Feline Oncogenomics: What Do We Know about the Genetics of Cancer in Domestic Cats?
Abstract
Simple Summary
Abstract
1. Introduction
2. Lymphoma
3. Mammary Tumours
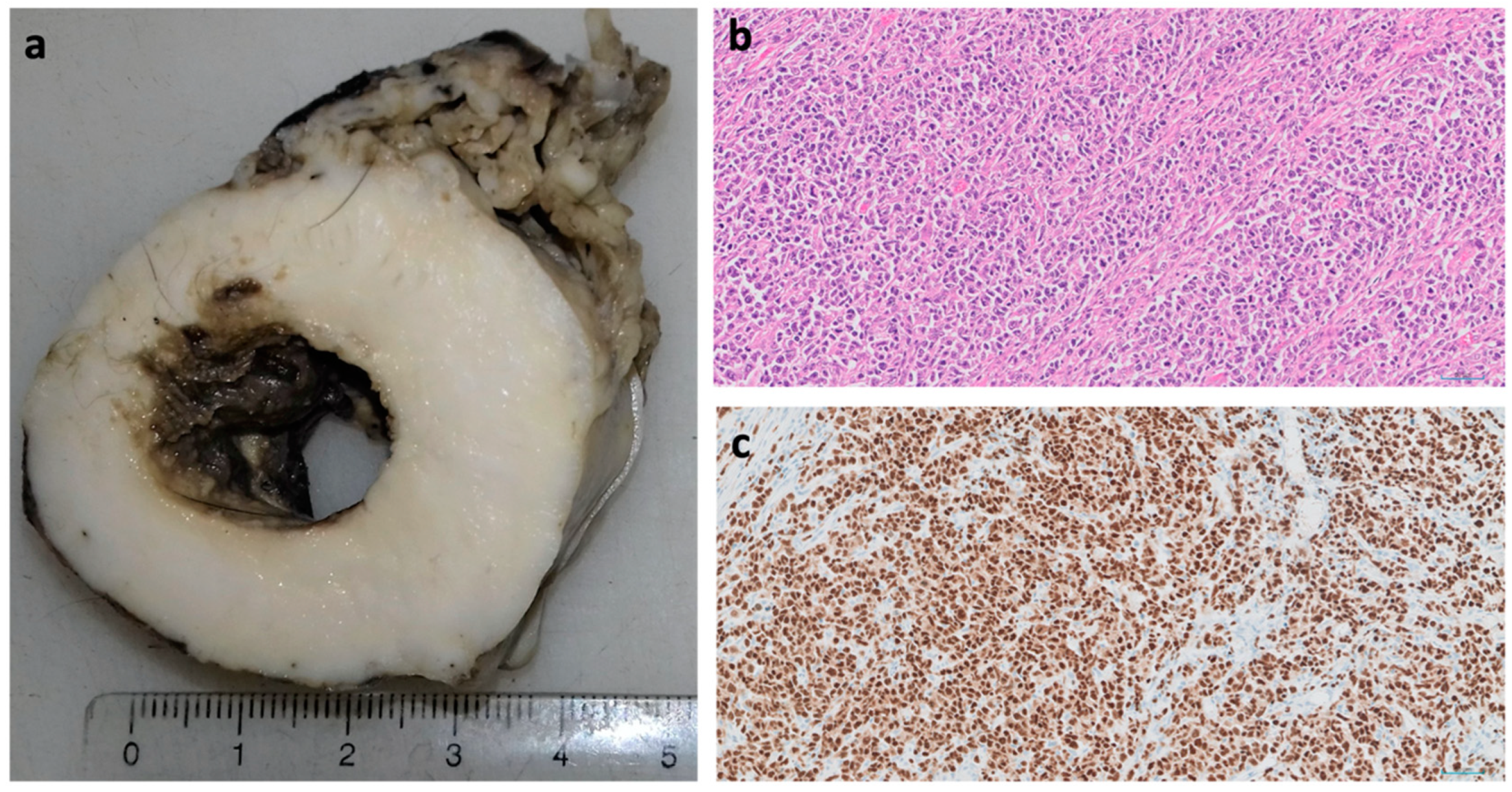
4. Squamous Cell Carcinoma
5. Soft Tissue Tumours
6. Mast Cell Tumours
7. Haemangiosarcoma
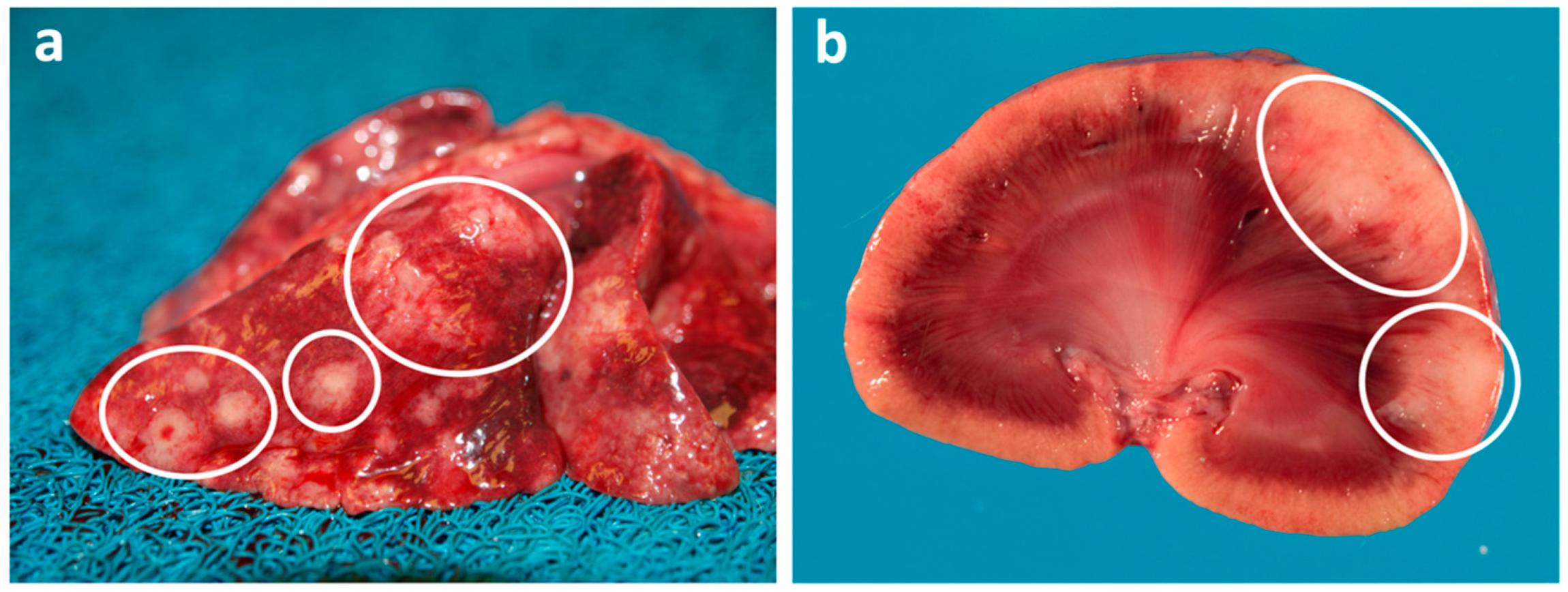
8. Pulmonary Carcinoma
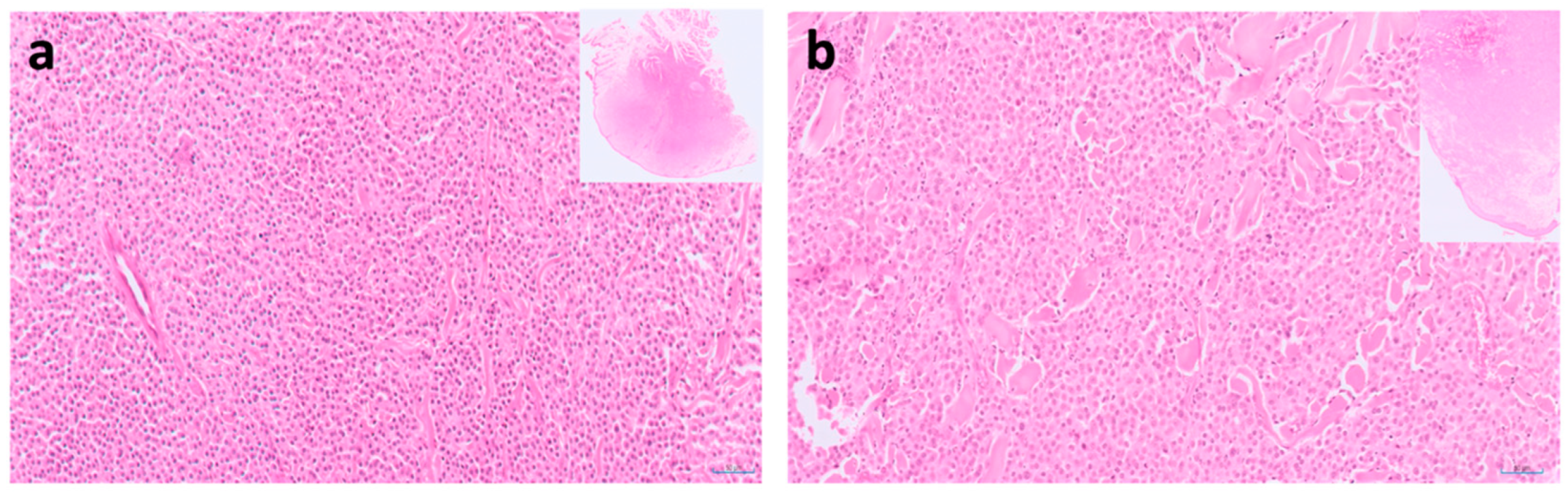
9. Pancreatic Carcinoma
10. Osteosarcoma
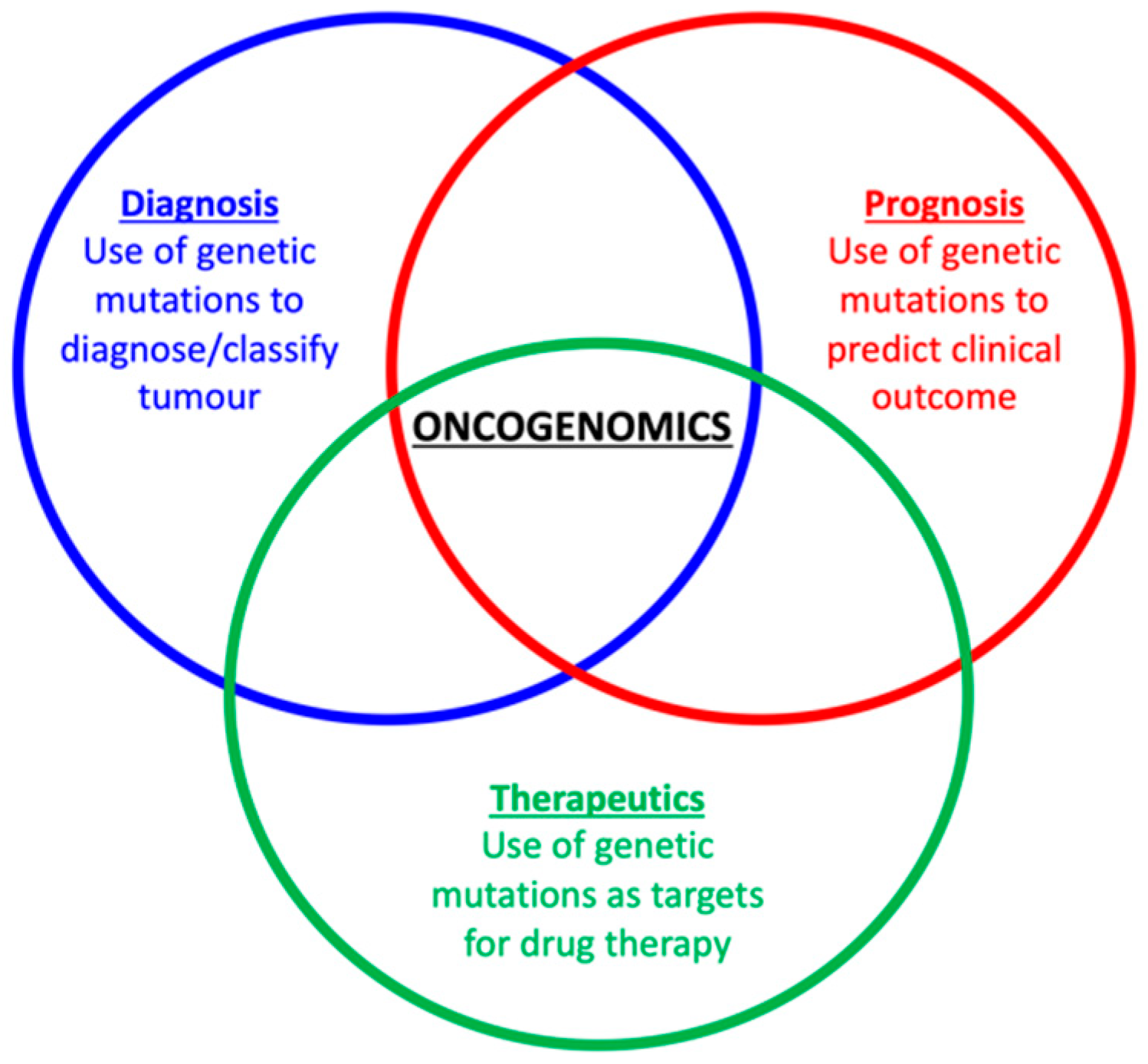
11. Looking towards a Future of Greater Understanding of the Feline Oncogenome
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perrin, T. The Business of Urban Animals Survey: The facts and statistics on companion animals in Canada. Can. Vet. J 2009, 50, 48–52. [Google Scholar] [PubMed]
- Bir, C.; Ortez, M.; Olynk Widmar, N.J.; Wolf, C.A.; Hansen, C.; Ouedraogo, F.B. Familiarity and Use of Veterinary Services by US Resident Dog and Cat Owners. Animals 2020, 10, 483. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.K.; Gruffydd-Jones, T.J.; Roberts, M.A.; Browne, W.J. Assessing changes in the UK pet cat and dog populations: Numbers and household ownership. Vet. Rec. 2015, 177, 259. [Google Scholar] [CrossRef] [PubMed]
- MacVean, D.W.; Monlux, A.W.; Anderson, P.S., Jr.; Silberg, S.L.; Roszel, J.F. Frequency of canine and feline tumors in a defined population. Vet. Pathol. 1978, 15, 700–715. [Google Scholar] [CrossRef] [PubMed]
- Graf, R.; Gruntzig, K.; Hassig, M.; Axhausen, K.W.; Fabrikant, S.; Welle, M.; Meier, D.; Guscetti, F.; Folkers, G.; Otto, V.; et al. Swiss Feline Cancer Registry: A Retrospective Study of the Occurrence of Tumours in Cats in Switzerland from 1965 to 2008. J Comp. Pathol. 2015, 153, 266–277. [Google Scholar] [CrossRef]
- Ho, N.T.; Smith, K.C.; Dobromylskyj, M.J. Retrospective study of more than 9000 feline cutaneous tumours in the UK: 2006–2013. J. Feline Med. Surg. 2018, 20, 128–134. [Google Scholar] [CrossRef]
- Fuentes-Pananá, E.M.; Alonso-Morales, R.A.; Romero-Romero, L.; Pérez-Enriquez, J.M. Tumor prevalence in cats: Experience from a reference diagnostic center in Mexico City (2006–2018). Vet. Méx. OA 2020, 7, 837. [Google Scholar] [CrossRef]
- Soares, M.; Marques, C.; Catarino, J.; Batista, M.R.; Catita, J.; Faisca, P. National survey of cat tumors in 2019: A retrospective study. Rev. Lusófona Ciênc. Med. Vet. 2021, 11, 14–19. [Google Scholar]
- Pinello, K.; Pires, I.; Castro, A.F.; Carvalho, P.T.; Santos, A.; de Matos, A.; Queiroga, F.; Canadas-Sousa, A.; Dias-Pereira, P.; Catarino, J.; et al. Cross Species Analysis and Comparison of Tumors in Dogs and Cats, by Age, Sex, Topography and Main Morphologies. Data from Vet-OncoNet. Vet. Sci. 2022, 9, 167. [Google Scholar] [CrossRef]
- Kastenhuber, E.R.; Lowe, S.W. Putting p53 in Context. Cell 2017, 170, 1062–1078. [Google Scholar] [CrossRef]
- Pontius, J.U.; Mullikin, J.C.; Smith, D.R.; Agencourt Sequencing, T.; Lindblad-Toh, K.; Gnerre, S.; Clamp, M.; Chang, J.; Stephens, R.; Neelam, B.; et al. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007, 17, 1675–1689. [Google Scholar] [CrossRef] [PubMed]
- Montague, M.J.; Li, G.; Gandolfi, B.; Khan, R.; Aken, B.L.; Searle, S.M.; Minx, P.; Hillier, L.W.; Koboldt, D.C.; Davis, B.W.; et al. Comparative analysis of the domestic cat genome reveals genetic signatures underlying feline biology and domestication. Proc. Natl. Acad. Sci. USA 2014, 111, 17230–17235. [Google Scholar] [CrossRef] [PubMed]
- Tamazian, G.; Simonov, S.; Dobrynin, P.; Makunin, A.; Logachev, A.; Komissarov, A.; Shevchenko, A.; Brukhin, V.; Cherkasov, N.; Svitin, A.; et al. Annotated features of domestic cat—Felis catus genome. Gigascience 2014, 3, 13. [Google Scholar] [CrossRef] [PubMed]
