, 41, 399–407. [Google Scholar] [CrossRef]
- Chen, W.H.; Han, Y.F.; Liang, Z.Q.; Zou, X. Morphological traits, DELTA system, and molecular analysis for Gibellula clavispora sp. nov. from China. Mycotaxon 2016, 131, 111–121. [Google Scholar] [CrossRef]
- Chen, M.; Wang, T.; Lin, Y.; Huang, B. Morphological and molecular analyses reveal two new species of Gibellula (Cordycipitaceae, Hypocreales) from China. MycoKeys 2022, 90, 53–69. [Google Scholar] [CrossRef]
- Zou, X.; Chen, W.H.; Han, Y.F.; Liang, Z.Q. A new species of the genus Gibellula. Mycosystema 2016, 35, 1161–1168. [Google Scholar] [CrossRef]
- Wang, T.; Chang, X.Y.; Huang, B.; Li, Z.Z.; Chen, M.J. Species diversity of spider-pathogenic fungi in China. Mycosystema 2024, 43, 230–243. [Google Scholar]
- Sun, T.; Zou, W.Q.; Dong, Q.Y.; Huang, O.; Tang, D.X.; Yu, H. Morphology, phylogeny, mitogenomics and metagenomics reveal a new entomopathogenic fungus Ophiocordyceps nujiangensis (Hypocreales, Ophiocordycipitaceae) from Southwestern China. MycoKeys 2022, 94, 91–108. [Google Scholar] [CrossRef]
- Wang, Y.B.; Wang, Y.; Fan, Q.; Duan, D.E.; Zhang, G.D.; Dai, R.Q.; Dai, Y.D.; Zeng, W.B.; Chen, Z.H.; Li, D.D.; et al. Multigene phylogeny of the family Cordycipitaceae (Hypocreales): New taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Divers. 2020, 103, 1–46. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Rehner, S.A.; Samuels, G.J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 1994, 98, 625–634. [Google Scholar] [CrossRef]
- Bischoff, J.F.; Rehner, S.A.; Humber, R.A. Metarhizium frigidum sp. nov.: A cryptic species of M. anisopliae and a member of the M. flavoviride complex. Mycologia 2006, 98, 737–745. [Google Scholar] [CrossRef]
- Sung, G.H.; Hywel-Jones, N.L.; Sung, J.M.; Luangsa-ard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Castlebury, L.A.; Rossman, A.Y.; Sung, G.H.; Hyten, A.S.; Spatafora, J.W. Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycol. Res. 2004, 108, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.B.; Yu, H.; Dai, Y.D.; Chen, Z.H.; Zeng, W.B.; Yuan, F.; Liang, Z.Q. Polycephalomyces yunnanensis (Hypocreales), a new species of Polycephalomyces parasitizing Ophiocordyceps nutans and stink bugs (hemipteran adults). Phytotaxa 2015, 208, 34–44. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Nei, M.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, X.C.; Wu, L.X.; Wang, Y.; Xu, A.; Lin, W.F. Blackwellomyces kaihuaensis and Metarhizium putuoense (Hypocreales), two new entomogenous fungi from subtropical forests in Zhejiang Province, Eastern China. Forests 2023, 14, 2333. [Google Scholar] [CrossRef]
