1. Introduction
Urine is an important biological test material for nutritional research as well as medical diagnostics. It is advantageous due to its non-invasive, painless collection, and contains substances that provide information on the health status of the organism [
1]. In this context, urinary proteome analyses are of increasing interest, as specific proteins have been demonstrated to be associated with several diseases [
1]. These studies, however, have predominantly been performed in human medicine research, while investigations related to veterinary medicine are relatively scarce. To our best knowledge, only three studies have been published on the urine proteome of cats so far, with their focus especially on animals with chronic kidney disease [
2,
3] and urinary tract diseases [
4]. Jepson et al. [
2] found differences in the urine proteome clusters of cats with a chronic kidney disease that developed either azotemia within one year after sample collection or remained nonazotemic. In this study, surface-enhanced laser desorption–ionization time-of-flight mass spectrometry (SELDI-TOF-MS) was used for the proteome analyses [
2]. Ferlizza et al. [
3] compared the urine proteome of healthy cats and cats with a chronic kidney disease (IRIS stage 2–4). They observed 13 over- or underrepresented proteins in the urine of the kidney patients compared to the control animals, making them potential candidates as biomarkers of renal function [
3]. The methods used for this investigation included one-dimensional sodium-dodecyl-sulfate polyacrylamide gel electrophoresis and two-dimensional electrophoresis (2DE), followed by mass spectrometry [
3]. Finally, Lemberger et al. [
4] evaluated the urine proteome of healthy control cats and cats with idiopathic cystitis, bacterial urinary tract infection and urolithiasis, using 1-dimensional gel electrophoresis. The urinary fibronectin concentrations were further evaluated by Western blot and immunohistochemical measurements [
4]. The authors found that the urinary fibronectin concentrations were higher in the cats with idiopathic cystitis when compared to the control cats, and assumed that fibronectin could, therefore, act as a potential biomarker for this disease [
4]. In addition, the protein concentrations in the urine of the cats with urinary tract diseases were higher than in the urine of the control animals, and patients with urolithiasis had higher urinary protein concentrations compared to cats with idiopathic cystitis [
4].
Although the available studies have focused on the effects of diseases on the urine proteome of cats, dietetic interventions might also be an influencing factor, as nutrition substantially modulates metabolic pathways [
5]. Several nutritional factors might be of interest for proteomic research, among others the protein or amino acid supply. For the present investigation, we used urine samples of a study evaluating the impact of a high-protein diet supplemented without or with arginine, ornithine and zeolite in healthy adult cats [
6]. As the main findings of this study, the postprandial blood urea increase in the animals was enhanced by the dietary arginine and ornithine supplementation, but the renal urea excretion did not increase simultaneously. Thus, it can be speculated that the supplements promoted the hepatic ammonia detoxification in felines, but that the renal excretory mechanisms might have been impaired by the dietetic interventions. In addition, the dietary zeolite supplementation seemed to affect the nitrogen metabolism as well, as demonstrated by decreased concentrations of biogenic amines in the feces of the cats and a reduced renal ammonia excretion [
6]. In order to evaluate the impact of the dietary supplements in the feline organism in more detail, we aimed to perform a proteomics approach, using the urine of the animals. We hypothesized that the analyses would reveal a dietary modulation of the cats’ protein metabolism or kidney function. The objectives of the present study were, therefore, two-fold: (I) to generate basic data on the urine proteome of healthy adult cats, and (II) to assess the effects of a dietary arginine, ornithine and zeolite supplementation on the feline urine proteome.
2. Materials and Methods
The study was approved by the relevant authority in Berlin, Germany (Landesamt für Gesundheit und Soziales; approval number G 0120/15).
The design of the feeding study has been previously published in detail [
6]. In the present investigation, urine samples of a part of this study were used for the proteome analyses.
2.1. Feeding Study
Four healthy adult cats (2 male neutered, 2 female intact, aged 80.3 ± 20.8 months at the beginning of this study) received a high-protein basic diet (60.3% crude protein in dry matter) supplemented without (w/o) or with arginine (+100% compared to the arginine concentration in the basic diet), ornithine (+200% compared to the arginine concentration in the basic diet) or zeolite (0.375 g/kg body weight/day) for 11 days each. On the last four days of the feeding periods, the cats were housed in metabolic cages to collect urine.