- Buckley, R.M.; Davis, B.W.; Brashear, W.A.; Farias, F.H.G.; Kuroki, K.; Graves, T.; Hillier, L.W.; Kremitzki, M.; Li, G.; Middleton, R.P.; et al. A new domestic cat genome assembly based on long sequence reads empowers feline genomic medicine and identifies a novel gene for dwarfism. PLoS Genet. 2020, 16, e1008926. [Google Scholar] [CrossRef]
- Vail, D.M.; Thamm, D.H.; Liptak, J. Withrow and MacEwen’s Small Animal Clinical Oncology, 6th ed.; Elselvier: Edinburgh, UK, 2020; p. 864. [Google Scholar]
- Louwerens, M.; London, C.A.; Pedersen, N.C.; Lyons, L.A. Feline lymphoma in the post-feline leukemia virus era. J. Vet. Intern. Med. 2005, 19, 329–335. [Google Scholar] [CrossRef]
- Leiet-Filho, R.V.; Panziera, W.; Bandinelli, M.B.; Henker, L.C.; da Conceição Monteiro, K.; Corbellini, L.G.; Driemeier, D.; Sonne, L.; Pavarini, S.P. Epidemiological, pathological and immunohistochemical aspects of 125 cases of feline lymphoma in Southern Brazil. Vet. Comp. Oncol. 2020, 18, 224–230. [Google Scholar] [CrossRef]
- Paulin, M.V.; Couronne, L.; Beguin, J.; Le Poder, S.; Delverdier, M.; Semin, M.O.; Bruneau, J.; Cerf-Bensussan, N.; Malamut, G.; Cellier, C.; et al. Feline low-grade alimentary lymphoma: An emerging entity and a potential animal model for human disease. BMC Vet. Res. 2018, 14, 306. [Google Scholar] [CrossRef]
- Stein, T.J.; Pellin, M.; Steinberg, H.; Chun, R. Treatment of feline gastrointestinal small-cell lymphoma with chlorambucil and glucocorticoids. J. Am. Anim. Hosp. Assoc. 2010, 46, 413–417. [Google Scholar] [CrossRef]
- Kiselow, M.A.; Rassnick, K.M.; McDonough, S.P.; Goldstein, R.E.; Simpson, K.W.; Weinkle, T.K.; Erb, H.N. Outcome of cats with low-grade lymphocytic lymphoma: 41 cases (1995–2005). J. Am. Vet. Med. Assoc. 2008, 232, 405–410. [Google Scholar] [CrossRef]
- Collette, S.A.; Allstadt, S.D.; Chon, E.M.; Vernau, W.; Smith, A.N.; Garrett, L.D.; Choy, K.; Rebhun, R.B.; Rodriguez, C.O., Jr.; Skorupski, K.A. Treatment of feline intermediate- to high-grade lymphoma with a modified university of Wisconsin-Madison protocol: 119 cases (2004–2012). Vet. Comp. Oncol. 2016, 14 (Suppl. S1), 136–146. [Google Scholar] [CrossRef]
- Fabrizio, F.; Calam, A.E.; Dobson, J.M.; Middleton, S.A.; Murphy, S.; Taylor, S.S.; Schwartz, A.; Stell, A.J. Feline mediastinal lymphoma: A retrospective study of signalment, retroviral status, response to chemotherapy and prognostic indicators. J. Feline Med. Surg. 2014, 16, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Teske, E.; van Straten, G.; van Noort, R.; Rutteman, G.R. Chemotherapy with cyclophosphamide, vincristine, and prednisolone (COP) in cats with malignant lymphoma: New results with an old protocol. J. Vet. Intern. Med. 2002, 16, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Vail, D.M.; Moore, A.S.; Ogilvie, G.K.; Volk, L.M. Feline lymphoma (145 cases): Proliferation indices, cluster of differentiation 3 immunoreactivity, and their association with prognosis in 90 cats. J. Vet. Intern. Med. 1998, 12, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.G.; Hohenhaus, A.E.; Lamb, K.E. Incidence and treatment of feline renal lymphoma: 27 cases. J. Feline Med. Surg. 2021, 23, 936–944. [Google Scholar] [CrossRef] [PubMed]
- Okuda, M.; Umeda, A.; Sakai, T.; Ohashi, T.; Momoi, Y.; Youn, H.Y.; Watari, T.; Goitsuka, R.; Tsujimoto, H.; Hasegawa, A. Cloning of feline p53 tumor-suppressor gene and its aberration in hematopoietic tumors. Int. J. Cancer 1994, 58, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Miki, R.; Okuda, M.; Oikawa, T.; Watanabe, M.; Ma, Z.; Matsumoto, K.; Iwata, H.; Inokuma, H. Centrosome amplification and chromosomal instability in feline lymphoma cell lines. J. Vet. Med. Sci. 2004, 66, 797–805. [Google Scholar] [CrossRef][Green Version]