- Hyde, K.D.; Tennakoon, D.S.; Jeewon, R.D.; Bhat, J.; Maharachchikumbura, S.S.N.; Rossi, W.; Leonardi, M.; Lee, H.B.; Mun, H.Y.; Houbraken, J.; et al. Fungal diversity notes 1036–1150: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2019, 96, 1–242. [Google Scholar] [CrossRef]
- Alves, J.E.R.; Santos, A.C.S.; Belo Pedroso, S.K.; Ribeiro Melo, R.F.; Tiago, P.V. Untangling a web of spider fungi: Gibellula agroflorestalis (Hypocreales, Ascomycota), a new species of spider parasite from Brazil. J. Invertebr. Pathol. 2025, 209, 108278. [Google Scholar] [CrossRef]
- Evans, H.C.; Fogg, T.; Buddie, A.G.; Yeap, Y.T.; Araújo, J.P.M. The araneopathogenic genus Gibellula (Cordycipitaceae: Hypocreales) in the British Isles, including a new zombie species on orb-weaving cave spiders (Metainae: Tetragnathidae). Fungal Syst. Evol. 2025, 15, 153–178. [Google Scholar] [CrossRef]
- Mendes-Pereira, T.; de Araújo, J.P.M.; Mendes, F.; Fonseca, E. Gibellula aurea sp. nov. (Ascomycota, Cordycipitaceae): A new golden spider-devouring fungus from a Brazilian Atlantic Rainforest. Phytotaxa 2022, 573, 85–102. [Google Scholar] [CrossRef]
- Kuephadungphan, W.; Tasanathai, K.; Petcharad, B.; Khonsanit, A.; Stadler, M.; Luangsa-ard, J.J. Phylogeny- and morphology-based recognition of new species in the spider-parasitic genus Gibellula (Hypocreales, Cordycipitaceae) from Thailand. MycoKeys 2020, 72, 17–42. [Google Scholar] [CrossRef] [PubMed]
- Kepler, R.M.; Sung, G.H.; Ban, S.; Nakagiri, A.; Chen, M.J.; Huang, B.; Li, Z.; Spatafora, J.W. New teleomorph combinations in the entomopathogenic genus Metacordyceps. Mycologia 2012, 104, 182–197. [Google Scholar] [CrossRef]
- Spatafora, J.W.; Sung, G.H.; Sung, J.M.; Hywel-Jones, N.L.; White, J.F. Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Mol. Ecol. 2007, 16, 1701–1711. [Google Scholar] [CrossRef] [PubMed]
- Kuephadungphan, W.; Macabeo, A.P.G.; Luangsa-Ard, J.J.; Tasanathai, K.; Thanakitpipattana, D.; Phongpaichit, S.; Yuyama, K.; Stadler, M. Studies on the biologically active secondary metabolites of the new spider parasitic fungus Gibellula gamsii. Mycol. Prog. 2019, 18, 135–146. [Google Scholar] [CrossRef]
- Kepler, R.M.; Luangsa-Ard, J.J.; Hywel-Jones, N.L.; Quandt, C.A.; Sung, G.H.; Rehner, S.A.; Aime, M.C.; Henkel, T.W.; Sanjuan, T.; Zare, R.; et al. A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 2017, 8, 335–353. [Google Scholar] [CrossRef]
- Liu, Z.L.; Wei, D.P.; Chen, J.H.; Wu, W.J.; Liu, Z.H.; Zhang, W.; Chen, H.; Peng, X.C.; Kang, J.C.; Qian, Y.X.; et al. A new spider-pathogenic species Gibellula liaoningensis (Cordycipitaceae) from Liaoning Province, China. Phytotaxa 2025, 702, 48–60. [Google Scholar] [CrossRef]
