Crocodylus porosus Sera a Potential Source to Identify Novel Epigenetic Targets: In Silico Analysis
Abstract
:1. Introduction
2. Methods
2.1. In-Silico Analysis
2.2. Literature Review
3. Results
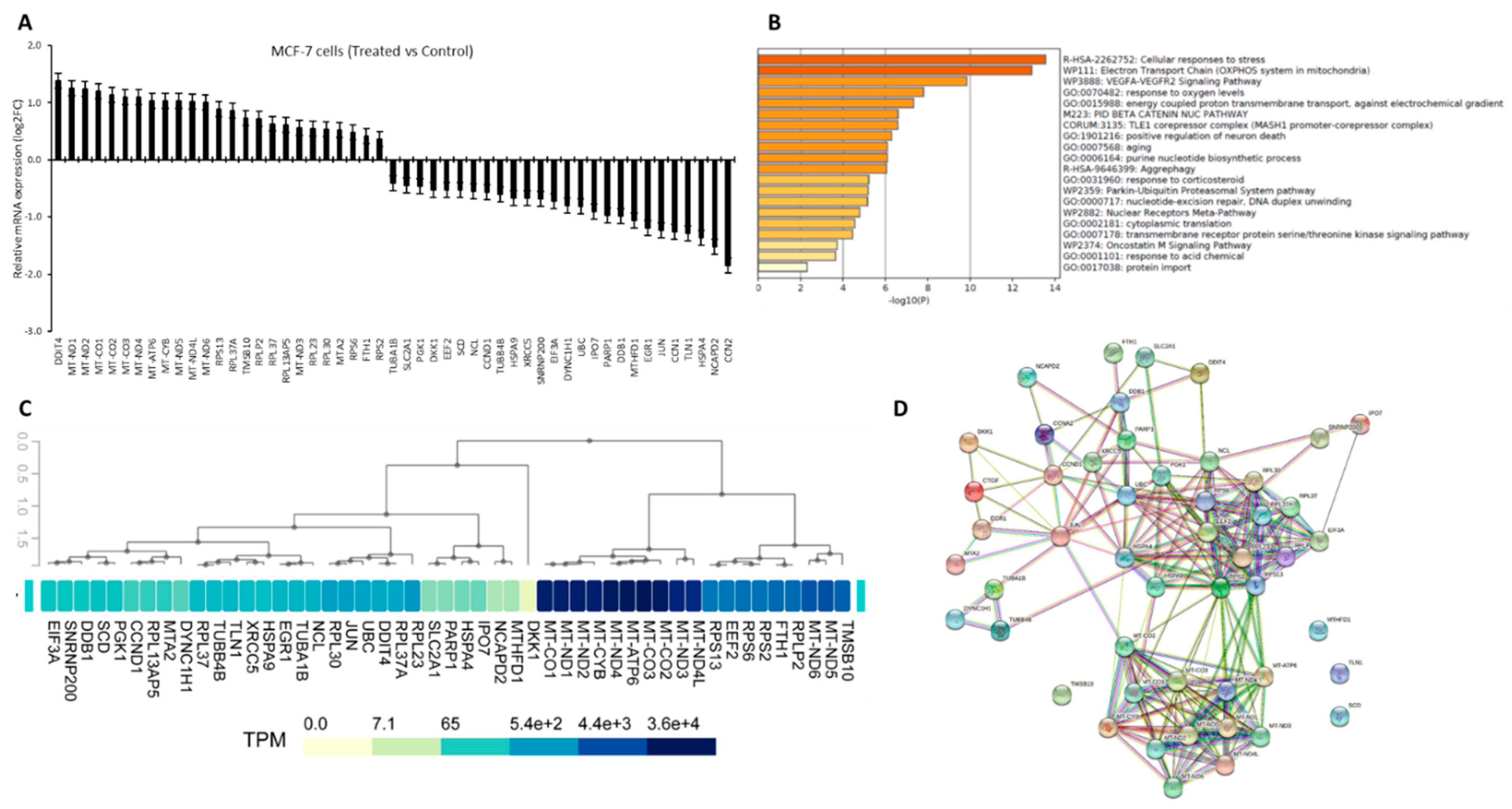
3.1. Crocodylus Porosus Serum-Induced Differentially Expressed Targets Genes in MCF-7 Cells
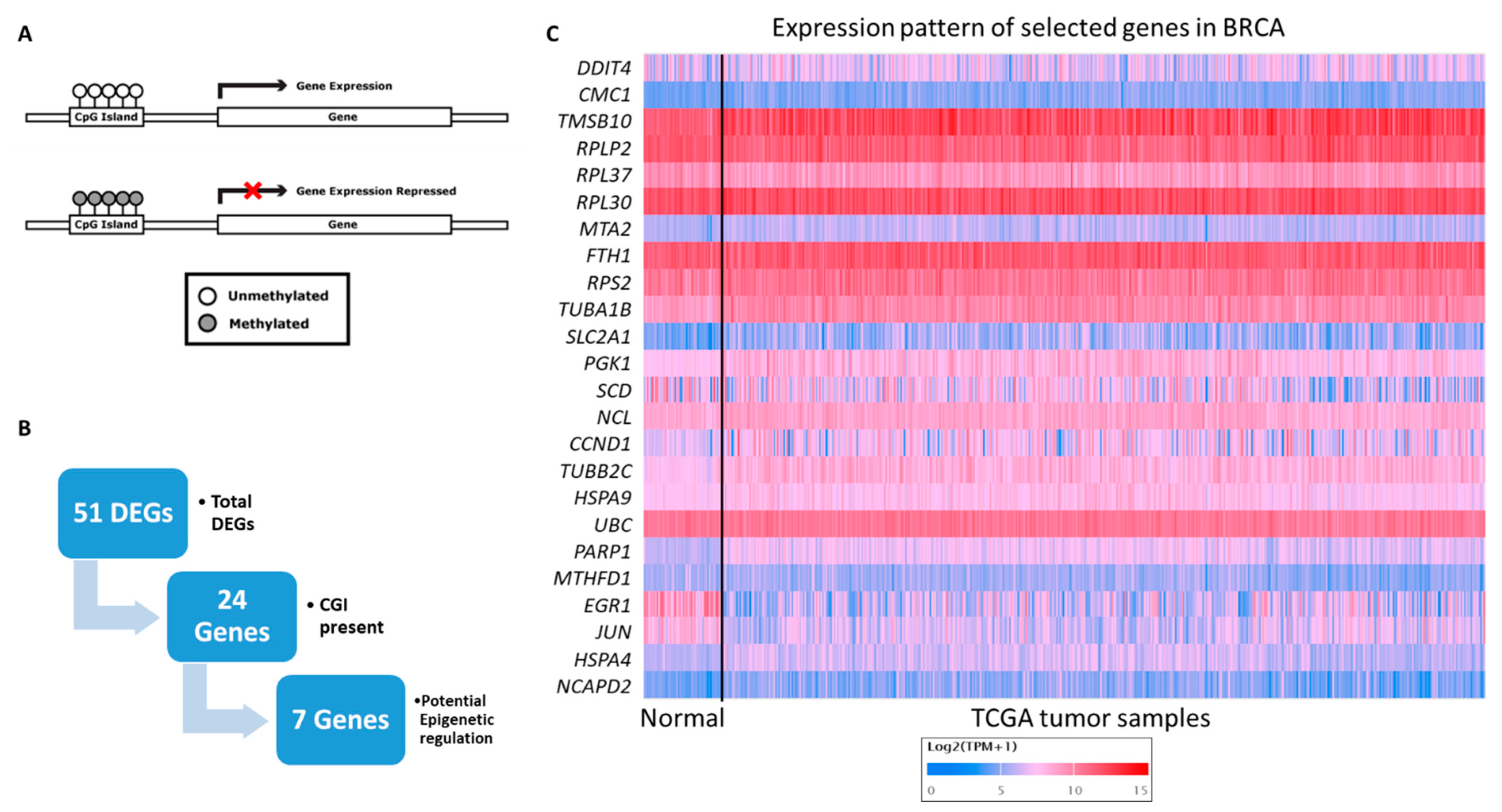
3.2. Gene Filtration for Potential Epigenetic Regulation in Breast Cancer
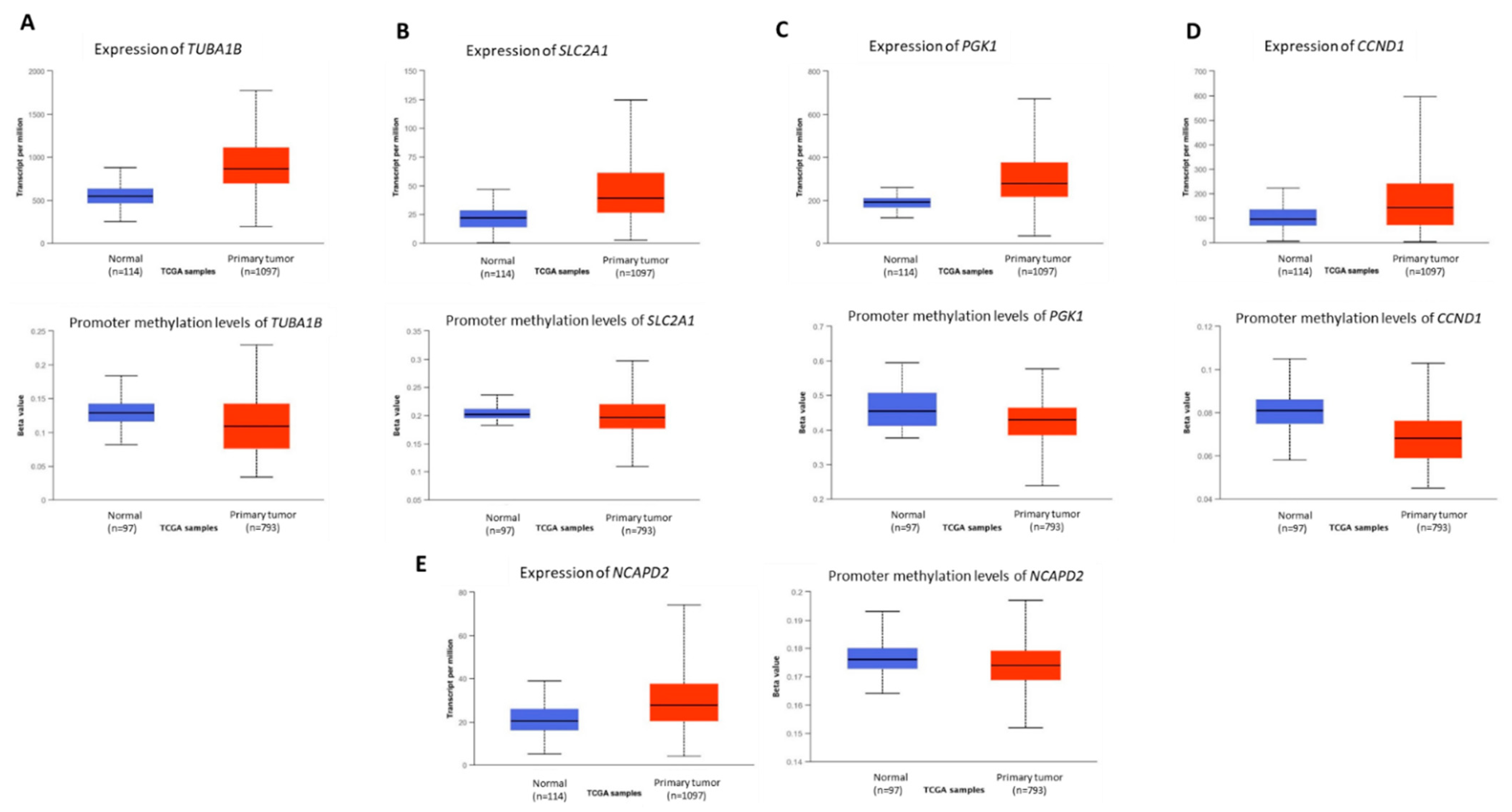
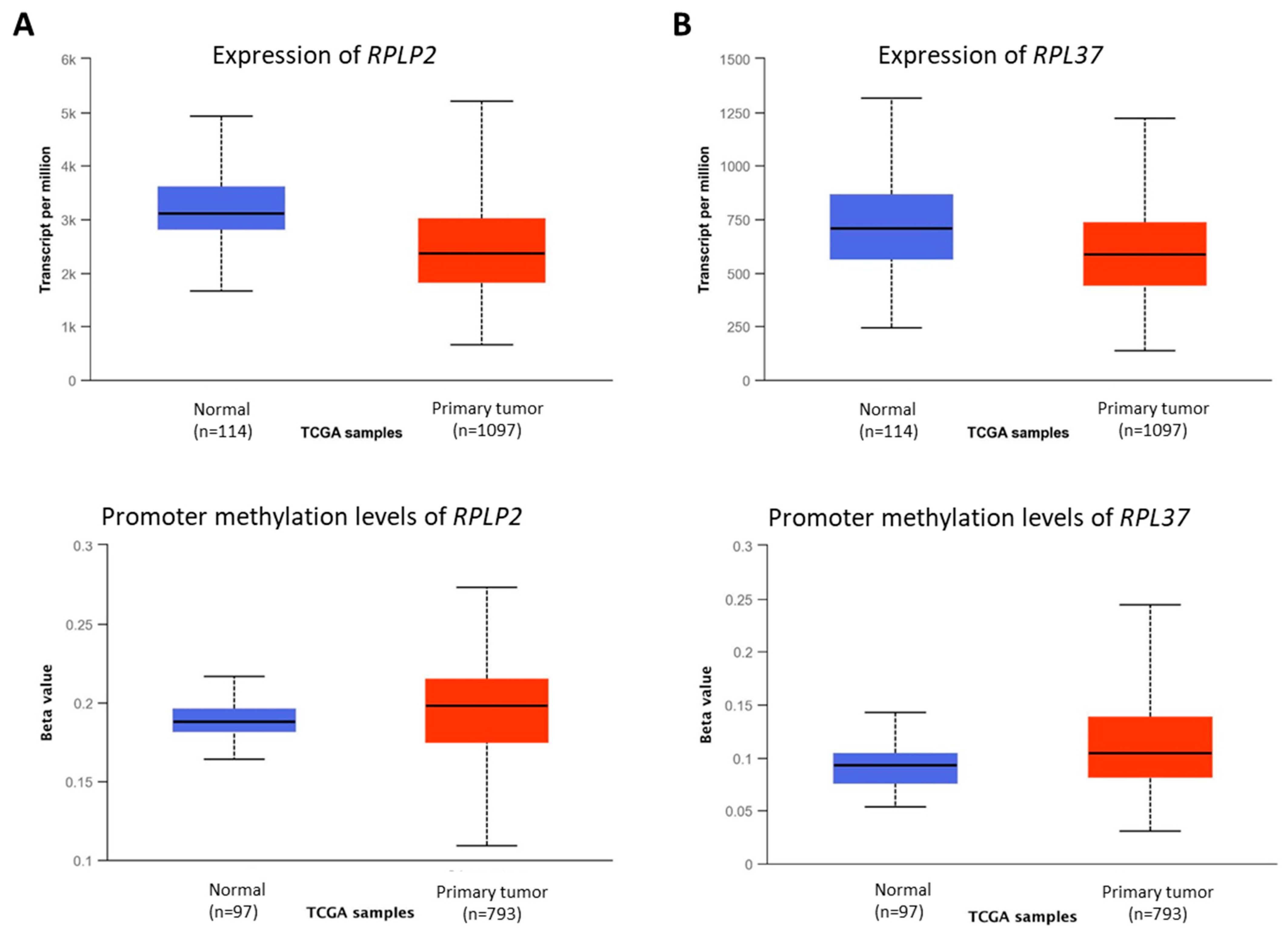
3.3. Novel Genes Which Might Be the Target of Crocodile Serum
| Gene Symbol | Genamee n | Function (genecards.org) | Reference(s) Related to Breast Cancer |
|---|---|---|---|
| TUBA1B | Tubulin Alpha 1b | Involved in cell cycle spindle assembly and chromosome separation | [11,12,13,14,15] |
| SLC2A1 | Solute Carrier Family 2 Member 1 | Glucose transporter responsible for constitutive glucose uptake | [16,17,18,19,20,21,22] |
| PGK1 | Phosphoglycerate Kinase 1 | Participates in energy production via glycolysis and tumor cell angiogenesis | [23,24,25,26,27] |
| NCAPD2 | Non-SMC Condensin I Complex Subunit D2 | Required for conversion of interphase chromatin into mitotic-like condense chromosomes | [28,29] |
| CCND1 | Cyclin D1 | Progression of the cell cycle and to induce the Warburg effect in cancer cells | [30,31] |
| RPLP2 | Ribosomal Protein Lateral Stalk Subunit P2 | An important role in the elongation step of protein synthesis | [32] |
| RPL37 | Ribosomal Protein L37 | Involved in rRNA processing in the nucleus and cytosol | [33] |
