Seven-Year Evolution of β-Lactam Resistance Phenotypes in Escherichia coli Isolated from Young Diarrheic and Septicaemic Calves in Belgium
Abstract
:1. Introduction
2. Material and Methods
2.1. E. coli Isolation
2.2. Disk Diffusion Assay
2.3. Statistical Analysis
3. Results
3.1. Evolution of β-Lactam Resistance
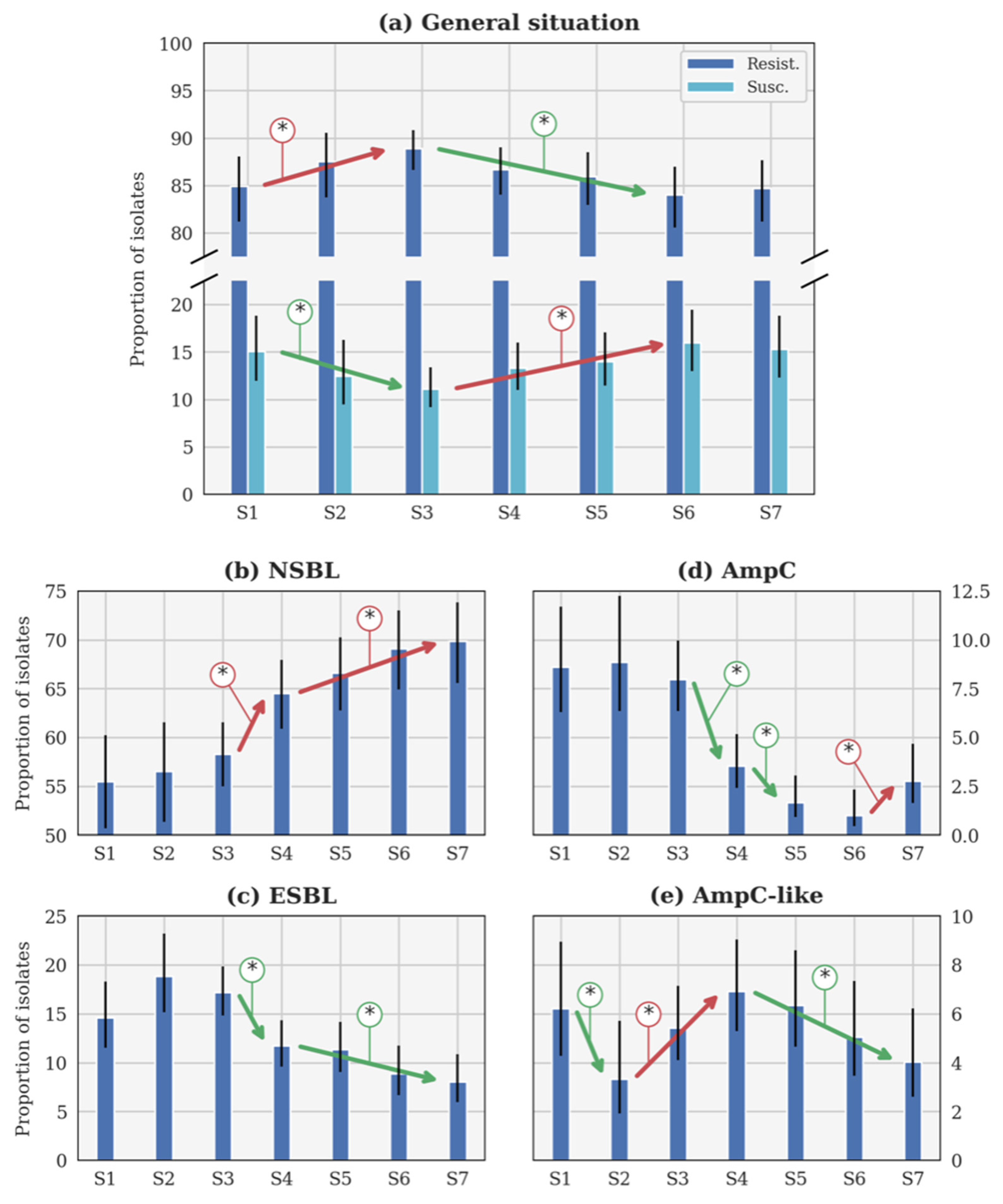
3.1.1. General Evolution of β-Lactam Resistance
3.1.2. Evolution of the NSBL Profile
3.1.3. Evolution of the ESBL Profile
3.1.4. Evolution of the AmpC Profile
3.1.5. Evolution of the AmpC-like Profile
3.2. Comparison of β-Lactam Resistance Profiles between the Escherichia coli Isolates by Origin
3.3. Comparison of β-Lactam Resistance Profiles between the Escherichia coli Virulotypes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Sykes, R.B.; Matthew, M. The ß-lactamases of gram-negative bacteria and their role in resistance to ß-lactam antibiotics. J. Antimicrob. Chemother. 1976, 2, 115–157. [Google Scholar] [CrossRef] [PubMed]
- Guérin, V.; Thiry, D.; Lucas, P.; Blanchard, Y.; Cawez, F.; Mercuri, P.S.; Galleni, M.; Saulmont, M.; Mainil, J. Identification of β-Lactamase-Encoding (bla) Genes in Phenotypically β-Lactam-Resistant Escherichia coli Isolated from Young Calves in Belgium. Microb. Drug Resist. 2021, 27, 1578–1584. [Google Scholar] [CrossRef] [PubMed]
- Courvalin, P.; LeClercq, R.; Rice, L.B. Antibiogramme, 3rd ed.; Eska: Paris, France, 2012; pp. 143–162. (In French) [Google Scholar]
- Abraham, E.P.; Chain, E. An enzyme from bacteria able to destroy penicillin. Nature 1940, 146, 837. [Google Scholar] [CrossRef]
- Tenaillon, O.; Skurnik, D.; Picard, B.; Denamur, E. The population genetics of commensal Escherichia coli. Nat. Rev. 2010, 8, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Allocati, N.; Masulli, M.; Alexeyev, M.F.; Di Ilio, C. Escherichia coli in Europe: An overview. Int. J. Environ. Res. Public Health 2013, 10, 6235–6254. [Google Scholar] [CrossRef] [PubMed]
- Van de Sande-Bruinsma, N.; Grundmann, H.; Verloo, D.; Tiemersma, E.; Monen, J.; Goossens, H.; Ferech, M. European Antimicrobial Resistance Surveillance System Group; European Surveillance of Antimicrobial Consumption Project Group. Antimicrobial drug use and resistance in Europe. Emerg. Infect. Dis. 2008, 14, 1722–1730. [Google Scholar] [CrossRef] [PubMed]