2.2. Proteome Analyses
For the label-free quantitative shotgun proteomic approach, the concentration of total protein in the urine samples was determined using the Pierce™ 660 nm Protein Assay Reagent (Thermo Fisher Scientific, Waltham, MA, USA), following the manufacturer’s protocol. Different volumes of urine, corresponding to 30 µg protein, were filled up to 500 µL with 8 M urea in 50 mM TRIS (both Roth, Karlsruhe, Germany). These samples were loaded onto a Pall 10 kDa filter (Pall Corporation, Port Washington, NY, USA) for a FASP digest and subsequent liquid chromatography coupled to mass spectrometry (LC-MS) analysis, as described by Soler et al. [
7]. In brief, the digested protein extracts were desalted using C18 spin columns (Pierce, Thermo Fisher Scientific), dried in an Eppendorf concentrator plus (Eppendorf, Hamburg, Germany) and solved in 0.1% TFA (Thermo Fisher Scientific) to a peptide concentration of 0.1 µg/µL. The separation of 300 ng of the peptides was performed on a 25 cm Acclaim PepMap C18 column (75 µm inner diameter, 2 µm particle size and 100 Å pore size) with a flow rate of 300 nL/min on a Thermo nano-HPLC system, coupled to a Q Exactive HF Orbitrap mass spectrometer (both Thermo Fisher Scientific) using a 60 min gradient [
7]. The LC-MS analyses were performed in triple determination per sample.
2.3. Database Searches
For a first evaluation of the proteome data, the Proteome Discoverer Software 2.4.0.305 (Thermo Fisher Scientific) was used with the following settings: Protein database: NCBI_cat_tx9685_190612.fasta; accessed on 12 June 2019; crap.fasta (
https://www.thegpm.org/crap/; accessed on 20 November 2016); enzyme name: trypsin (full); max. missed cleavage sites: 2; precursor mass tolerance: 10 ppm; fragment mass tolerance: 0.02 Da; dynamic modification: oxidation/+15.995 Da (M); N-terminal modification: Acetyl/+42.011 Da (N-terminus); static modification: carbamidomethyl/+57.021 Da (C); decoy database search: target FDR (strict): 0.01; target FDR (relaxed): 0.05; validation based on: q-value.
After bioinformatical analysis, the number of identified proteins in the urine samples could be determined for the three LC-MS injections. The accessions of the identified proteins were then submitted to the STRING database (
https://string-db.org; accessed from 17 March 2022 to 11 September 2022), which was performed separately for each cat and each feeding group. The settings for the database search were: multiple proteins; list of names: protein accession; organisms:
Felis catus.
As the identified proteins of the three injections were submitted together to the STRING database, doublings were removed by the system, resulting in the output of all urinary proteins found for one cat per feeding group. The urinary proteins of the cats of the same feeding group were again submitted to the STRING database to receive all urinary proteins found for each feeding group.