- Oikawa, T.; Okuda, M.; Kaneko, N.; Watanabe, M.; Hiraoka, H.; Itamoto, K.; Nakaichi, M.; Mizuno, T.; Inokuma, H. Cloning of the feline GADD45 cDNA and analysis of its mutation in feline lymphoma cell lines. J. Vet. Med. Sci. 2006, 68, 297–301. [Google Scholar] [CrossRef][Green Version]
- Rydzewski, L.; Scheffold, S.; Hecht, W.; Burkhardt, E.; Kerner, K.; Klymiuk, M.C.; Deinzer, R.; Reinacher, M.; Henrich, M. Identification of a novel feline large granular lymphoma cell line (S87) as non-MHC-restricted cytotoxic T-cell line and assessment of its genetic instability. Vet. Immunol. Immunopathol. 2016, 177, 24–34. [Google Scholar] [CrossRef]
- Fujita, M.; Kaneda, M. DNA methylation inhibitor causes cell growth retardation and gene expression changes in feline lymphoma cells. J. Vet. Med. Sci. 2017, 79, 1352–1358. [Google Scholar] [CrossRef][Green Version]
- Yamazaki, J.; Jelinek, J.; Yokoyama, S.; Takiguchi, M. Genome-wide DNA methylation profile in feline haematological tumours: A preliminary study. Res. Vet. Sci. 2021, 140, 221–228. [Google Scholar] [CrossRef]
- Mayr, B.; Heczko, U.; Schellander, K.; Schleger, W.; Reifinger, M. Sequence of an exon of the feline p53 gene--mutation in a lymphosarcoma. Br. Vet. J. 1993, 149, 387–390. [Google Scholar] [CrossRef]
- Okuda, M.; Minehata, K.; Setoguchi, A.; Watari, T.; Goitsuka, R.; Tsujimoto, H.; Hasegawa, A. Cloning of feline p21WAF1 and p27Kip1 cDNAs and search for their aberration in leukemias and lymphomas in cats. Leukemia 1997, 11, 372–375. [Google Scholar] [PubMed]
- Mayr, B.; Winkler, G.; Schaffner, G.; Reifinger, M.; Brem, G. N-ras mutation in a feline lymphoma. Low frequency of N-ras mutations in a series of feline, canine and bovine lymphomas. Vet. J. 2002, 163, 326–328. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Baba, K.; Goto, Y.; Masuda, K.; Ohno, K.; Tsujimoto, H. Alternatively spliced transcripts of Fas mRNAs in feline lymphoid cells. Eur. J. Immunogenet. 2004, 31, 159–166. [Google Scholar] [CrossRef]
- Roberti, A.; Dobay, M.P.; Bisig, B.; Vallois, D.; Boechat, C.; Lanitis, E.; Bouchindhomme, B.; Parrens, M.C.; Bossard, C.; Quintanilla-Martinez, L.; et al. Type II enteropathy-associated T-cell lymphoma features a unique genomic profile with highly recurrent SETD2 alterations. Nat. Commun. 2016, 7, 12602. [Google Scholar] [CrossRef]
- Kieslinger, M.; Swoboda, A.; Kramer, N.; Freund, P.; Pratscher, B.; Neubauer, H.A.; Steinborn, R.; Wolfesberger, B.; Fuchs-Baumgartinger, A.; Moriggl, R.; et al. A Recurrent STAT5B(N642H) Driver Mutation in Feline Alimentary T Cell Lymphoma. Cancers 2021, 13, 5238. [Google Scholar] [CrossRef]
- MacEwen, E. Spontaneous tumors in dogs and cats: Models for the study of cancer biology and treatment. Cancer Metastasis Rev. 1990, 9, 125–136. [Google Scholar] [CrossRef]
- Zapulli, V.; Peña, L.; Rasotto, R.; Kiupel, M.; Goldschmidt, M.H.; Gama, A.; Scruggs, J.L. Surgical Pathology of Tumors of Domestic Animals Volume 2: Mammary Tumors: Mammary Tumors; Davis Thompson Foundation: Pullman, WA, USA, 2019. [Google Scholar]
- Petrucci, G.N.; Henriques, J.; Lobo, L.; Vilhena, H.; Figueira, A.C.; Canadas-Sousa, A.; Dias-Pereira, P.; Prada, J.; Pires, I.; Queiroga, F.L. Adjuvant doxorubicin vs metronomic cyclophosphamide and meloxicam vs surgery alone for cats with mammary carcinomas: A retrospective study of 137 cases. Vet. Comp. Oncol. 2021, 19, 714–723. [Google Scholar] [CrossRef]
- Soares, M.; Madeira, S.; Correia, J.; Peleteiro, M.; Cardoso, F.; Ferreira, F. Molecular based subtyping of feline mammary carcinomas and clinicopathological characterization. Breast 2016, 27, 44–51. [Google Scholar] [CrossRef]