- Singer, R.S.; Finch, R.; Wegener, H.C.; Bywater, R.; Walters, J.; Lipsitch, M. Antibiotic resistance—The interplay between antibiotic use in animals and human beings. Lancet Infect. Dis. 2003, 3, 47–51. [Google Scholar] [CrossRef]
- Naas, T.; Oueslati, S.; Bonnin, R.A.; Dabos, M.L.; Zavala, A.; Dortet, L.; Retailleau, P.; Iorga, B.I. Beta-Lactamase DataBase (BLDB)—Structure and Function. J. Enzym. Inhib. Med. Chem. 2017, 32, 917–919. [Google Scholar] [CrossRef] [PubMed]
- Naseer, U.; Sundsfjord, A. The CTX-M conundrum: Dissemination of plasmids and Escherichia coli clones. Microb. Drug Resist. 2011, 17, 83–97. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Critically Important Antimicrobials for Human Medicine, 6th Revision ed; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- World Health Organization (WHO). Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017. Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf?ua=1 (accessed on 27 April 2021).
- European Centre for Disease Prevention and Control (ECDC). Antimicrobial resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report 2019. 2020. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/surveillance-antimicrobial-resistance-Europe-2019.pdf (accessed on 27 April 2021).
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA J. 2020, 18, e06007. [Google Scholar] [CrossRef] [Green Version]
- Royal Decree of Belgium. Arrêté Royal du 21 Juillet 2016. Arrêté Royal Relatif aux Conditions d’Utilisation des Médicaments par les Médecins Vétérinaires et par les Responsables des Animaux. Available online: http://www.etaamb.be/fr/arrete-royal-du-21-juillet-2016_n2016024152.html (accessed on 29 April 2021). (In French).
- Association Régionale de Santé et d’Identification Animal (ARSIA). Antibiogrammes, Rapport d’Activité et Résultats de l’Arsia. 2020. Available online: https://www.arsia.be/wp-content/uploads/documents-telechargeables/RA-Antibiogrammes-2020.pdf (accessed on 3 December 2021).
- Dieren Gezondsheid Zorg (DGZ). Antibioticaresistentie—Evolutie tot Eind 2019, Rundvee. 2019. Available online: https://www.dgz.be/media/fq3lkx3z/rundvee_antibioticaresistentie_2015-2019.pdf (accessed on 3 December 2021). (In Dutch).