- Johnson, D.; Sung, G.H.; Hywel-Jones, N.L.; Luangsa-Ard, J.J.; Bischoff, J.F.; Kepler, R.M.; Spatafora, J.W. Systematics and evolution of the genus Torrubiella (Hypocreales, Ascomycota). Mycol. Res. 2009, 113, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Mendes-Pereira, T.; de Araújo, J.P.M.; Kloss, T.G.; Costa-Rezende, D.H.; de Carvalho, D.S.; Góes-Neto, A. Disentangling the taxonomy, systematics, and life history of the spider-parasitic fungus Gibellula (Cordycipitaceae, Hypocreales). J. Fungi 2023, 9, 457. [Google Scholar] [CrossRef]
- Tan, Y.P.; Shivas, R.G. Nomenclatural novelties. Index Aust. Fungi 2023, 10, 1–17. [Google Scholar]
- Mongkolsamrit, S.; Noisripoom, W.; Tasanathai, K.; Khonsanit, A.; Thanakitpipattana, D.; Himaman, W.; Kobmoo, N.; Luangsa-Ard, J.J. Molecular phylogeny and morphology reveal cryptic species in Blackwellomyces and Cordyceps (Cordycipitaceae) from Thailand. Mycol. Prog. 2020, 19, 957–983. [Google Scholar] [CrossRef]
- Chen, M.J.; Wang, T.; Lin, Y.A.N.; Huang, B.O. Gibellula flava sp. nov. (Cordycipitaceae, Hypocreales), a new pathogen of spider from China. Phytotaxa 2021, 527, 125–133. [Google Scholar] [CrossRef]
- Gao, R.X. Description of a parasitic fungus Gibellula suffulta on spiders in Fujian. Acta Microbiol. Sin. 1981, 21, 308–310. [Google Scholar]
- Tzean, S.S.; Hsieh, L.S.; Wu, W.J. Torrubiella dimorpha, a new species of spider parasite from Taiwan. Mycol. Res. 1998, 102, 1350–1354. [Google Scholar] [CrossRef]
- Kuephadungphan, W.; Phongpaichit, S.; Luangsa-Ard, J.J.; Rukachaisirikul, V. Antimicrobial activity of invertebrate-pathogenic fungi in the genera Akanthomyces and Gibellula. Mycoscience 2013, 55, 127–133. [Google Scholar] [CrossRef]
- Helaly, S.E.; Kuephadungphan, W.; Phainuphong, P.; Ibrahim, M.A.A.; Tasanathai, K.; Mongkolsamrit, S.; Luangsa-Ard, J.J.; Phongpaichit, S.; Rukachaisirikul, V.; Stadler, M. Pigmentosins from Gibellula sp. as antibiofilm agents and a new glycosylated asperfuran from Cordyceps javanica. Beilstein J. Org. Chem. 2019, 15, 2968–2981. [Google Scholar] [CrossRef] [PubMed]



| Species | Voucher/Information | GenBank Accession Number | References | |||||
|---|---|---|---|---|---|---|---|---|
| nrSSU | ITS | nrLSU | tef-1α | rpb1 | rpb2 | |||
| Blackwellomyces kaihuaensis | HMAS 285455T | OQ981975 | OQ981961 | OQ981968 | OQ980401 | OQ980409 | OQ980408 | [35] |
| Blackwellomyces lateris | MFLU 18-0663T | MK086057 | MK086059 | MK086061 | MK069471 | MK084615 | MK079354 | [36] |
| Gibellula agrofloretalis | A30 | PP958494 | N/A | N/A | PP965288 | N/A | N/A | [37] |
| Gibellula agrofloretalis | C11 | PP958496 | N/A | N/A | PP965293 | N/A | N/A | [37] |
| Gibellula agrofloretalis | D7 | PP958504 | N/A | PP958435 | PP965304 | N/A | N/A | [37] |
| Gibellula attenboroughii | IMI 507230T | PQ036924 | N/A | PQ036929 | PQ046101 | N/A | N/A | [38] |