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siddiqui, R.; Jeyamogan, S.; Ali, S.M.; Abbas, F.; Sagathevan, K.A.; Khan, N.A. Crocodiles and alligators: Antiamoebic and antitumor compounds of crocodiles. Exp. Parasitol. 2017, 183, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Jeyamogan, S.; Khan, N.A.; Siddiqui, R. Animals living in polluted environments are a potential source of anti-tumor molecule (s). Cancer Chemother. Pharmacol. 2017, 80, 919–924. [Google Scholar] [CrossRef] [PubMed]
- Jeyamogan, S.; Khan, N.A.; Sagathevan, K.; Siddiqui, R. Sera/organ lysates of selected animals living in polluted environments exhibit cytotoxicity against cancer cell lines. Anti-Cancer Agents Med. Chem. Former. Curr. Med. Chem. Anti-Cancer Agents 2019, 19, 2251–2268. [Google Scholar] [CrossRef] [PubMed]
- Akbar, N.; Siddiqui, R.; Sagathevan, K.; Iqbal, M.; Khan, N.A. Gut bacteria of water monitor lizard (Varanus salvator) are a potential source of antibacterial compound (s). Antibiotics 2019, 8, 164. [Google Scholar] [CrossRef] [Green Version]
- Heerboth, S.; Lapinska, K.; Snyder, N.; Leary, M.; Rollinson, S.; Sarkar, S. Use of epigenetic drugs in disease: An overview. Genet. Epigenet. 2014, 6, 9–19. [Google Scholar] [CrossRef]
- Dworkin, A.M.; Huang, T.H.; Toland, A.E. Epigenetic alterations in the breast: Implications for breast cancer detection, prognosis and treatment. Semin. Cancer Biol. 2009, 19, 165–171. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, J.S.; Guimei, M.; Jayakumar, M.N.; Shafarin, J.; Janeeh, A.S.; AbuJabal, R.; Jabal, R.; Eladl, M.; Ranade, A.; Ali, A.; et al. Estrogen-induced hypomethylation and overexpression of YAP1 facilitate breast cancer cell growth and survival. Neoplasia 2021, 23, 68–79. [Google Scholar] [CrossRef]