- Markey, B.K.; Leonard, F.C.; Archambault, M.; Cullinane, A.; Maguire, D. Clinical Veterinary Microbiology, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 252–255. [Google Scholar]
- Beutin, L.; Montenegro, M.A.; Orskov, I.; Orskov, F.; Prada, J.; Zimmermann, S.; Stephan, R. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J. Clin. Microbiol. 1989, 27, 2559–2564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belgian Veterinary Surveillance on Antimicrobial Consumption Report 2019 (BELVETSAC 2019). Federal Agency for Medicines and Health Products: Brussels, Belgium. Available online: https://belvetsac.ugent.be/BelvetSac_report_2019.pdf (accessed on 3 December 2021).
- Mader, R.; Damborg, P.; Amat, J.P.; Bengtsson, B.; Bourély, C.; Broens, E.M.; Busani, L.; Crespo-Robledo, P.; Filippitzi, M.E.; Fitzgerald, W.; et al. Building the European Antimicrobial Resistance Surveillance network in veterinary medicine (EARS-Vet). Euro Surveill. 2021, 26, 2001359. [Google Scholar] [CrossRef] [PubMed]
- Bodendoerfer, E.; Marchesi, M.; Imkamp, F.; Courvalin, P.; Böttger, E.C.; Mancini, S. Co-occurrence of aminoglycoside and β-lactam resistance mechanisms in aminoglycoside-non-susceptible Escherichia coli isolated in the Zurich area, Switzerland. Int. J. Antimicrob. Agents 2020, 56, 106019. [Google Scholar] [CrossRef] [PubMed]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djebala, S.; Moula, N.; Bayrou, C.; Sartelet, A.; Bossaert, P. Prophylactic antibiotic usage by Belgian veterinarians during elective caesarean section in Belgian blue cattle. Prev. Vet. Med. 2019, 172, 104785. [Google Scholar] [CrossRef] [PubMed]
- Belgium Center of Pharmacotherapeutic Information—Veterinary (CBIPvet). Available online: https://www.vetcompendium.be/fr/database?f%5B0%5D=type%3Acommentaar&f%5B1%5D=field_groep%3A1839 (accessed on 5 September 2021).
- Maynou, G.; Migura-Garcia, L.; Chester-Jones, H.; Ziegler, D.; Bach, A.; Terré, M. Effects of feeding pasteurized waste milk to dairy calves on phenotypes and genotypes of antimicrobial resistance in fecal Escherichia coli isolates before and after weaning. J. Dairy Sci. 2017, 100, 7967–7979. [Google Scholar] [CrossRef] [Green Version]
- European Centre for Disease Prevention and Control (ECDC). Antimicrobial consumption in the EU/EEA—Annual Epidemiological Report 2019. Stockholm. 2020. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/Antimicrobial-consumption-in-the-EU-Annual-Epidemiological-Report-2019.pdf (accessed on 10 September 2021).
- World Organisation for Animal Health (OIE). OIE Annual Report on the Use of Antimicrobial Agents in Animals. 2020. Available online: https://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/docs/pdf/A_Fourth_Annual_Report_AMU.pdf (accessed on 10 September 2021).