- Soares, M.; Correia, J.; Peleteiro, M.C.; Ferreira, F. St Gallen molecular subtypes in feline mammary carcinoma and paired metastases-disease progression and clinical implications from a 3-year follow-up study. Tumour. Biol. 2016, 37, 4053–4064. [Google Scholar] [CrossRef]
- Granados-Soler, J.L.; Bornemann-Kolatzki, K.; Beck, J.; Brenig, B.; Schutz, E.; Betz, D.; Junginger, J.; Hewicker-Trautwein, M.; Murua Escobar, H.; Nolte, I. Analysis of Copy-Number Variations and Feline Mammary Carcinoma Survival. Sci. Rep. 2020, 10, 1003. [Google Scholar] [CrossRef] [PubMed]
- Sommerville, L.; Howard, J.; Evans, S.; Kelly, P.; McCann, A. Comparative gene expression study highlights molecular similarities between triple negative breast cancer tumours and feline mammary carcinomas. Vet. Comp. Oncol. 2022, 20, 535–538. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, L.; Derose, Y.S.; Lin, Y.C.; Bieniasz, M.; Eyob, H.; Buys, S.S.; Neumayer, L.; Welm, A.L. Short-Form Ron Promotes Spontaneous Breast Cancer Metastasis through Interaction with Phosphoinositide 3-Kinase. Genes Cancer 2011, 2, 753–762. [Google Scholar] [CrossRef]
- Ishikawa, T.; Ichikawa, Y.; Shimizu, D.; Sasaki, T.; Tanabe, M.; Chishima, T.; Takabe, K.; Endo, I. The role of HER-2 in Breast Cancer. J. Surg. Sci. 2014, 2, 4–9. [Google Scholar] [PubMed]
- Maniscalco, L.; Guil-Luna, S.; Iussich, S.; Gattino, F.; Trupia, C.; Millan, Y.; de Las Mulas, J.M.; Cespedez, R.S.; Saeki, K.; Accornero, P.; et al. Expression of the Short Form of RON/STK in Feline Mammary Carcinoma. Vet. Pathol. 2019, 56, 220–229. [Google Scholar] [CrossRef] [PubMed]
- De Maria, R.; Olivero, M.; Iussich, S.; Nakaichi, M.; Murata, T.; Biolatti, B.; Di Renzo, M.F. Spontaneous feline mammary carcinoma is a model of HER2 overexpressing poor prognosis human breast cancer. Cancer Res. 2005, 65, 907–912. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, D.; Soares, M.; Correia, J.; Adega, F.; Ferreira, F.; Chaves, R. Assessment of ERBB2 and TOP2α gene status and expression profile in feline mammary tumors: Findings and guidelines. Aging 2019, 11, 4688–4705. [Google Scholar] [CrossRef]
- Santos, S.; Baptista, C.S.; Abreu, R.M.; Bastos, E.; Amorim, I.; Gut, I.G.; Gartner, F.; Chaves, R. ERBB2 in cat mammary neoplasias disclosed a positive correlation between RNA and protein low expression levels: A model for erbB-2 negative human breast cancer. PLoS ONE 2013, 8, e83673. [Google Scholar] [CrossRef]
- Muscatello, L.V.; Di Oto, E.; Sarli, G.; Monti, V.; Foschini, M.P.; Benazzi, C.; Brunetti, B. HER2 Amplification Status in Feline Mammary Carcinoma: A Tissue Microarray-Fluorescence In Situ Hydridization-Based Study. Vet. Pathol. 2019, 56, 230–238. [Google Scholar] [CrossRef]
- Ferreira, D.; Martins, B.; Soares, M.; Correia, J.; Adega, F.; Ferreira, F.; Chaves, R. Gene expression association study in feline mammary carcinomas. PLoS ONE 2019, 14, e0221776. [Google Scholar] [CrossRef]
- Baptista, C.S.; Santos, S.; Laso, A.; Bastos, E.; Ávila, S.; Guedes-Pinto, H.; Gärtner, F.; Gut, I.G.; Castrillo, J.L.; Chaves, R. Sequence variation and mRNA expression of the TWIST1 gene in cats with mammary hyperplasia and neoplasia. Vet. J. 2012, 191, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Mayr, B.; Schaffner, G.; Kurzbauer, R.; Reifinger, M.; Schellander, K. Sequence of an exon of tumour suppressor p53 gene—A comparative study in domestic animals: Mutation in a feline solid mammary carcinoma. Br. Vet. J. 1995, 151, 325–329. [Google Scholar] [CrossRef]
- Mayr, B.; Reifinger, M.; Loupal, G. Polymorphisms in feline tumour suppressor gene p53. Mutations in an osteosarcoma and a mammary carcinoma. Vet. J. 1998, 155, 103–106. [Google Scholar] [CrossRef]