2.4. Biological Processes
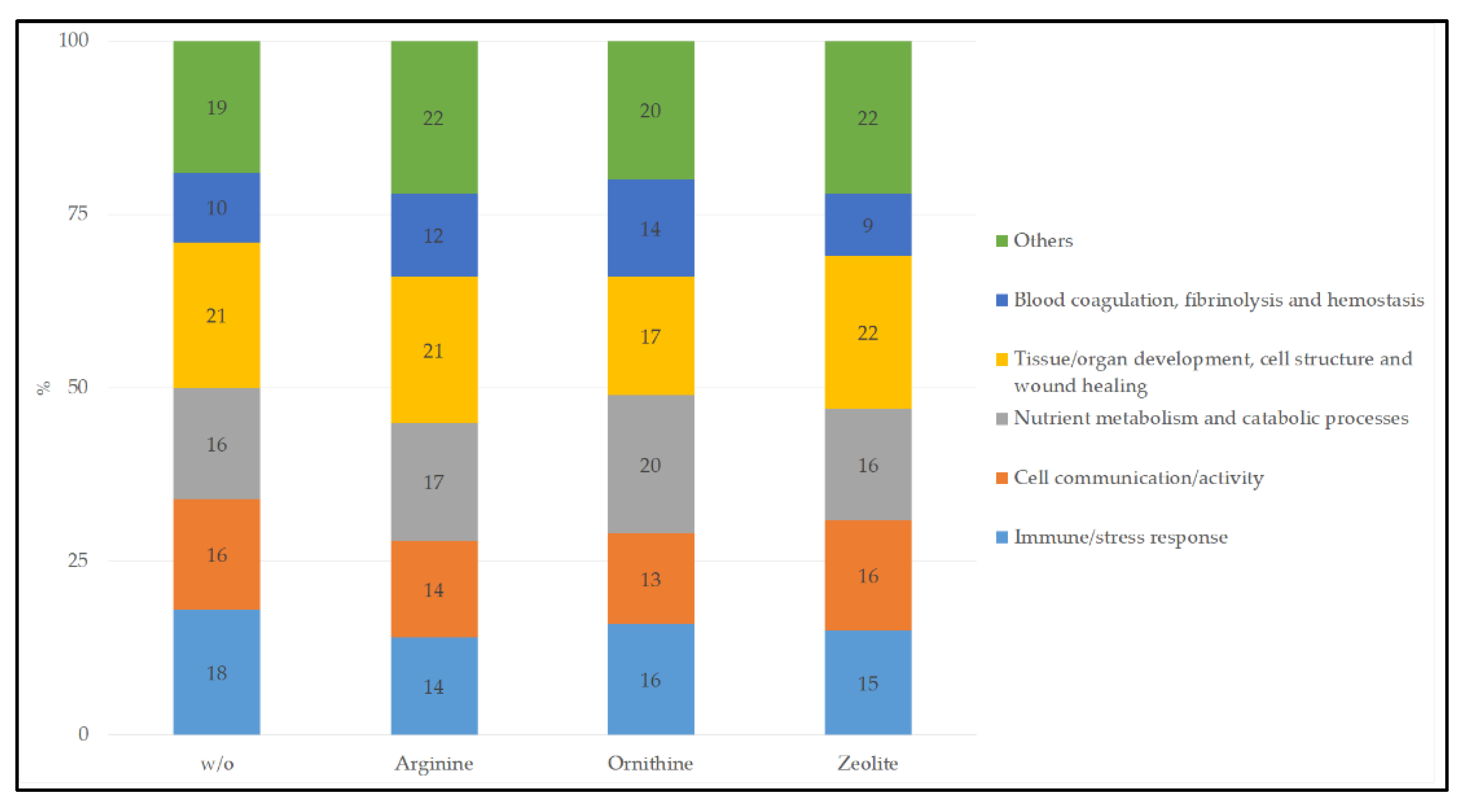
The urinary proteins are associated with several biological processes, as specified by the STRING database. In a first step, the biological processes were summarized to the following groups: immune/stress response; cell communication/activity; nutrient metabolism and catabolic processes; tissue/organ development, cell structure and wound healing; blood coagulation, fibrinolysis and hemostasis; others. This analysis was a qualitative evaluation, i.e., if a biological process was found for different cats of the same feeding group, it was counted as one. In a second step, the number of proteins associated with a biological process was considered to compare the feeding groups.
2.5. KEGG (Kyoto Encyclopedia of Genes and Genomes) Pathways
The up- and downregulated urinary proteins between the arginine, ornithine or zeolite group and the w/o group (see statistical data analyses) were submitted to the STRING database (search for multiple proteins, list of names: protein accession, organism: Felis catus) to detect the associated KEGG pathways.
2.6. Statistical Data Analyses
A first statistical data analysis was carried out by the t-test (Proteome Discoverer Software 2.4.0.305, Thermo Fisher Scientific) to detect up- and downregulated proteins between the supplemented groups versus the w/o group (fold change ± 2). An FDR adjusted p-value < 0.05 was considered to be statistically significant. In addition, a principal component analysis was carried out in the course of the protein quantification (Proteome Discoverer Software 2.4.0.305, Thermo Fisher Scientific; formatting with Microsoft PowerPoint 2016).