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S.; et al. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef]
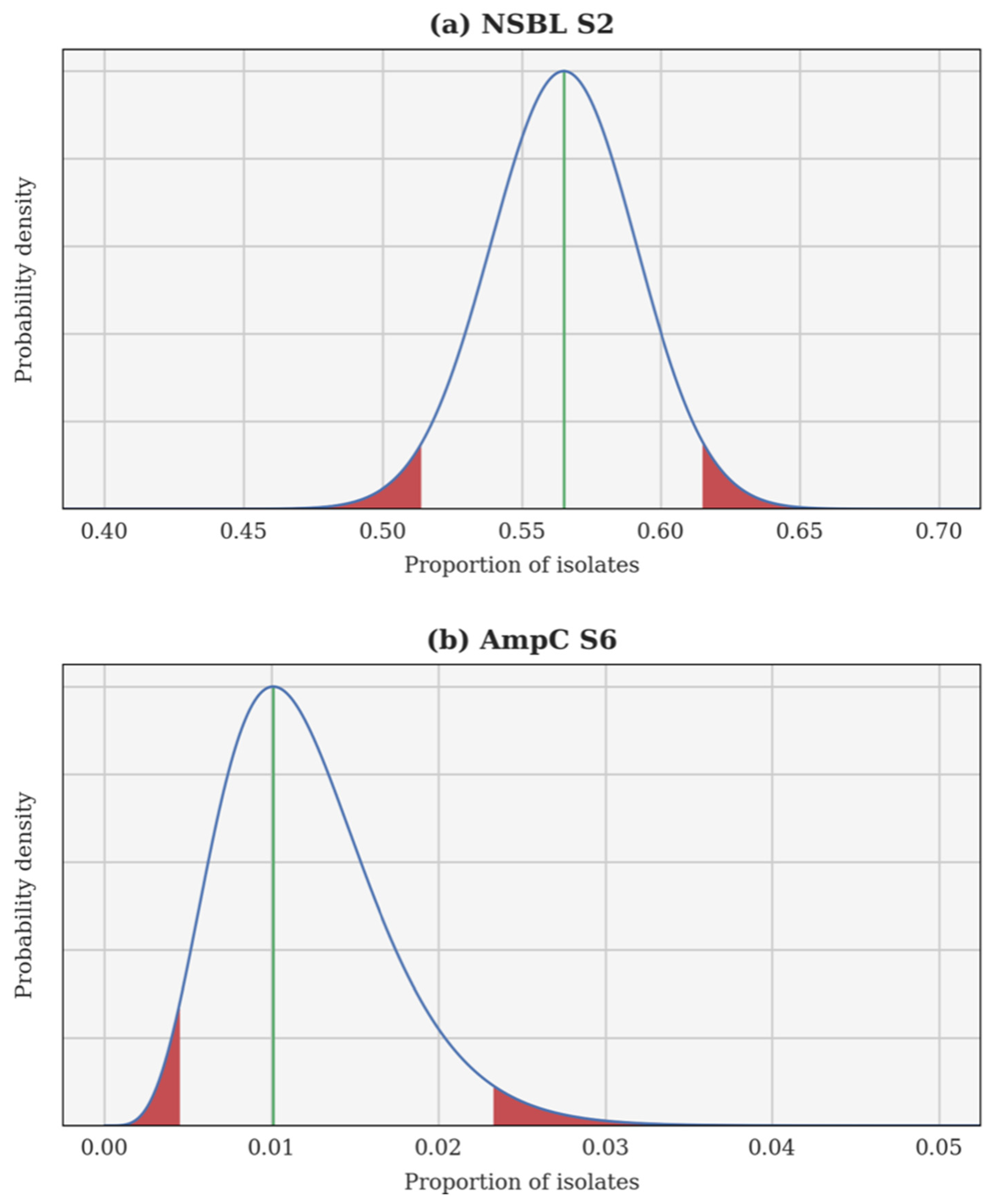
| Calving Seasons | Number of E. coli Isolates | Number of Calves |
|---|---|---|
| S1: 2014–2015 | 418 | 384 |
| S2: 2015–2016 | 361 | 334 |
| S3: 2016–2017 | 866 | 740 |
| S4: 2017–2018 | 707 | 646 |
| S5: 2018–2019 | 599 | 545 |
| S6: 2019–2020 | 495 | 464 |
| S7: 2020–2021 | 471 | 424 |
| Total overall calving seasons | 3917 | 3537 |
| Calving Season and Origin of Escherichia coli | Resistance Profiles (%) | Total | ||||
|---|---|---|---|---|---|---|
| Susceptible | NSBL | ESBL | AmpC | AmpC-like | ||
| S1 | 63 | 232 | 61 | 36 | 26 | 418 |
| (15.1) | (55.5) | (14.6) | (8.6) | (6.2) | (100.0) | |
| Faeces | 28 | 147 | 28 | 12 | 13 | 228 |
| (6.7) | (35.2) | (6.7) | (2.9) | (3.1) | (54.5) | |
| Intestinal content + organs | 35 | 85 | 33 | 24 | 13 | 190 |
| (8.4) | (20.3) | (7.9) | (5.7) | (3.1) | (45.5) | |
| S2 | 45 | 204 | 68 | 32 | 12 | 361 |
| (12.5) | (56.5) | (18.8) | (8.9) | (3.3) | (100.0) | |
| Faeces | 20 | 114 | 33 | 11 | 6 | 184 |
| (5.5) | (31.6) | (9.1) | (3.0) | (1.7) | (51.0) | |
| Intestinal content + organs | 25 | 90 | 35 | 21 | 6 | 177 |
| (6.9) | (24.9) | (9.7) | (5.8) | (1.7) | (49.0) | |
| S3 | 96 | 505 | 149 | 69 | 47 | 866 |
| (11.1) | (58.3) | (17.2) | (8.0) | (5.4) | (100.0) | |
| Faeces | 55 | 301 | 85 | 31 | 20 | 492 |
| (6.4) | (34.8) | (9.8) | (3.6) | (2.3) | (56.8) | |
| Intestinal content + organs | 41 | 204 | 64 | 38 | 27 | 374 |
| (4.7) | (23.6) | (7.4) | (4.4) | (3.1) | (43.2) | |
| S4 | 94 | 456 | 83 | 25 | 49 | 707 |
| (13.3) | (64.5) | (11.7) | (3.5) | (6.9) | (100.0) | |
| Faeces | 51 | 304 | 51 | 11 | 34 | 451 |
| (7.2) | (43.0) | (7.2) | (1.6) | (4.8) | (63.8) | |
| Intestinal content + organs | 43 | 152 | 32 | 14 | 15 | 256 |
| (6.1) | (21.5) | (4.5) | (2.0) | (2.1) | (36.2) | |
| S5 | 84 | 399 | 68 | 10 | 38 | 599 |
| (14.0) | (66.6) | (11.4) | (1.7) | (6.3) | (100.0) | |
| Faeces | 57 | 309 | 42 | 5 | 29 | 442 |
| (9.5) | (51.6) | (7.0) | (0.8) | (4.8) | (73.8) | |
| Intestinal content + organs | 27 | 90 | 26 | 5 | 9 | 157 |
| (4.5) | (15.0) | (4.3) | (0.8) | (1.5) | (26.2) | |
| S6 | 79 | 342 | 44 | 5 | 25 | 495 |