- Mayr, B.; Blauensteiner, J.; Edlinger, A.; Reifinger, M.; Alton, K.; Schaffner, G.; Brem, G. Presence of p53 mutations in feline neoplasms. Res. Vet. Sci. 2000, 68, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Govoni, V.M.; Da Silva, S.T.; Guerra, J.M.; Pereira, I.V.A.; Queiroga, F.L.; Cogliati, B. Genetic variants of BRCA1 and BRCA2 genes in cats with mammary gland carcinoma. Vet. Comp. Oncol. 2021, 19, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Kuchenbaecker, K.B.; Hopper, J.L.; Barnes, D.R.; Phillips, K.A.; Mooij, T.M.; Roos-Blom, M.J.; Jervis, S.; van Leeuwen, F.E.; Milne, R.L.; Andrieu, N.; et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA 2017, 317, 2402–2416. [Google Scholar] [CrossRef]
- Wiese, D.A.; Thaiwong, T.; Yuzbasiyan-Gurkan, V.; Kiupel, M. Feline mammary basal-like adenocarcinomas: A potential model for human triple-negative breast cancer (TNBC) with basal-like subtype. BMC Cancer 2013, 13, 403. [Google Scholar] [CrossRef]
- Sahlin, P.; Windh, P.; Lauritzen, C.; Emanuelsson, M.; Grönberg, H.; Stenman, G. Women with Saethre-Chotzen syndrome are at increased risk of breast cancer. Genes Chromosomes Cancer 2007, 46, 656–660. [Google Scholar] [CrossRef]
- Miller, M.A.; Nelson, S.; Turk, J.R.; Pace, L.W.; Brown, T.P.; Shaw, D.P.; Fischer, J.R.; Gosser, H.S. Cutaneous neoplasia in 340 cats. Vet Pathol. 1991, 28, 389–395. [Google Scholar] [CrossRef]
- Bilgic, O.; Duda, L.; Sànchez, M.D.; Lewis, J.R. Feline Oral Squamous Cell Carcinoma: Clinical Manifestations and Literature Review. J. Vet. Dent. 2015, 32, 30–40. [Google Scholar] [CrossRef]
- Withrow, S.J.; Liptak, J. Cancer of the gastrointestinal tract. In Small Animal Clinical Oncology; Withrow, S.J., Vail, D., Page, R.L., Eds.; Elsevier: St. Louis, MO, USA, 2013; pp. 381–383. [Google Scholar]
- Lana, S.E.; Ogilvie, G.; Withrow, S.J.; Straw, R.C.; Rogers, K.S. Feline cutaneous squamous cell carcinoma of the nasal planum and the pinnae: 61 cases. J. Am. Anim. Hosp. Assoc. 1997, 33, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Hayes, A.M.; Adams, V.J.; Scase, T.J.; Murphy, S. Survival of 54 cats with oral squamous cell carcinoma in United Kingdom general practice. J. Small Anim. Pract. 2007, 48, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Goldschmidt, M.H.; Goldschmidt, K.H. Chapter 4—Epithelial and melanocytic tumors of the skin. In Tumors in Domestic Animals, 5th ed.; Meuten, D.J., Ed.; John Wiley & Sons, Inc: Ames, IA, USA, 2017; pp. 88–141. [Google Scholar]
- Teifke, J.P.; Lohr, C.V. Immunohistochemical detection of P53 overexpression in paraffin wax-embedded squamous cell carcinomas of cattle, horses, cats and dogs. J. Comp. Pathol. 1996, 114, 205–210. [Google Scholar] [CrossRef]
- Renzi, A.; De Bonis, P.; Morandi, L.; Lenzi, J.; Tinto, D.; Rigillo, A.; Bettini, G.; Bellei, E.; Sabattini, S. Prevalence of p53 dysregulations in feline oral squamous cell carcinoma and non-neoplastic oral mucosa. PLoS ONE 2019, 14, e0215621. [Google Scholar] [CrossRef]
- Renzi, A.; Morandi, L.; Lenzi, J.; Rigillo, A.; Bettini, G.; Bellei, E.; Giacomini, A.; Tinto, D.; Sabattini, S. Analysis of DNA methylation and TP53 mutational status for differentiating feline oral squamous cell carcinoma from non-neoplastic mucosa: A preliminary study. Vet. Comp. Oncol. 2020, 18, 825–837. [Google Scholar] [CrossRef]
- Renzi, A.; Morandi, L.; Bellei, E.; Marconato, L.; Rigillo, A.; Aralla, M.; Lenzi, J.; Bettini, G.; Tinto, D.; Sabattini, S. Validation of oral brushing as a non-invasive technique for the identification of feline oral squamous cell carcinoma by DNA methylation and TP53 mutation analysis. Vet. Comp. Oncol. 2021, 19, 501–509. [Google Scholar] [CrossRef]
- Munday, J.S. Papillomaviruses in felids. Vet. J. 2014, 199, 340–347. [Google Scholar] [CrossRef]