| Gibellula aurea | LBMCF0003 | OK329880 | N/A | N/A | OK392618 | N/A | OL117022 | [39] |
| Gibellula aurea | LBMCF0006 | N/A | N/A | OK329875 | OK392624 | N/A | OK315662 | [39] |
| Gibellula aurea | LBMCF0007 | N/A | OK329885 | OK329876 | OK392622 | N/A | OK315663 | [39] |
| Gibellula baishanensis | GMBC 3152T | PX425053 | PX425062 | PX425069 | PX434402 | PX434411 | PX434420 | This study |
| Gibellula baishanensis | GMBC 3153 | PX425054 | PX425063 | PX425070 | PX434403 | PX434412 | PX434421 | This study |
| Gibellula brevistipitata | BCC 45580T | N/A | OK040729 | OK040706 | OK040697 | OK040715 | N/A | [7] |
| Gibellula cebrennini | BCC 53605T | N/A | MT477069 | MT477062 | MT503328 | MT503321 | MT503336 | [40] |
| Gibellula cebrennini | BCC 39705 | N/A | MH532874 | MH394673 | MH521895 | MH521822 | MH521859 | [40] |
| Gibellula clavulifera var. alba | ARSEF 1915T | DQ522562 | JN049837 | DQ518777 | DQ522360 | DQ522408 | DQ522467 | [41,42] |
| Gibellula dimorpha | BCC 47518 | N/A | MH532884 | MH394679 | MH521892 | MH521819 | MH521863 | [7] |
| Gibellula flava | GNJ20200814-46 | MW969660 | N/A | MW969673 | MW961413 | MW980146 | N/A | [18] |
| Gibellula flava | WFS20190625-25T | MW036749 | N/A | MW084343 | MW091325 | MW384883 | N/A | [18] |
| Gibellula fusiformispora | BCC 56802T | N/A | MT477070 | MT477063 | MT503329 | MT503322 | MT503337 | [40] |
| Gibellula fusiformispora | BCC 45076 | N/A | MH532882 | N/A | N/A | MH521823 | MH521860 | [40] |
| Gibellula gamsii | BCC 27968T | N/A | MH152529 | MH152539 | MH152560 | MH152547 | N/A | [43] |
| Gibellula gamsii | BCC 29228 | N/A | MH152533 | MH152543 | MH152564 | MH152551 | MH152558 | [43] |
| Gibellula gamsii | EPF034 | N/A | JX192720 | JX192753 | JX192817 | N/A | N/A | [43] |
| Gibellula jilinensis | GMBC 3154 | PX425050 | PX425059 | PX425066 | PX434399 | PX434408 | PX434417 | This study |
| Gibellula jilinensis | GMBC 3157 | PX425051 | PX425060 | PX425067 | PX434400 | PX434409 | PX434418 | This study |
| Gibellula jilinensis | GMBC 3160T | PX425052 | PX425061 | PX425068 | PX434401 | PX434410 | PX434419 | This study |
| Gibellula kunmingensis | GMBC 3148T | PX425057 | PX425064 | PX425073 | PX434406 | PX434415 | PX434424 | This study |
| Gibellula kunmingensis | GMBC 3149 | PX425058 | PX425065 | PX425074 | PX434407 | PX434416 | PX434425 | This study |
| Gibellula leiopus | BCC 16025 | N/A | N/A | MF416548 | MF416492 | MF416649 | N/A | [44] |
| Gibellula leiopus | BCC 49250 | N/A | OK070780 | OK070781 | OK070782 | OK070783 | OK070784 | [7] |
| Gibellula liaoningensis | HKAS 145357 | PQ817100 | PQ817098 | PQ817102 | PQ815114 | PQ815116 | PQ815118 | [45] |
| Gibellula liaoningensis | HKAS 145358T | PQ817099 | PQ817097 | PQ817101 | PQ815113 | PQ815115 | PQ815117 | [45] |