| (16.0) | (69.1) | (8.9) | (1.0) | (5.1) | (100.0) | |
| Faeces | 48 | 251 | 31 | 3 | 18 | 351 |
| (9.7) | (50.7) | (6.3) | (0.6) | (3.6) | (70.9) | |
| Intestinal content + organs | 31 | 91 | 13 | 2 | 7 | 144 |
| (6.3) | (18.4) | (2.6) | (0.4) | (1.4) | (29.1) | |
| S7 | 72 | 329 | 38 | 13 | 19 | 471 |
| (15.3) | (69.9) | (8.1) | (2.8) | (4.0) | (100.0) | |
| Faeces | 35 | 220 | 28 | 7 | 16 | 306 |
| (7.4) | (46.7) | (5.9) | (1.5) | (3.4) | (65.0) | |
| Intestinal content + organs | 37 | 109 | 10 | 6 | 3 | 165 |
| (7.9) | (23.1) | (2.1) | (1.3) | (0.6) | (35.0) | |
| Total of 7 seasons | 533 | 2467 | 511 | 190 | 216 | 3917 |
| (13.6) | (63.0) | (13.0) | (4.9) | (5.5) | (100.0) | |
| Sensitive | NSBL | AmpC-like | AmpC | ESBL | Overall Resistance | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Season Comparison | p-Value | Season Comparison | p-Value | Season Comparison | p-Value | Season Comparison | p-Value | Season Comparison | p-Value | Season Comparison | p-Value |
| S1 > S2 | 0.17220 | S1 < S2 | 0.41691 | S1 > S2 | 0.04287 | S1 < S2 | 0.50021 | S1 < S2 | 0.06798 | S1 < S2 | 0.17220 |
| S2 > S3 | 0.27458 | S2 < S3 | 0.30130 | S2 < S3 | 0.07414 | S2 > S3 | 0.33818 | S2 > S3 | 0.27266 | S2 < S3 | 0.27458 |
| S3 < S4 | 0.10395 | S3 < S4 | 0.00708 | S3 < S4 | 0.12870 | S3 > S4 | 0.00013 | S3 > S4 | 0.00140 | S3 > S4 | 0.10395 |
| S4 < S5 | 0.38117 | S4 < S5 | 0.22915 | S4 > S5 | 0.37838 | S4 > S5 | 0.02655 | S4 > S5 | 0.44848 | S4 > S5 | 0.38117 |
| S5 < S6 | 0.20884 | S5 < S6 | 0.20953 | S5 > S6 | 0.21715 | S5 > S6 | 0.25295 | S5 > S6 | 0.10761 | S5 > S6 | 0.20884 |
| S6 > S7 | 0.42125 | S6 < S7 | 0.42607 | S6 > S7 | 0.27372 | S6 < S7 | 0.03709 | S6 > S7 | 0.36649 | S6 < S7 | 0.42125 |
| Supplementary comparison tests | |||||||||||
| S1 < S7 | 0.50232 | S1 < S7 | 0.00001 | S1 > S7 | 0.09170 | S1 > S7 | 0.00010 | S1 > S7 | 0.00143 | S1 > S7 | 0.50232 |
| S1 > S3 | 0.02725 | S1 < S3 | 0.18548 | S2 < S4 | 0.00968 | NN | NN | S1 < S3 | 0.13417 | S1 < S3 | 0.02725 |
| S3 < S6 | 0.00664 | S4 < S7 | 0.03217 | S4 > S7 | 0.02329 | NN | NN | S4 > S7 | 0.02538 | S3 > S6 | 0.00664 |
| Escherichia coli from Diarrheic vs. Necropsied Calves | Resistance Profiles | Global Resistance | ||||
|---|---|---|---|---|---|---|
| Susceptible | NSBL | ESBL | AmpC | AmpC-like | ||
