Explorative Field Study on the Use of Oral Fluids for the Surveillance of Actinobacillus pleuropneumoniae Infections in Fattening Farms by an Apx-Real-Time PCR
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Pigs/Farms
2.2. Data Collection
2.2.1. Sampling of OFs
2.2.2. Evaluation of Lung Lesions
2.2.3. Molecular Biological Examinations of OFs
2.3. Statistical Analysis
3. Results
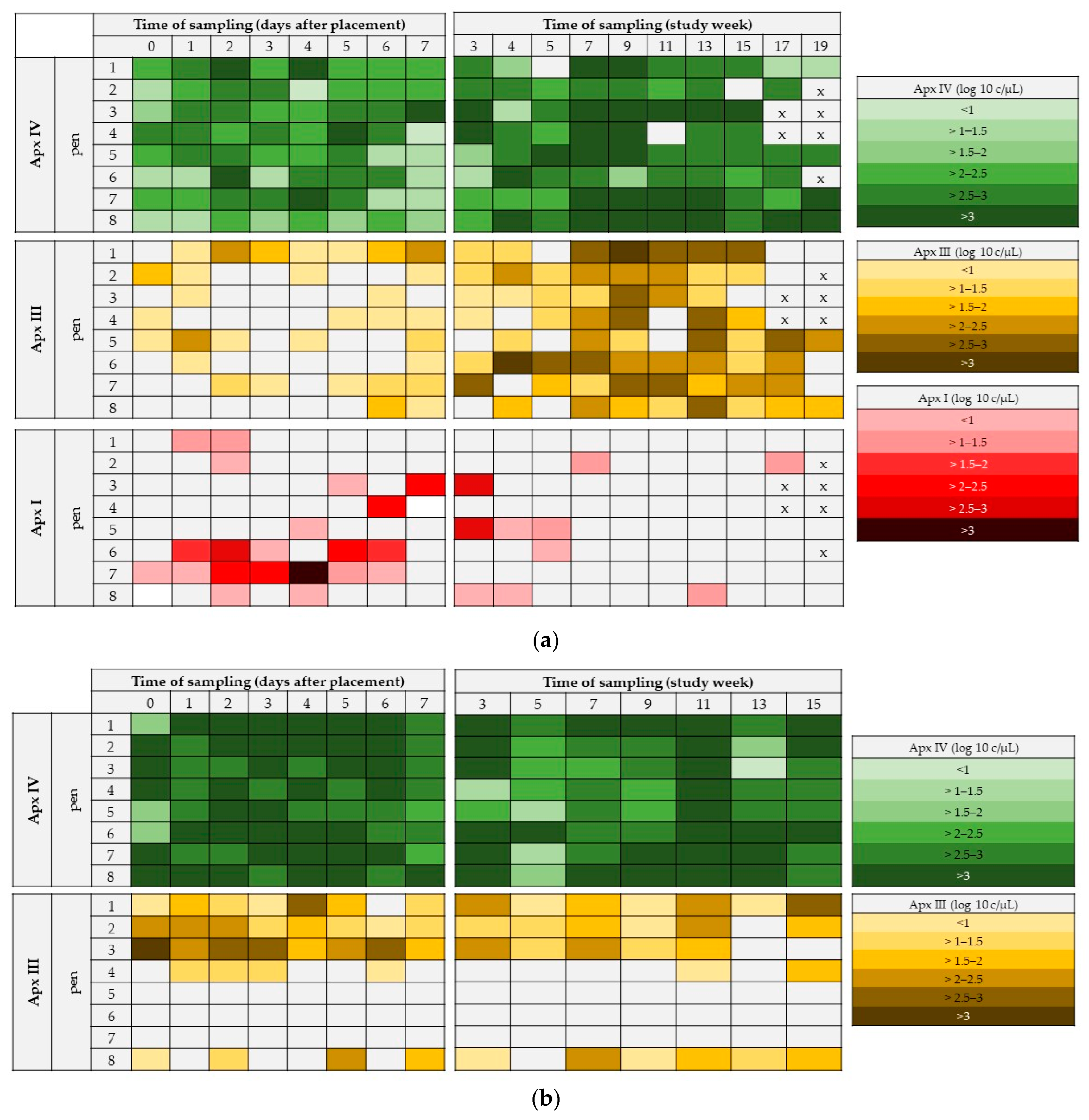
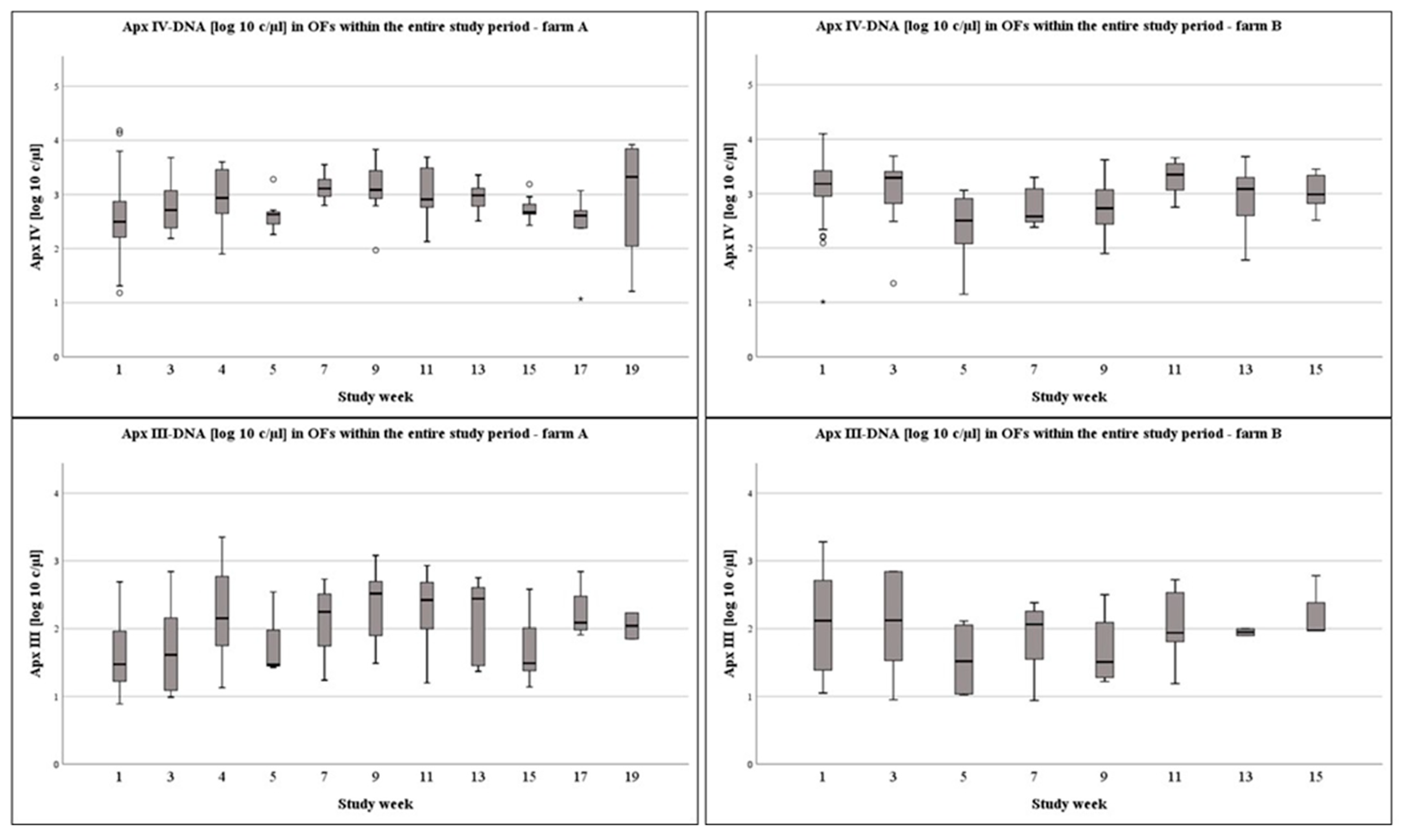
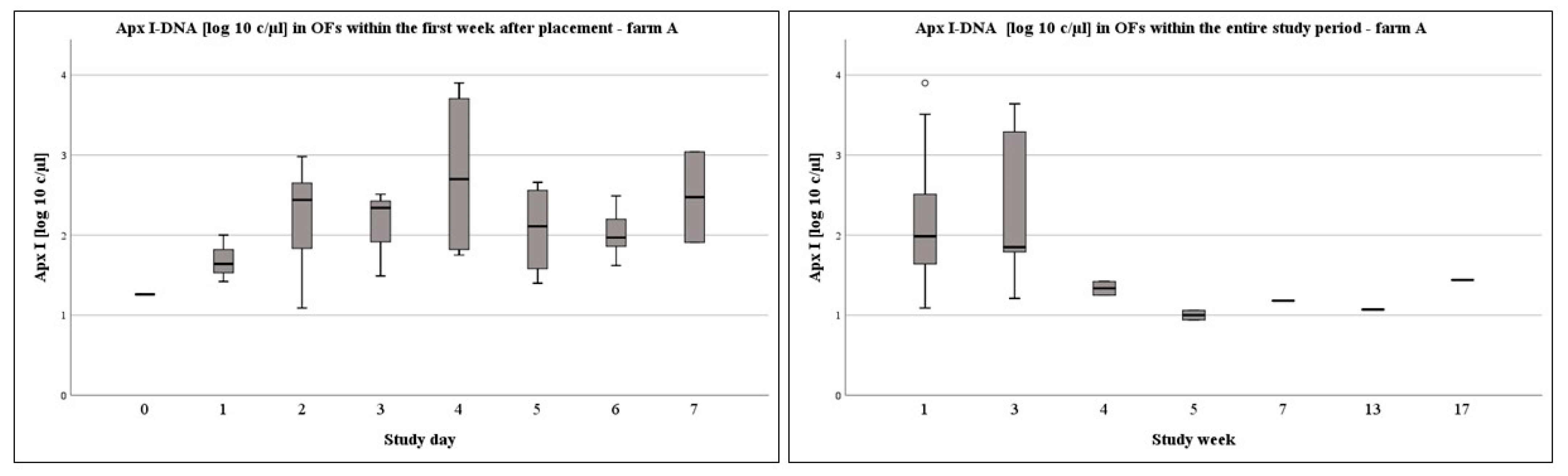
3.1. OFs on Pen-Level at Different Times of Sampling
3.2. Lung Lesion Scoring at Slaughter
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hernández-García, J.; Robben, N.; Magnée, D.; Eley, T.; Dennis, I.; Kayes, S.M.; Thomson, J.R.; Tucker, A.W. The use of oral fluids to monitor key pathogens in porcine respiratory disease complex. Porc. Health Manag. 2017, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, A.; Wang, C.; Prickett, J.R.; Pogranichniy, R.; Yoon, K.-J.; Main, R.; Johnson, J.K.; Rademacher, C.; Hoogland, M.; Hoffmann, P.; et al. Efficient surveillance of pig populations using oral fluids. Prev. Veter. Med. 2012, 104, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Prickett, J.; Simer, R.; Christopher-Hennings, J.; Yoon, K.-J.; Evans, R.B.; Zimmerman, J.J. Detection of Porcine reproductive and respiratory syndrome virus infection in porcine oral fluid samples: A longitudinal study under experimental conditions. J. Veter. Diagn. Investig. 2008, 20, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Kittawornrat, A.; Prickett, J.; Chittick, W.; Wang, C.; Engle, M.; Johnson, J.; Patnayak, D.; Schwartz, T.; Whitney, D.; Olsen, C.; et al. Porcine reproductive and respiratory syndrome virus (PRRSV) in serum and oral fluid samples from individual boars: Will oral fluid replace serum for PRRSV surveillance? Virus Res. 2010, 154, 170–176. [Google Scholar] [CrossRef]