The distribution of the biological processes associated with the proteins found in the urine of the dietary treatment groups was qualitatively compared by Microsoft Excel (2016). Additionally, the number of proteins associated with a single biological process was evaluated. For this, the feeding groups were again compared by the t-test (SPSS 28, IBM Corp., Armonk, NY, USA), with a p-value < 0.05 being statistically significant.
3. Results
3.1. Urinary Protein Concentration and Number of Identified Proteins
The analyzed protein concentration in the urine samples of the cats was 64.7 ± 26.2, 80.8 ± 39.4, 49.6 ± 33.0 and 53.6 ± 41.7 µg/mL for the w/o, arginine, ornithine and zeolite treatment, respectively.
The LC-MS/MS method identified 516 ± 45 (w/o group), 512 ± 18 (arginine group), 399 ± 162 (ornithine group) and 455 ± 146 (zeolite group) urinary proteins. However, as demonstrated by the high standard deviations, the numbers markedly differed among the cats within each group. When the feeding groups were compared, no differences in total protein numbers could be detected between the supplemented groups versus the w/o group.
3.2. Biological Processes
When the identified proteins were submitted to the STRING database, 365 proteins of the w/o group could be found for Felis catus, which are associated with 115 biological processes. For the arginine group, 286 proteins were found and are involved in 88 biological processes. Comparable values were observed for the ornithine group, with 260 identified proteins in 84 biological processes. In the zeolite group, 342 proteins were found in the STRING database, which are associated with 115 biological processes.
When the biological processes were summarized to groups, a comparable distribution could be observed between the four feeding groups (
Figure 1).
The mean number of urinary proteins associated with the detected biological processes is presented in
Table 1,
Table 2,
Table 3,
Table 4,
Table 5 and
Table 6. The comparison between the supplemented groups versus the w/o treatment revealed only marginal differences, with a lower number of urinary proteins related to a “regulation of protein metabolic process” in the ornithine group, and a higher number of urinary proteins associated with a “regulation of body fluid levels” in the arginine group.
3.3. Up- and Downregulated Urinary Proteins and Associated KEGG Pathways
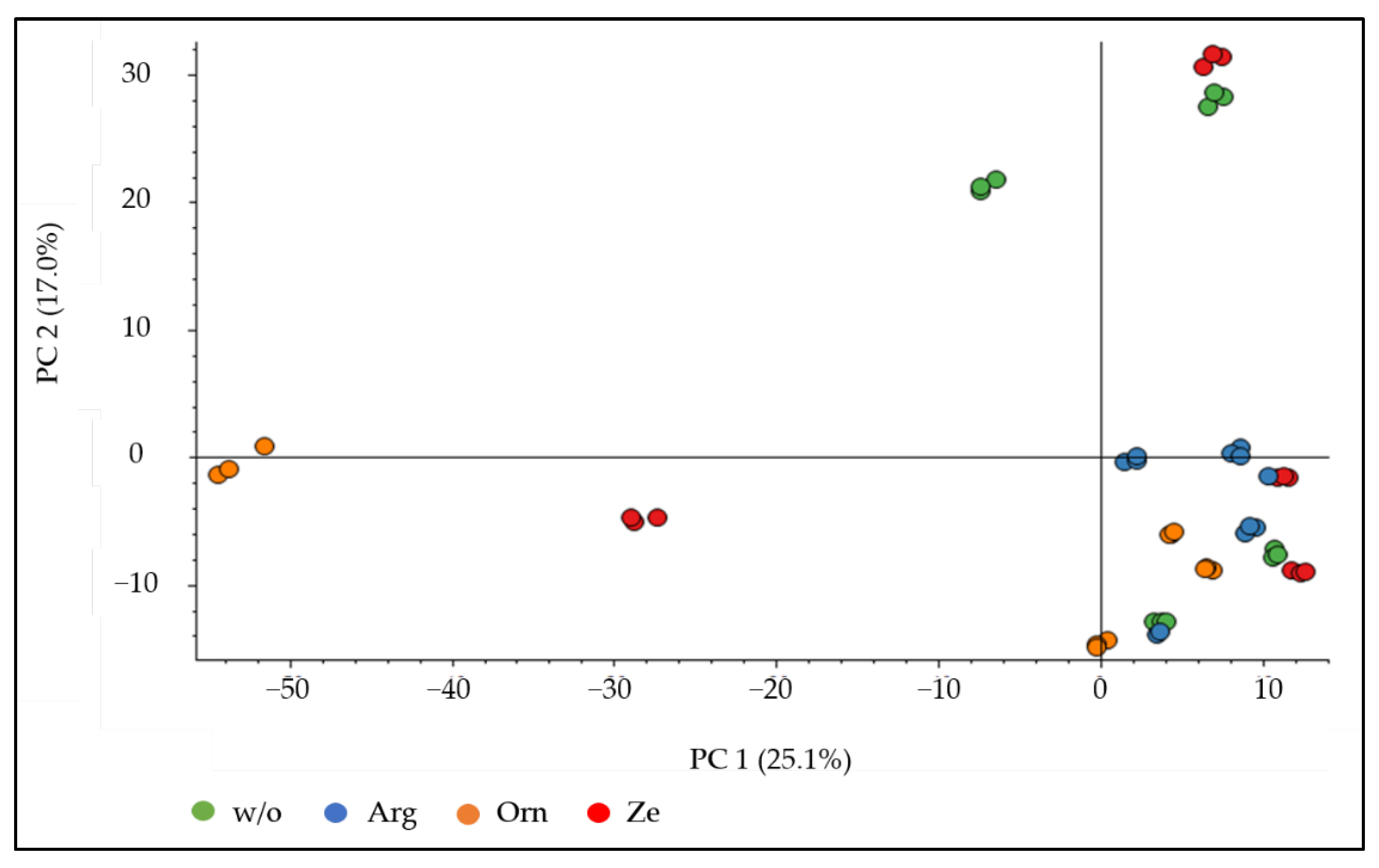