| Gibellula longicaudata | BCC 40861 | N/A | OK040730 | OK040707 | OK040698 | OK040716 | OK040724 | [7] |
| Gibellula longispora | NHJ 12014 | EU369098 | N/A | N/A | EU369017 | EU369055 | EU369075 | [46] |
| Gibellula longispora | GNJ20200813-16 | N/A | N/A | N/A | MW961414 | MW980145 | N/A | [18] |
| Gibellula longispora | GNJ20210710-02 | OL854201 | N/A | OL854212 | OL981628 | N/A | OL981635 | [18] |
| Gibellula mainsii | LBMCF2022.96 | OQ585789 | OQ589484 | N/A | OQ658392 | N/A | N/A | [47] |
| Gibellula mirabilis | LBMCF2021.70 | OQ585786 | OQ589481 | OQ585976 | OQ658389 | N/A | N/A | [47] |
| Gibellula mirabilis | LBMCF2021.80 | OQ585787 | OQ589482 | OQ585977 | OQ658390 | N/A | N/A | [47] |
| Gibellula mirabilis | LBMCF2022.107 | OQ585792 | N/A | OQ585979 | OQ658395 | N/A | N/A | [47] |
| Gibellula nigelii | NHJ 10808T | EU369099 | N/A | EU369035 | EU369018 | EU369056 | EU369076 | [46] |
| Gibellula paralongispora | GMBC 3162T | PX425055 | N/A | PX425071 | PX434404 | PX434413 | PX434422 | This study |
| Gibellula paralongispora | GMBC 3163 | PX425056 | N/A | PX425072 | PX434405 | PX434414 | PX434423 | This study |
| Gibellula parvula | BCC 48888 | N/A | OK040731 | OK040708 | OK040699 | OK040717 | OK040725 | [7] |
| Gibellula parvula | BCC 49748T | N/A | OK040732 | OK040709 | OK040700 | OK040718 | OK040726 | [7] |
| Gibellula penicillioides | GNJ20200814-11 | MW969650 | MW969669 | MW969661 | MW961415 | MZ215998 | N/A | [18] |
| Gibellula penicillioides | GNJ20200814-14T | MW969651 | MW969670 | MW969662 | MW961416 | MZ215999 | N/A | [18] |
| Gibellula penicillioides | GNJ20200814-17 | MW969652 | MW969671 | MW969663 | MW961417 | N/A | N/A | [18] |
| Gibellula pigmentosinum | BCC 38246 | N/A | MH532872 | MH394672 | MH521893 | MH521800 | MH521855 | [40] |
| Gibellula pigmentosinum | BCC 41203T | N/A | MT477071 | MT477064 | MT503330 | MT503323 | N/A | [40] |
| Gibellula pilosa | BCC 57817T | N/A | OK040733 | OK040710 | OK040701 | OK040719 | N/A | [7] |
| Gibellula pulchra | BCC 47555 | N/A | MH532885 | N/A | MH521897 | MH521804 | N/A | [7] |
| Gibellula pulchra | LBMCF2020.02 | OQ585783 | N/A | OQ585973 | OQ658386 | N/A | N/A | [47] |
| Gibellula pulchra | LBMCF2020.03 | OQ585784 | N/A | OQ585974 | OQ658387 | N/A | N/A | [47] |
| Gibellula pulchra | LBMCF2020.07 | OQ585785 | N/A | OQ585975 | OQ658388 | N/A | N/A | [47] |
| Gibellula pulchra | LBMCF2022.GA | OQ585780 | N/A | OQ585970 | OQ658383 | N/A | N/A | [47] |
| Gibellula pulchra | LBMCF2022.GB | OQ585781 | OQ589487 | OQ585971 | OQ658384 | N/A | N/A | [47] |
| Gibellula queenslandica | BRIP 72767aT | N/A | OR452099 | OR452103 | OR459912 | N/A | OR459907 | [48] |
| Gibellula scorpioides | BCC 47976T | N/A | MT477078 | MT477066 | MT503335 | MT503325 | MT503339 | [40] |