- Biernacka, K.; Karbowiak, P.; Wróbel, P.; Charęza, T.; Czopowicz, M.; Balka, G.; Goodell, C.; Rauh, R.; Stadejek, T. Detection of porcine reproductive and respiratory syndrome virus (PRRSV) and influenza A virus (IAV) in oral fluid of pigs. Res. Veter. Sci. 2016, 109, 74–80. [Google Scholar] [CrossRef]
- Nielsen, G.B.; Nielsen, J.P.; Haugegaard, J.; Leth, S.C.; Larsen, L.E.; Kristensen, C.S.; Pedersen, K.S.; Stege, H.; Hjulsager, C.K.; Houe, H. Comparison of serum pools and oral fluid samples for detection of porcine circovirus type 2 by quantitative real-time PCR in finisher pigs. Porc. Health Manag. 2018, 4, 2. [Google Scholar] [CrossRef]
- Prickett, J.R.; Johnson, J.; Murtaugh, M.P.; Puvanendiran, S.; Wang, C.; Zimmerman, J.J.; Opriessnig, T. Prolonged Detection of PCV2 and Anti-PCV2 Antibody in Oral Fluids Following Experimental Inoculation. Transbound. Emerg. Dis. 2011, 58, 121–127. [Google Scholar] [CrossRef]
- Dietze, K.; Tucakov, A.; Engel, T.; Wirtz, S.; Depner, K.; Globig, A.; Kammerer, R.; Mouchantat, S. Rope-based oral fluid sampling for early detection of classical swine fever in domestic pigs at group level. BMC Veter. Res. 2016, 13, 5. [Google Scholar] [CrossRef]
- Goonewardene, K.B.; Chung, C.J.; Goolia, M.; Blakemore, L.; Fabian, A.; Mohamed, F.; Nfon, C.; Clavijo, A.; Dodd, K.A.; Ambagala, A. Evaluation of oral fluid as an aggregate sample for early detection of African swine fever virus using four independent pen-based experimental studies. Transbound. Emerg. Dis. 2021, 68, 2867–2877. [Google Scholar] [CrossRef]