- Munday, J.S.; Gibson, I.; French, A.F. Papillomaviral DNA and increased p16CDKN2A protein are frequently present within feline cutaneous squamous cell carcinomas in ultraviolet-protected skin. Vet. Dermatol. 2011, 22, 360–366. [Google Scholar] [CrossRef]
- Chu, S.; Wylie, T.; Wylie, K.M.; Johnson, G.C.; Skidmore, Z.L.; Fleer, M.; Griffith, O.L.; Bryan, J.N. A virome sequencing approach to feline oral squamous cell carcinoma to evaluate viral causative factors. Vet. Microbiol. 2020, 240, 108491. [Google Scholar] [CrossRef]
- Roccabianca, P.; Schulman, Y.; Avallone, G.; Foster, R.A.; Scruggs, J.L.; Dittmer, K.E.; Kiupel, M. Surgical Pathology of Tumors of Domestic Animals. 3: Tumors of Soft Tissue; Davis-Thompson DVM Foundation: Formoor Ln Gurnee, IL, USA, 2020. [Google Scholar]
- Avallone, G.; Boracchi, P.; Stefanello, D.; Ferrari, R.; Rebughini, A.; Roccabianca, P. Canine perivascular wall tumors: High prognostic impact of site, depth, and completeness of margins. Vet. Pathol. 2014, 51, 713–721. [Google Scholar] [CrossRef]
- Stefanello, D.; Avallone, G.; Ferrari, R.; Roccabianca, P.; Boracchi, P. Canine cutaneous perivascular wall tumors at first presentation: Clinical behavior and prognostic factors in 55 cases. J. Vet. Intern. Med. 2011, 25, 1398–1405. [Google Scholar] [CrossRef] [PubMed]
- Dobromylskyj, M.J.; Richards, V.; Smith, K.C. Prognostic factors and proposed grading system for cutaneous and subcutaneous soft tissue sarcomas in cats, based on a retrospective study. J. Feline Med. Surg. 2021, 23, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Davidson, E.B.; Gregory, C.; Kass, P.H. Surgical excision of soft tissue fibrosarcomas in cats. Vet. Surg. 2007, 26, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Shaw, S.C.; Kent, M.S.; Gordon, I.K.; Collins, C.J.; Greasby, T.A.; Beckett, L.A.; Hammond, G.M.; Skorupski, K.A. Temporal changes in characteristics of injection-site sarcomas in cats: 392 cases (1990–2006). J. Am. Vet. Med. Assoc. 2009, 234, 376–380. [Google Scholar] [CrossRef]
- Hartmann, K.; Day, M.J.; Thiry, E.; Lloret, A.; Frymus, T.; Addie, D.; Boucraut-Baralon, C.; Egberink, H.; Gruffydd-Jones, T.; Horzinek, M.C.; et al. Feline injection-site sarcoma: ABCD guidelines on prevention and management. J. Feline Med. Surg. 2015, 17, 606–613. [Google Scholar] [CrossRef]
- Sorensen, K.C.; Kitchell, B.E.; Schaeffer, D.J.; Mardis, P.E. Expression of matrix metalloproteinases in feline vaccine site-associated sarcomas. Am. J. Vet. Res. 2004, 65, 373–379. [Google Scholar] [CrossRef]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef]
- Banerji, N.; Kapur, V.; Kanjilal, S. Association of germ-line polymorphisms in the feline p53 gene with genetic predisposition to vaccine-associated feline sarcoma. J. Hered. 2007, 98, 421–427. [Google Scholar] [CrossRef]
- Mucha, D.; Laberke, S.; Meyer, S.; Hirschberger, J. Lack of association between p53 SNP and FISS in a cat population from Germany. Vet. Comp. Oncol. 2014, 12, 130–137. [Google Scholar] [CrossRef]
- Nambiar, P.R.; Haines, D.M.; Ellis, J.A.; Kidney, B.A.; Jackson, M.L. Mutational analysis of tumor suppressor gene p53 in feline vaccine site-associated sarcomas. Am. J. Vet. Res. 2000, 61, 1277–1281. [Google Scholar] [CrossRef]
- Banerji, N.; Kanjilal, S. Somatic alterations of the p53 tumor suppressor gene in vaccine-associated feline sarcoma. Am. J. Vet. Res. 2006, 67, 1766–1772. [Google Scholar] [CrossRef] [PubMed]
- Mayr, B.; Reifinger, M.; Alton, K.; Schaffner, G. Novel p53 tumour suppressor mutations in cases of spindle cell sarcoma, pleomorphic sarcoma and fibrosarcoma in cats. Vet. Res. Commun. 1998, 22, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Mayr, B.; Schaffner, G.; Kurzbauer, R.; Schneider, A.; Reifinger, M.; Loupal, G. Mutations in tumour suppressor gene p53 in two feline fibrosarcomas. Br. Vet. J. 1995, 151, 707–713. [Google Scholar] [CrossRef]