| Gibellula solita | BCC 45574T | N/A | OK040736 | OK040712 | OK040703 | OK040721 | N/A | [7] |
| Gibellula sp. | NHJ 5401 | EU369102 | N/A | N/A | N/A | EU369059 | EU369079 | [46] |
| Gibellula sp. | NHJ 10788 | EU369101 | N/A | EU369036 | EU369019 | EU369058 | EU369078 | [46] |
| Gibellula trimorpha | BCC 36526T | N/A | OK040737 | N/A | OK040704 | OK040722 | OK040728 | [7] |
| Gibellula trimorpha | BCC 36538 | N/A | MH532867 | MH394668 | MH521890 | MH521817 | MH521861 | [7] |
| Gibellula unica | BCC 45112 | N/A | OK040738 | N/A | OK040705 | OK040723 | N/A | [7] |
| Gibellula unica | BCC 46590 | N/A | MH532883 | MH394678 | N/A | MH521803 | MH521866 | [7] |
| Gibellula yunnanensis | GMB 3142T | PX354537 | PX354531 | PX354543 | PX370035 | PX371911 | PX371921 | This study |
| Gibellula yunnanensis | GMB 3143 | PX354538 | PX354532 | PX354544 | PX370036 | PX371912 | PX371922 | This study |
| Hevansia arachnophila | NHJ 2633 | N/A | MH532900 | GQ249978 | MH521917 | MH521843 | MH521884 | [43] |
| Hevansia minula | BCC 47519T | N/A | MZ684087 | MZ684002 | MZ707811 | MZ707826 | MZ707833 | [9] |
| Hevansia minula | BCC 47520 | N/A | MZ684088 | MZ684003 | MZ707812 | MZ707827 | MZ707834 | [9] |
| Hevansia nelumboides | TNS 16306 | MF416585 | N/A | N/A | MF416475 | N/A | MF416438 | [44] |
| Hevansia novoguineensis | BCC 42675 | N/A | MZ684089 | MZ684004 | MZ707814 | N/A | MZ707835 | [9] |
| Hevansia novoguineensis | CBS 610.80T | N/A | MH532831 | MH394646 | MH521885 | N/A | MH521844 | [49] |
| Jenniferia cinerea | NHJ 03510T | N/A | N/A | N/A | EU369009 | EU369048 | EU369070 | [46] |
| Jenniferia cinerea | BCC 2191 | GQ249956 | GQ250000 | GQ249971 | GQ250029 | N/A | N/A | [43] |
| Jenniferia griseocinerea | BCC 42062T | N/A | MZ684091 | MZ684006 | MZ707815 | MZ707828 | MZ707837 | [9] |
| Jenniferia griseocinerea | BCC 42063 | N/A | MZ684092 | MZ684007 | MZ707816 | MZ707829 | MZ707838 | [9] |
| Jenniferia thomisidarum | BCC 37881T | N/A | MZ684099 | MZ684010 | MZ707823 | MZ707830 | MZ707843 | [9] |
| Jenniferia thomisidarum | BCC 37882 | N/A | MZ684100 | MZ684011 | MZ707824 | MZ707831 | MZ707844 | [9] |
| Species | Conidiophore (μm) | Metulae (μm) | Phialide (μm) | Conidia (μm) | References |
|---|---|---|---|---|---|
| Gibellula attenboroughii | Verrucose, 80–120 × 5–8 | Borne on vesicle, 10–12 × 6–8 | Cylindrical to narrowly clavate, 7.5–9.5 × 2.5–3.5 | Hyaline, smooth, 4–6 × 1.5–2 | [38] |
| G. baishangensis | Verrucose to globose, 69–89 × 8–11 | Broadly ovoid to broadly ellipsoid, 5–11 × 6–7 | Spherical to ovoid, 10–14 × 2–3 | Hyaline, smooth, 5.5–7 × 2–4 | This study |
| G. flava | Verrucose, 33.5–123.5 × 4–9.5 | Obovoid to broadly obovoid, 5.5–7 × 3.5–5.5 | Narrowly obovate to clavate, 5.5–7 × 1.5–2.5 | Fusiform, 3–4 × 1–2 | [50] |