- Pieters, M.; Daniels, J.; Rovira, A. Comparison of sample types and diagnostic methods for in vivo detection of Mycoplasma hyopneumoniae during early stages of infection. Veter. Microbiol. 2017, 203, 103–109. [Google Scholar] [CrossRef]
- Deffner, P.; Maurer, R.; Cvjetković, V.; Sipos, W.; Krejci, R.; Ritzmann, M.; Eddicks, M. Cross-sectional study on the in-herd prevalence of Mycoplasma hyopneumoniae at different stages of pig production. Veter. Rec. 2022, 191, e1317. [Google Scholar] [CrossRef] [PubMed]
- Schaller, A.; Kuhn, R.; Kuhnert, P.; Nicolet, J.; Anderson, T.J.; Maclnnes, J.I.; Segers, R.P.A.M.; Frey, J. Characterization of apxlVA, a new RTX determinant of Actinobacillus pleuropneumoniae. Microbiology 1999, 145, 2105–2116. [Google Scholar] [CrossRef] [PubMed]
- Bosse, J.; Janson, H.; Sheehan, B.J.; Beddek, A.J.; Rycroft, A.N.; Kroll, J.S.; Langford, P. Actinobacillus pleuropneumoniae: Pathobiology and pathogenesis of infection. Microbes. Infect. 2002, 4, 225–235. [Google Scholar] [CrossRef]
- Rayamajhi, N.; Shin, S.J.; Kang, S.G.; Lee, D.Y.; Ahn, J.M.; Yoo, H.S. Development and Use of a Multiplex Polymerase Chain Reaction Assay Based on Apx Toxin Genes for Genotyping of Actinobacillus Pleuropneumoniae Isolates. J. Veter. Diagn. Investig. 2005, 17, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Gram, T.; Ahrens, P.; Andreasen, M.; Nielsen, J.P. An Actinobacillus pleuropneumoniae PCR typing system based on the apx and omlA genes—Evaluation of isolates from lungs and tonsils of pigs. Veter. Microbiol. 2000, 75, 43–57. [Google Scholar] [CrossRef]