- Jeyamogan, S.K.N.; Sagathevan, K.; Siddiqui, R. Crocodylus porosus: A potential source of anticancer molecules. BMJ Open Sci. 2020, 4, e100040. [Google Scholar] [CrossRef]
- Muhammad, J.S.; Bajbouj, K.; Shafarin, J.; Hamad, M. Estrogen-induced epigenetic silencing of FTH1 and TFRC genes reduces liver cancer cell growth and survival. Epigenetics 2020, 15, 1302–1318. [Google Scholar] [CrossRef]
- Muhammad, J.S.; Khan, M.R.; Ghias, K. DNA methylation as an epigenetic regulator of gallbladder cancer: An overview. Int. J. Surg. 2018, 53, 178–183. [Google Scholar] [CrossRef]
- Kim, N.D.; Park, E.S.; Kim, Y.H.; Moon, S.K.; Lee, S.S.; Ahn, S.K.; Yu, D.Y.; No, K.T.; Kim, K.H. Structure-based virtual screening of novel tubulin inhibitors and their characterization as anti-mitotic agents. Bioorganic Med. Chem. 2010, 18, 7092–7100. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Zhang, J.; He, S.; Wan, C.; Shan, A.; Wang, Y.; Yu, L.; Liu, G.; Chen, K.; Shi, J.; et al. Increased alpha-tubulin1b expression indicates poor prognosis and resistance to chemotherapy in hepatocellular carcinoma. Dig. Dis. Sci. 2013, 58, 2713–2720. [Google Scholar] [CrossRef] [PubMed]
- Blenk, S.; Engelmann, J.C.; Pinkert, S.; Weniger, M.; Schultz, J.; Rosenwald, A.; Müller-Hermelink, H.; Müller, T.; Dandeka, T. Explorative data analysis of MCL reveals gene expression networks implicated in survival and prognosis supported by explorative CGH analysis. BMC Cancer 2008, 8, 106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lou, W.; Ding, B.; Zhong, G.; Yao, J.; Fan, W.; Fu, P. RP11-480I12.5-004 Promotes Growth and Tumorigenesis of Breast Cancer by Relieving miR-29c-3p-Mediated AKT3 and CDK6 Degradation. Mol. Ther. Nucleic Acids 2020, 21, 916–931. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Arai, E.; Makiuchi, S.; Tsuda, N.; Kuramoto, J.; Ohara, K.; Takahashi, Y.; Ito, N.; Ojima, H.; Hiraoka, N.; et al. Aberrant DNA methylation results in altered gene expression in non-alcoholic steatohepatitis-related hepatocellular carcinomas. J. Cancer Res. Clin. Oncol. 2020, 146, 2461–2477. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Ba-Alawi, W.; Deblois, G.; Cruickshank, J.; Duan, S.; Lima-Fernandes, E.; Haight, J.; Tonekaboni, S.; Fortier, A.; Kuasne, H.; et al. GLUT1 inhibition blocks growth of RB1-positive triple negative breast cancer. Nat. Commun. 2020, 11, 4205. [Google Scholar] [CrossRef]
- de Castro, T.B.; Mota, A.L.; Bordin-Junior, N.A.; Neto, D.S.; Zuccari, D. Immunohistochemical Expression of Melatonin Receptor MT1 and Glucose Transporter GLUT1 in Human Breast Cancer. Anticancer Agents Med. Chem. 2018, 18, 2110–2116. [Google Scholar] [CrossRef]
- Hamann, I.; Krys, D.; Glubrecht, D.; Bouvet, V.; Marshall, A.; Vos, L.; Mackey, J.R.; Wuest, M.; Wuest, F. Expression and function of hexose transporters GLUT1, GLUT2, and GLUT5 in breast cancer-effects of hypoxia. FASEB J. 2018, 32, 5104–5118. [Google Scholar] [CrossRef]
- Wellberg, E.A.; Johnson, S.; Finlay-Schultz, J.; Lewis, A.S.; Terrell, K.L.; Sartorius, C.A.; Abel, E.; Muller, W.; Anderson, S. The glucose transporter GLUT1 is required for ErbB2-induced mammary tumorigenesis. Breast Cancer Res. 2016, 18, 131. [Google Scholar] [CrossRef] [Green Version]