- Thomas, R.; Valli, V.E.; Ellis, P.; Bell, J.; Karlsson, E.K.; Cullen, J.; Lindblad-Toh, K.; Langford, C.F.; Breen, M. Microarray-based cytogenetic profiling reveals recurrent and subtype-associated genomic copy number aberrations in feline sarcomas. Chromosome Res. 2009, 17, 987–1000. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Pontius, J.U.; Borst, L.B.; Breen, M. Development of a Genome-Wide Oligonucleotide Microarray Platform for Detection of DNA Copy Number Aberrations in Feline Cancers. Vet. Sci. 2020, 7, 88. [Google Scholar] [CrossRef]
- Wei, Q.; Ramsey, S.A.; Larson, M.K.; Berlow, N.E.; Ochola, D.; Shiprack, C.; Kashyap, A.; Séguin, B.; Keller, C.; Löhr, C.V. Elucidating the transcriptional program of feline injection-site sarcoma using a cross-species mRNA-sequencing approach. BMC Cancer 2019, 19, 311. [Google Scholar] [CrossRef]
- Rissetto, K.; Villamil, J.A.; Selting, K.A.; Tyler, J.; Henry, C.J. Recent trends in feline intestinal neoplasia: An epidemiologic study of 1129 cases in the veterinary medical database from 1964 to 2004. J. Am. Anim. Hosp. Assoc. 2011, 47, 28–36. [Google Scholar] [CrossRef]
- Patnaik, A.K.; Ehler, W.J.; MacEwen, E.G. Canine cutaneous mast cell tumor: Morphologic grading and survival time in 83 dogs. Vet. Pathol. 1984, 21, 469–474. [Google Scholar] [CrossRef]
- Kiupel, M.; Webster, J.D.; Bailey, K.L.; Best, S.; DeLay, J.; Detrisac, C.J.; Fitzgerald, S.D.; Gamble, D.; Ginn, P.E.; Goldschmidt, M.H.; et al. Proposal of a 2-tier histologic grading system for canine cutaneous mast cell tumors to more accurately predict biological behavior. Vet. Pathol. 2011, 48, 147–155. [Google Scholar] [CrossRef]
- de Nardi, A.B.; Dos Santos Horta, R.; Fonseca-Alves, C.E.; de Paiva, F.N.; Linhares, L.C.M.; Firmo, B.F.; Ruiz Sueiro, F.A.; de Oliveira, K.D.; Lourenço, S.V.; De Francisco Strefezzi, R.; et al. Diagnosis, Prognosis and Treatment of Canine Cutaneous and Subcutaneous Mast Cell Tumors. Cells 2022, 11, 618. [Google Scholar] [CrossRef]
- Thompson, J.J.; Pearl, D.L.; Yager, J.A.; Best, S.J.; Coomber, B.L.; Foster, R.A. Canine subcutaneous mast cell tumor: Characterization and prognostic indices. Vet. Pathol. 2011, 48, 156–168. [Google Scholar] [CrossRef] [PubMed]
- Wilcock, B.P.; Yager, J.A.; Zink, M.C. The morphology and behaviour of feline cutaneous mastocytomas. Vet. Pathol. 1986, 23, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Barrett, L.E.; Skorupski, K.; Brown, D.C.; Weinstein, N.; Clifford, C.; Szivek, A.; Haney, S.; Kraiza, S.; Krick, E.L. Outcome following treatment of feline gastrointestinal mast cell tumours. Vet. Comp. Oncol. 2018, 16, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Sabattini, S.; Giantin, M.; Barbanera, A.; Shahidan, L.Z.; Docasto, M.; Zancanella, V.; Prata, D.; Trivigno, E.; Bettini, G. Feline intestinal mast cell tumours: Clinicopathological characterisation and KIT mutation analysis. J. Feline Med. Surg. 2016, 18, 280–289. [Google Scholar] [CrossRef]
- Tsai, M.; Takeishi, T.; Thompson, H.; Langley, K.E.; Zsebo, K.M.; Metcalfe, D.D.; Geissler, E.N.; Galli, S.J. Induction of mast cell proliferation, maturation, and heparin synthesis by the rat c-kit ligand, stem cell factor. Proc. Natl. Acad. Sci. USA 1991, 88, 6382–6386. [Google Scholar] [CrossRef]
- Bodemer, C.; Hermine, O.; Palmérini, F.; Yang, Y.; Grandpeix-Guyodo, C.; Leventhal, P.S.; Hadj-Rabia, S.; Nasca, L.; Georgin-Lavialle, S.; Cohen-Akenine, A.; et al. Pediatric mastocytosis is a clonal disease associated with D816V and other activating c-KIT mutations. J. Investig. Dermatol. 2010, 130, 804–815. [Google Scholar] [CrossRef]