- Ito, H. Development of a cps-Based Multiplex PCR for Typing of Actinobacillus pleuropneumoniae Serotypes 1,2 and 5. J. Veter. Med. Sci. 2010, 72, 653–655. [Google Scholar] [CrossRef] [PubMed]
- Jessing, S.G.; Angen, Ø.; Inzana, T.J. Evaluation of a Multiplex PCR Test for Simultaneous Identification and Serotyping of Actinobacillus pleuropneumoniae Serotypes 2, 5, and 6. J. Clin. Microbiol. 2003, 41, 4095–4100. [Google Scholar] [CrossRef]
- Bossé, J.T.; Li, Y.; Crespo, R.F.; Lacouture, S.; Gottschalk, M.; Sárközi, R.; Fodor, L.; Amoribieta, M.C.; Angen, Ø.; Nedbalcova, K.; et al. Comparative sequence analysis of the capsular polysaccharide loci of Actinobacillus pleuropneumoniae serovars 1–18, and development of two multiplex PCRs for comprehensive capsule typing. Veter. Microbiol. 2018, 220, 83–89. [Google Scholar] [CrossRef]
- Stringer, O.W.; Bossé, J.T.; Lacouture, S.; Gottschalk, M.; Fodor, L.; Angen, Ø.; Velazquez, E.; Penny, P.; Lei, L.; Langford, P.R.; et al. Proposal of Actinobacillus pleuropneumoniae serovar 19, and reformulation of previous multiplex PCRs for capsule-specific typing of all known serovars. Veter. Microbiol. 2021, 255, 109021. [Google Scholar] [CrossRef]
- Xiao, G.; Cao, S.; Huang, X.; Wen, X. DNA microarray-based identification and typing of Actinobacillus pleuropneumoniae. Can. J. Vet. Res. 2009, 73, 190–199. [Google Scholar]
- Tobias, T.; Bouma, A.; Klinkenberg, D.; Daemen, A.; Stegeman, J.; Wagenaar, J.; Duim, B. Detection of Actinobacillus pleuropneumoniae in pigs by real-time quantitative PCR for the apxIVA gene. Veter. J. 2012, 193, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Savoye, C.; Jobert, J.; Berthelot-Hérault, F.; Keribin, A.; Cariolet, R.; Morvan, H.; Madec, F.; Kobisch, M. A PCR assay used to study aerosol transmission of Actinobacillus pleuropneumoniae from samples of live pigs under experimental conditions. Veter. Microbiol. 2000, 73, 337–347. [Google Scholar] [CrossRef]