- Oh, S.; Kim, H.; Nam, K.; Shin, I. Glut1 promotes cell proliferation, migration and invasion by regulating epidermal growth factor receptor and integrin signaling in triple-negative breast cancer cells. BMB Rep. 2017, 50, 132–137. [Google Scholar] [CrossRef]
- Zhao, F.; Ming, J.; Zhou, Y.; Fan, L. Inhibition of Glut1 by WZB117 sensitizes radioresistant breast cancer cells to irradiation. Cancer Chemother. Pharmacol. 2016, 77, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zou, J.; Deng, T.; Liu, J. Clinicopathological and prognostic significance of GLUT1 in breast cancer: A meta-analysis. Med. Baltim. 2018, 97, e12961. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Wu, C.; Wang, M.; Wei, K.; Wang, J. Identification of novel cell glycolysis related gene signature predicting survival in patients with breast cancer. Sci. Rep. 2021, 11, 3986. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, J.; Zhuang, J.; Liu, C.; Gao, C.; Li, H.; Ma, X.; Li, J.; Sun, C. A Novel Glycolysis-Related Four-mRNA Signature for Predicting the Survival of Patients with Breast Cancer. Front. Genet. 2021, 12, 606937. [Google Scholar] [CrossRef]
- Fu, D.; He, C.; Wei, J.; Zhang, Z.; Luo, Y.; Tan, H.; Ren, C. PGK1 is a Potential Survival Biomarker and Invasion Promoter by Regulating the HIF-1alpha-Mediated Epithelial-Mesenchymal Transition Process in Breast Cancer. Cell Physiol. Biochem. 2018, 51, 2434–2444. [Google Scholar] [CrossRef]
- Sun, S.; Liang, X.; Zhang, X.; Liu, T.; Shi, Q.; Song, Y.; Jiang, Y.; Wu, H.; Jiang, Y.; Lu, X.; et al. Phosphoglycerate kinase-1 is a predictor of poor survival and a novel prognostic biomarker of chemoresistance to paclitaxel treatment in breast cancer. Br. J. Cancer 2015, 112, 1332–1339. [Google Scholar] [CrossRef] [Green Version]