The principal component analysis plot demonstrated a clustering of the technical replicates, but not for a specific dietary treatment (
Figure 2). However, when the urine proteome of the arginine group was statistically compared with the w/o group, 80 downregulated and 27 upregulated proteins were detected. Out of the 80 downregulated proteins, 35 were found in the STRING database, where 4 proteins are associated with the KEGG pathway “glycolysis/gluconeogenesis” and 5 proteins with “
Salmonella infection” (
Table 7). Only 8 out of the 27 upregulated urinary proteins were found in the STRING database, and no associated KEGG pathways could be identified.
In the ornithine group, 125 urinary proteins were downregulated and 30 proteins were upregulated compared to the w/o group. Fifty-three of the downregulated proteins were identified in the STRING database, with 5 being associated with the KEGG pathway “Ribosome” and 5 with “
Salmonella infection” (
Table 8). Fourteen upregulated proteins were found in the STRING database, where 3 of them are related to the “Estrogen signaling pathway” (
Table 8).
Eighty-five urinary proteins were downregulated and 21 urinary proteins were upregulated in the cats receiving zeolite to their diet compared to the w/o treatment. Thirty-eight of the downregulated proteins and 9 of the upregulated proteins were identified in the STRING database, but no associated KEGG pathways could be detected.
4. Discussion
In the present study, urine samples of cats were analyzed by a label-free quantitative approach to evaluate the feline urine proteome. While proteome analyses are especially used in human medicine to identify potential biomarkers for health and disease [
1], investigations in cats remain scarce. To our knowledge, only three studies have been conducted in this context so far, focusing on cats with kidney and urinary tract diseases [
2,
3,
4]. However, different methods were applied for the proteome analyses, some resulting in a detection of only 5 proteins [
4], 17–23 protein clusters that were common to all array spectra [
2], and 13–14 proteins [
3] in the urine samples investigated. In the present study, the proteome approach by high resolution LC-MS/MS revealed a high number of 399–516 proteins in the urine samples of the four feeding groups, making this analytical method advantageous for an untargeted urine proteome evaluation in cats.