| S1 | 0.09914 | 0.00007 | 0.16452 | 0.00856 | 0.68683 | 0.09914 |
| S2 | 0.42590 | 0.03446 | 0.68770 | 0.06327 | 1.00000 | 0.42590 |
| S3 | 1.00000 | 0.05172 | 1.00000 | 0.04268 | 0.04899 | 1.00000 |
| S4 | 0.04960 | 0.03377 | 0.62907 | 0.05420 | 0.44391 | 0.04960 |
| S5 | 0.18336 | 0.00569 | 0.01947 | 0.13785 | 0.84952 | 0.18336 |
| S6 | 0.04195 | 0.08637 | 1.00000 | 0.63112 | 1.00000 | 0.04195 |
| S7 | 0.00200 | 0.20700 | 0.28907 | 0.39208 | 0.08722 | 0.00200 |
| Escherichia coli Virulotype | Resistance Profiles in % | Total | ||||
|---|---|---|---|---|---|---|
| Susceptible | NSBL | ESBL | AmpC | AmpC-like | ||
| F5 | 0.2 | 5.8 | 0.3 | 0.1 | − | 6.3 |
| F17a | 3.0 | 12.5 | 2.3 | 1.8 | 1.7 | 21.4 |
| CS31A | 4.1 | 34.5 | 7.9 | 1.5 | 2.8 | 50.9 |
| Ehly+ | 2.3 | 1.5 | 0.2 | 0.1 | 0.1 | 4.1 |
| ND | 4.0 | 8.6 | 2.4 | 1.4 | 0.9 | 17.3 |
| Total | 13.6 | 63.0 | 13.1 | 4.9 | 5.5 | 100.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guérin, V.; Farchi, A.; Thiry, D.; Cawez, F.; Mercuri, P.S.; Galleni, M.; Mainil, J.; Saulmont, M. Seven-Year Evolution of β-Lactam Resistance Phenotypes in Escherichia coli Isolated from Young Diarrheic and Septicaemic Calves in Belgium. Vet. Sci. 2022, 9, 45. https://doi.org/10.3390/vetsci9020045
Guérin V, Farchi A, Thiry D, Cawez F, Mercuri PS, Galleni M, Mainil J, Saulmont M. Seven-Year Evolution of β-Lactam Resistance Phenotypes in Escherichia coli Isolated from Young Diarrheic and Septicaemic Calves in Belgium. Veterinary Sciences. 2022; 9(2):45. https://doi.org/10.3390/vetsci9020045
Chicago/Turabian StyleGuérin, Virginie, Alban Farchi, Damien Thiry, Frédéric Cawez, Paola Sandra Mercuri, Moreno Galleni, Jacques Mainil, and Marc Saulmont. 2022. "Seven-Year Evolution of β-Lactam Resistance Phenotypes in Escherichia coli Isolated from Young Diarrheic and Septicaemic Calves in Belgium" Veterinary Sciences 9, no. 2: 45. https://doi.org/10.3390/vetsci9020045
APA StyleGuérin, V., Farchi, A., Thiry, D., Cawez, F., Mercuri, P. S., Galleni, M., Mainil, J., & Saulmont, M. (2022). Seven-Year Evolution of β-Lactam Resistance Phenotypes in Escherichia coli Isolated from Young Diarrheic and Septicaemic Calves in Belgium. Veterinary Sciences, 9(2), 45. https://doi.org/10.3390/vetsci9020045