- Shashni, B.; Sakharkar, K.R.; Nagasaki, Y.; Sakharkar, M.K. Glycolytic enzymes PGK1 and PKM2 as novel transcriptional targets of PPARgamma in breast cancer pathophysiology. J. Drug Target 2013, 21, 161–174. [Google Scholar] [CrossRef]
- Ball, A.R., Jr.; Schmiesing, J.A.; Zhou, C.; Gregson, H.C.; Okada, Y.; Doi, T.; Yokomri, K. Identification of a chromosome-targeting domain in the human condensin subunit CNAP1/hCAP-D2/Eg7. Mol. Cell Biol. 2002, 22, 5769–5781. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, F.; Zhang, C.; Ren, M.; Kuang, M.; Xiao, T.; Di, X.; Feng, L.; Fu, L.; Cheng, S. Non-SMC Condensin I Complex Subunit D2 Is a Prognostic Factor in Triple-Negative Breast Cancer for the Ability to Promote Cell Cycle and Enhance Invasion. Am. J. Pathol. 2020, 190, 37–47. [Google Scholar] [CrossRef]
- Gan, S.; Dai, H.; Li, R.; Liu, W.; Ye, R.; Ha, Y.; Di, X.; Hu, W.; Zhang, Z.; Sun, Y. Identification of key differentially expressed genes between ER-positive/HER2-negative breast cancer and ER-negative/HER2-negative breast cancer using integrated bioinformatics analysis. Gland. Surg. 2020, 9, 661–675. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, A.; Bao, P.P.; Lin, L.; Wang, Y.; Wu, H.; Shu, X.; Liu, A.; Cai, Q. MicroRNA-374b inhibits breast cancer progression through regulating CCND1 and TGFA genes. Carcinogenesis 2021, 42, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Leong, S.; McKay, M.J.; Christopherson, R.I.; Baxter, R.C. Biomarkers of breast cancer apoptosis induced by chemotherapy and TRAIL. J. Proteome Res. 2012, 11, 1240–1250. [Google Scholar] [CrossRef]