| G. jilinensis | Verrucose, 78–109 × 6–8 | Broadly ovoid to broadly ellipsoid, 7–10 × 5–7 | Cylindrical to lageniform, 9–15 × 2–3 | Hyaline, smooth, 5.5–6.5 × 2–3 | This study |
| G. kunmingensis | Verrucose to globose, 52–120 × 14–16 | Broadly ovoid to broadly ellipsoid, 13–17 × 5–7 | Cylindrical to lageniform, borne on vesicles, 12–21 × 7–8 | Hyaline, smooth, 2–4 × 1.2–2 | This study |
| G. longispora | Multiseptate, minutely roughened, 159.5–290.5 × 8.5–11 | Broadly obovoid, 7.5–9.5 × 6–6.5 | Narrowly clavate to cylindrical, 9.5–11 × 3–3.5 | Bacilliform to cylindrical, 5.5–8 × 1–1.5 | [8] |
| G. paralongispora | Verrucose, 111–188 × 8–10 | Broadly obovate to oval, 6–11 × 4–7 | Clavate, 9–11 × 2–5 | Narrowly fusiform, 4.5–6 × 1.5–2 | This study |
| G. penicillioides | Penicillate, smooth, mostly biverticillate or terverticillate, 52.5–92 × 4.5–6 | Obovoid to cylindrical, 13–17.5 × 3.5–5 | Broadly cylindrical, 12.5–15.5 × 3–4 | 7.5–9 × 2.5–3.5 | [18] |
| G. pigmentosinum | Smooth to verrucose, 97.5–170 × 7–10 | Broadly obovoid, 6–8 × 4–6 | Obovoid to clavate, 5.5–8 × 2–3 | Obovoid with an acute Apex 3.5–5 × 1–2 | [40] |
| G. pilosa | Minutely roughened, 151–265 × 9–11 | Broadly obovoid, 9.5–11 × 7–8 | Narrowly clavate to cylindrical, 7–9 × 2.5–3 | Narrowly almond shaped, 3–4 × 1.5–2 | [8] |
| G. pulchra | Verrucose, 155–170 × 7.5–10 | Cylindrical, 6.2–7.5 × 5–6 | Clavate, 7.5–8 × 1.5–2.5 | Fusiform to fusiform ellipsoid, 3–5 × 1.5–2.5 | [17] |
| G. solita | Verrucose, 82–146 × 7.5–9.5 | Broadly obovoid, 7–7.5 × 5–6 | Narrowly clavate to cylindrical, 6–7 × 2–2.5 | Occasionally globose, 2–2.5 × 1–1.5 | [8] |
| G. yunnanensis | Verrucose, 27–58 × 3–8 | Broadly obovate to oval, 6.4–9.7 × 5–7 | Borne on vesicle, 6.4–9.7 × 5–7 | Narrowly fusiform, 6–9 × 2.2–3.1 | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, B.; Chen, H.; Zhang, X.; Guan, Y.-H.; Tang, D.-X.; Li, Q.-R.; Wang, Y. Five New Species of Gibellula (Hypocreales, Cordycipitaceae) from China. J. Fungi 2025, 11, 891. https://doi.org/10.3390/jof11120891
Tu B, Chen H, Zhang X, Guan Y-H, Tang D-X, Li Q-R, Wang Y. Five New Species of Gibellula (Hypocreales, Cordycipitaceae) from China. Journal of Fungi. 2025; 11(12):891. https://doi.org/10.3390/jof11120891
Chicago/Turabian StyleTu, Bo, Hui Chen, Xu Zhang, Yu-Hu Guan, De-Xiang Tang, Qi-Rui Li, and Yao Wang. 2025. "Five New Species of Gibellula (Hypocreales, Cordycipitaceae) from China" Journal of Fungi 11, no. 12: 891. https://doi.org/10.3390/jof11120891
APA StyleTu, B., Chen, H., Zhang, X., Guan, Y.-H., Tang, D.-X., Li, Q.-R., & Wang, Y. (2025). Five New Species of Gibellula (Hypocreales, Cordycipitaceae) from China. Journal of Fungi, 11(12), 891. https://doi.org/10.3390/jof11120891