- Chiers, K.; Van Overbeke, I.; Donné, E.; Baele, M.; Ducatelle, R.; De Baere, T.; Haesebrouck, F. Detection of Actinobacillus pleuropneumoniae in cultures from nasal and tonsillar swabs of pigs by a PCR assay based on the nucleotide sequence of a dsbE-like gene. Veter. Microbiol. 2001, 83, 147–159. [Google Scholar] [CrossRef]
- Fablet, C.; Renson, P.; Pol, F.; Dorenlor, V.; Mahé, S.; Eono, F.; Eveno, E.; Le Dimna, M.; Liegard-Vanhecke, D.; Eudier, S.; et al. Oral fluid versus blood sampling in group-housed sows and finishing pigs: Feasibility and performance of antibody detection for porcine reproductive and respiratory syndrome virus (PRRSV). Veter. Microbiol. 2017, 204, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Prickett, J.R.; Zimmerman, J.J. The development of oral fluid-based diagnostics and applications in veterinary medicine. Anim. Health Res. Rev. 2010, 11, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Sthitmatee, N.; Sirinarumitr, T.; Makonkewkeyoon, L.; Sakpuaram, T.; Tesaprateep, T. Identification of the Actinobacillus pleuropneumoniae serotype using PCR based-apx genes. Mol. Cell. Probes 2003, 17, 301–305. [Google Scholar] [CrossRef]
- Bundesministerium der Justiz. Verordnung zum Schutz landwirtschaftlicher Nutztiere und Anderer zur Erzeugung Tierischer Produkte Gehaltener Tiere bei ihrer Haltung (Tierschutz-Nutztierhaltungsverordnung—TierSchNutztV). Available online: https://www.gesetze-im-internet.de/tierschnutztv/BJNR275800001.html (accessed on 30 August 2022).