Nevertheless, when compared to data from human medicine, the number of detected proteins in the feline urine samples was markedly lower. Zhao et al. [
8] summarized six large-scale studies on the proteome of healthy human subjects, which identified between 1310–6085 urinary proteins. The number of proteins in urine of healthy dogs ranged between 176 [
9], 182 [
10] and 659 [
11], and is, therefore, more comparable to the data obtained for the cats of the present study. At this point, it can only be speculated why human subjects and those carnivorous species differ in their urine proteome. Besides different analytical methods used in the related studies, species-dependent differences might be relevant. In addition, individual factors, such as age, sex or dietary intake, might influence the kind and number of proteins in the urine. Research in humans is already evaluating the individual variation of the urine proteome [
12] and should be explored for animal-derived samples as well, since this might facilitate data interpretation and comparison. Given the small sample size of the present pilot study, however, this aspect cannot be determined for the feline urine proteome at this stage. Nevertheless, with increasing data deriving from larger scale investigations, it can be expected that potential interfering factors on the urine proteome of cats will be identified or disproved. The present data should, therefore, be considered as a first step towards a broader knowledge of the feline urine proteome, and as a starting point for future investigations.
The urine proteome of the four feeding groups did not markedly differ. Although a notable number of down- and upregulated proteins was detected in the urine of the cats receiving the dietary supplements compared to the control treatment, the physiological relevance of this finding, as evaluated by the associated biological processes and KEGG pathways, seemed to be low. However, one interesting result was the downregulation of four proteins related to the KEGG pathway “glycolysis/gluconeogenesis” in the arginine group. It might be speculated if mechanisms related to nitric oxide were responsible for this downregulation, as nitric oxide is synthesized from arginine and can affect the glucose metabolism [
13]. However, the pilot study character of the present investigation requires a careful data interpretation and follow-up studies to prove this hypothesis.
We used urine samples of a study that gained comprehensive data on the dietary effects of arginine, ornithine and zeolite on the formation and excretion of uremic toxins in cats [
6]. The results indicated an impact of arginine and ornithine on the hepatic ammonia detoxification, as these supplements enhanced the postprandial blood urea increase, but also potentially detrimental effects on the renal function, since the urea excretion by the kidneys was not enhanced in parallel. In addition, effects of the dietary zeolite supplementation on the intestinal nitrogen metabolism of the cats were observed [
6]. The proteome analyses of the urine samples aimed to provide more insights into the impact of the three supplements on the protein metabolism of the cats, and to identify potential indications for an impaired kidney function. However, as the proteome data were comparable among the feeding groups, including the control treatment, the results cannot support these objectives.
It should, however, be noted that the proteomics approach of the present study only allowed for a relative quantification of the urinary proteins (ratio of the identified proteins related to a given protein concentration in the samples [
14]). Thus, the individual differences in the urinary protein concentrations of the cats could not be considered for data evaluation. In addition, a certain number of the identified proteins has not been found in the STRING database. This implies missing information on the detected urinary proteins, and possibly also unrecognized differences between the feeding groups. Thus, there seems to be a potential for improvements in the data sets related to the feline urine proteome. This also includes information on the associated biological processes or KEGG pathways, which were partly hard to interpret in the present investigation. For instance, some downregulated urinary proteins of the arginine and ornithine group were related to the KEGG pathway “
Salmonella infection”, although no infection occurred in the cats enrolled in this study. It is more likely that the detected downregulated proteins are involved in more general defense mechanisms, which can, among others, be also relevant in the course of a
Salmonella infection. Future research should, therefore, also focus on an extension of the knowledge base and analytical tools to evaluate feline urine proteome data.
In the present study, voided urine samples were used. Although it has been discussed that voided urine could be contaminated by proteins from the genitourinary tract [
2], other collection methods also imply limitations. For instance, blood contamination of the urine due to cystocentesis is possible [
3], which might also apply to urine collection by catheterization. In addition, both cystocentesis and catheterization are invasive and mildly stressful, making urine collection for several days to receive pooled samples difficult. These, however, can be easily obtained by voided urine samples, and might, therefore, be potentially advantageous to provide data on the average urine proteome of an animal.