- Barros, F.M.C.; Katayama, M.L.; Brentani, H.; Abreu, A.P.; Barbosa, E.M.; Oliveira, C.T.; Góes, J.; Brentani, M.; Folgueira, M. Gene trio signatures as molecular markers to predict response to doxorubicin cyclophosphamide neoadjuvant chemotherapy in breast cancer patients. Braz. J. Med. Biol. Res. 2010, 43, 1225–1231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganesan, A.; Arimondo, B.; Rots, M.G.; Jeronimo, C.; Berdasco, M. The timeline of epigenetic drug discovery: From reality to dreams. Clin. Epigenetics 2019, 11, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siddiqui, R.; Muhammad, J.S.; Maciver, S.K.; Khan, N.A. Crocodylus porosus Sera a Potential Source to Identify Novel Epigenetic Targets: In Silico Analysis. Vet. Sci. 2022, 9, 210. https://doi.org/10.3390/vetsci9050210
Siddiqui R, Muhammad JS, Maciver SK, Khan NA. Crocodylus porosus Sera a Potential Source to Identify Novel Epigenetic Targets: In Silico Analysis. Veterinary Sciences. 2022; 9(5):210. https://doi.org/10.3390/vetsci9050210
Chicago/Turabian StyleSiddiqui, Ruqaiyyah, Jibran Sualeh Muhammad, Sutherland K. Maciver, and Naveed Ahmed Khan. 2022. "Crocodylus porosus Sera a Potential Source to Identify Novel Epigenetic Targets: In Silico Analysis" Veterinary Sciences 9, no. 5: 210. https://doi.org/10.3390/vetsci9050210
APA StyleSiddiqui, R., Muhammad, J. S., Maciver, S. K., & Khan, N. A. (2022). Crocodylus porosus Sera a Potential Source to Identify Novel Epigenetic Targets: In Silico Analysis. Veterinary Sciences, 9(5), 210. https://doi.org/10.3390/vetsci9050210