- Prickett, J.R.; Kim, W.; Simer, R.; Yoon, K.-J.; Zimmerman, J. Oral-fluid samples for surveillance of commercial growing pigs for porcine reproductive and respiratory syndrome virus and porcine circovirus type 2 infections. J. Swine Health Prod. 2008, 16, 86–91. [Google Scholar]
- Dottori, M.; Nigrelli, A.; Bonilauri, P.; Merialdi, G.; Gozio, S.; Cominotti, F. Proposta per un nuovo sistema di punteggiatura delle pleuriti suine in sede di macellazione: La griglia SPES (Slaughterhouse Pleurisy Evaluation System). Large Anim. Rev. 2007, 13, 161–165. [Google Scholar]
- Madec, F.; Kobisch, M. Lung lesion scoring of finisher pigs at the slaughterhouse. Journées De La Rech. Porc. 1982, 14, 405–412. [Google Scholar]
- Garibyan, L.; Avashia, N. Polymerase Chain Reaction. J. Investig. Dermatol. 2013, 133, 1–4. [Google Scholar] [CrossRef]
- Opriessnig, T.; Hemann, M.; Johnson, J.K.; Heinen, S.; Giménez-Lirola, L.G.; O’Neill, K.C.; Hoang, H.; Yoon, K.-J.; Gottschalk, M.; Halbur, P.G. Evaluation of diagnostic assays for the serological detection of Actinobacillus pleuropneumoniae on samples of known or unknown exposure. J. Veter. Diagn. Investig. 2013, 25, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Dreyfus, A.; Schaller, A.; Nivollet, S.; Segers, R.; Kobisch, M.; Mieli, L.; Soerensen, V.; Hüssy, D.; Miserez, R.; Zimmermann, W.; et al. Use of recombinant ApxIV in serodiagnosis of Actinobacillus pleuropneumoniae infections, development and prevalidation of the ApxIV ELISA. Veter. Microbiol. 2004, 99, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Giménez-Lirola, L.G.; Jiang, Y.-H.; Sun, D.; Hoang, H.; Yoon, K.-J.; Halbur, P.G.; Opriessnig, T. Simultaneous Detection of Antibodies against Apx Toxins ApxI, ApxII, ApxIII, and ApxIV in Pigs with Known and Unknown Actinobacillus pleuropneumoniae Exposure Using a Multiplexing Liquid Array Platform. Clin. Vaccine Immunol. 2013, 21, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M. The challenge of detecting herds sub-clinically infected with Actinobacillus pleuropneumoniae. Veter. J. 2015, 206, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M. Actinobacillosis. In Diseases of Swine; Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Eds.; Wiley-Blackwell: Chichest, UK, 2019; Volume 11, pp. 749–767. [Google Scholar]
- Chiers, K.; Donné, E.; Van Overbeke, I.; Ducatelle, R.; Haesebrouck, F. Actinobacillus pleuropneumoniae infections in closed swine herds: Infection patterns and serological profiles. Veter. Microbiol. 2002, 85, 343–352. [Google Scholar] [CrossRef]
- Cannon, R. Livestock Disease Surveys: A Field Manual for Veterinarians; Australian Government Publishing Service: Canbarra, Australia, 1982. [Google Scholar]
- Tobias, T.J.; Bouma, A.; Daemen, A.J.; Wagenaar, J.A.; Stegeman, A.; Klinkenberg, D. Association between transmission rate and disease severity for Actinobacillus pleuropneumoniae infection in pigs. Veter. Res. 2013, 44, 2. [Google Scholar] [CrossRef]
- Schaller, A.; Kuhnert, P.; de la Puente-Redondo, V.A.; Nicolet, J.; Frey, J. Apx toxins in Pasteurellaceae species from animals. Veter. Microbiol. 2000, 74, 365–376. [Google Scholar] [CrossRef]
- Freestone, P.P.; Sandrini, S.M.; Haigh, R.D.; Lyte, M. Microbial endocrinology: How stress influences susceptibility to infection. Trends Microbiol. 2008, 16, 55–64. [Google Scholar] [CrossRef]
- Peterson, P.K.; Chao, C.C.; Molitor, T.; Murtaugh, M.; Strgar, F.; Sharp, B.M. Stress and Pathogenesis of Infectious Disease. Clin. Infect. Dis. 1991, 13, 710–720. [Google Scholar] [CrossRef]
- Chiers, K.; De Waele, T.; Pasmans, F.; Ducatelle, R.; Haesebrouck, F. Virulence factors of Actinobacillus pleuropneumoniae involved in colonization, persistence and induction of lesions in its porcine host. Veter. Res. 2010, 41, 65. [Google Scholar] [CrossRef]
- Tobias, T.; Bouma, A.; Broek, J.V.D.; van Nes, A.; Daemen, A.; Wagenaar, J.; Stegeman, J.; Klinkenberg, D. Transmission of Actinobacillus pleuropneumoniae among weaned piglets on endemically infected farms. Prev. Veter. Med. 2014, 117, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, C.S.; Angen, Ø.; Andreasen, M.; Takai, H.; Nielsen, J.P.; Jorsal, S.E. Demonstration of airborne transmission of Actinobacillus pleuropneumoniae serotype 2 between simulated pig units located at close range. Veter. Microbiol. 2004, 98, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Jobert, J.L.; Savoye, C.; Cariolet, R.; Kobisch, M.; Madec, F. Experimental aerosol transmission of Actinobacillus pleuropneumoniae to pigs. Can. J. Veter. Res. 2000, 64, 21–26. [Google Scholar]

| Farm A | Farm B | |||||
|---|---|---|---|---|---|---|
| Time of Sampling | Pigs/Pen | Collected OFs/Pen | Total Collected | Pigs/Pen | Collected Ofs/Pen | Total Collected |
| D 0–7 | 35 | 2 | 128 | 27 | 2 | 128 |
| W 3 | 35 | 2 | 16 | 26–27 | 2 | 16 |
| W 4 | 35 | 2 | 16 | - | - | - |
| W 5–11 | 22–23 | 1 | 32 | 26–27 | 2 | 64 |
| W 13 | 23 | 1 | 8 | 0–26 | 1–2 * | 14 |
| W 15 | 8–21 | 1 | 8 | 1–15 | 1 * | 8 |
| W17 | 0–8 | 0–1 * | 6 | - | - | - |
| W 19 | 0–2 | 0–1 * | 4 | - | - | - |
| Total | 218 | 230 | ||||
| Farm | Time | Apx IV–Apx III | Apx IV–Apx I |
|---|---|---|---|
| A | total | rs: 0.355, p < 0.001 | rs: 0.218, p = 0.001 |
| W1 | - | rs: 0.377, p < 0.001 | |
| W4 | rs: 0.746, p = 0.001 | - | |
| W5 | - | rs: 0.764, p < 0.027 | |
| B | Total | rs: 0.340, p < 0.001 | - |
| W1 | rs: 0.364, p < 0.001 | - | |
| W15 | rs: 0.741, p < 0.036 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kleinmans, M.; Fiebig, K.; Tabeling, R.; Swam, H.; Duivelshof-Crienen, A.; Ritzmann, M.; Eddicks, M. Explorative Field Study on the Use of Oral Fluids for the Surveillance of Actinobacillus pleuropneumoniae Infections in Fattening Farms by an Apx-Real-Time PCR. Vet. Sci. 2022, 9, 552. https://doi.org/10.3390/vetsci9100552
Kleinmans M, Fiebig K, Tabeling R, Swam H, Duivelshof-Crienen A, Ritzmann M, Eddicks M. Explorative Field Study on the Use of Oral Fluids for the Surveillance of Actinobacillus pleuropneumoniae Infections in Fattening Farms by an Apx-Real-Time PCR. Veterinary Sciences. 2022; 9(10):552. https://doi.org/10.3390/vetsci9100552
Chicago/Turabian StyleKleinmans, Michael, Kerstin Fiebig, Robert Tabeling, Hanny Swam, Annelies Duivelshof-Crienen, Mathias Ritzmann, and Matthias Eddicks. 2022. "Explorative Field Study on the Use of Oral Fluids for the Surveillance of Actinobacillus pleuropneumoniae Infections in Fattening Farms by an Apx-Real-Time PCR" Veterinary Sciences 9, no. 10: 552. https://doi.org/10.3390/vetsci9100552
APA StyleKleinmans, M., Fiebig, K., Tabeling, R., Swam, H., Duivelshof-Crienen, A., Ritzmann, M., & Eddicks, M. (2022). Explorative Field Study on the Use of Oral Fluids for the Surveillance of Actinobacillus pleuropneumoniae Infections in Fattening Farms by an Apx-Real-Time PCR. Veterinary Sciences, 9(10), 552. https://doi.org/10.3390/vetsci9100552