- Letard, S.; Yang, Y.; Hanssens, K.; Palmérini, F.; Leventhal, P.S.; Guéry, S.; Moussy, A.; Kinet, J.P.; Hermine, O.; Dubreuil, P. Gain-of-function mutations in the extracellular domain of KIT are common in canine mast cell tumors. Mol. Cancer Res. 2008, 6, 1137–1145. [Google Scholar] [CrossRef]
- Isotani, M.; Yamada, O.; Lachowicz, J.L.; Tamura, K.; Yagihara, H.; Fujino, Y.; Ono, K.; Washizu, T.; Bonkobara, M. Mutations in the fifth immunoglobulin-like domain of kit are common and potentially sensitive to imatinib mesylate in feline mast cell tumours. Br. J. Haematol. 2010, 148, 144–153. [Google Scholar] [CrossRef]
- Hahn, K.A.; Ogilvie, G.; Rusk, T.; Devauchelle, P.; Leblanc, A.; Legendre, A.; Powers, B.; Leventhal, P.S.; Kinet, J.P.; Palmerini, F.; et al. Masitinib is safe and effective for the treatment of canine mast cell tumors. J. Vet. Intern. Med. 2008, 22, 1301–1309. [Google Scholar] [CrossRef]
- Mochizuki, H.; Thomas, R.; Moroff, S.; Breen, M. Genomic profiling of canine mast cell tumors identifies DNA copy number aberrations associated with KIT mutations and high histological grade. Chromosome Res. 2017, 25, 129–143. [Google Scholar] [CrossRef]
- Webster, J.D.; Yuzbasiyan-Gurkan, V.; Kaneene, J.B.; Miller, R.; Resau, J.H.; Kiupel, M. The role of c-KIT in tumorigenesis: Evaluation in canine cutaneous mast cell tumors. Neoplasia 2006, 8, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Tamlin, V.S.; Bottema, C.D.K.; Peaston, A.E. Comparative aspects of mast cell neoplasia in animals and the role of KIT in prognosis and treatment. Vet. Med. Sci. 2020, 6, 3–18. [Google Scholar] [CrossRef]
- Hadzijusufovic, E.; Peter, B.; Rebuzzi, L.; Baumgartner, C.; Gleixner, K.V.; Gruze, A.; Thaiwong, T.; Pickl, W.F.; Yuzbasiyan-Gurkan, V.; Willmann, M.; et al. Growth-inhibitory effects of four tyrosine kinase inhibitors on neoplastic feline mast cells exhibiting a Kit exon 8 ITD mutation. Vet. Immunol. Immunopathol. 2009, 132, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Isotani, M.; Tamura, K.; Yagihara, H.; Hikosaka, M.; Ono, K.; Washizu, T.; Bonkobara, M. Identification of a c-kit exon 8 internal tandem duplication in a feline mast cell tumor case and its favorable response to the tyrosine kinase inhibitor imatinib mesylate. Vet. Immunol. Immunopathol. 2006, 114, 168–172. [Google Scholar] [CrossRef] [PubMed]
- Berger, E.P.; Johannes, C.M.; Post, G.S.; Rothchild, G.; Shiu, K.B.; Wetzel, S.; Fox, L.E. Retrospective evaluation of toceranib phosphate (Palladia) use in cats with mast cell neoplasia. J. Feline Med. Surg. 2018, 20, 95–102. [Google Scholar] [CrossRef]
- Sabattini, S.; Guadagni Frizzon, M.; Gentilini, F.; Turba, M.E.; Capitani, O.; Bettini, G. Prognostic significance of Kit receptor tyrosine kinase dysregulations in feline cutaneous mast cell tumors. Vet. Pathol. 2013, 50, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Tamlin, V.S.; Bottema, C.D.K.; Campbell-Ward, M.L.; Hanshaw, D.; Peaston, A.E. KIT mutations in mast cell tumours from cheetahs (Acinonyx jubatus) and domestic cats (Felis catus). Vet. Comp. Oncol. 2021, 19, 381–392. [Google Scholar] [CrossRef]
- Dank, G.; Chien, M.B.; London, C.A. Activating mutations in the catalytic or juxtamembrane domain of c-kit in splenic mast cell tumors of cats. Am. J. Vet. Res. 2002, 63, 1129–1133. [Google Scholar] [CrossRef]
- Kim, J.-H.; Graef, A.J.; Dickerson, E.B.; Modiano, J.F. Pathobiology of Hemangiosarcoma in Dogs: Research Advances and Future Perspectives. Vet. Sci. 2015, 2, 388–405. [Google Scholar] [CrossRef]
- Kakiuchi-Kiyota, S.; Obert, L.A.; Crowell, D.M.; Xia, S.; Roy, M.D.; Coskran, T.M.; Kreeger, J.M.; Crabbs, T.A.; Cohen, S.M.; Cattley, R.C.; et al. Expression of Hematopoietic Stem and Endothelial Cell Markers in Canine Hemangiosarcoma. Toxicol. Pathol. 2020, 48, 481–493. [Google Scholar] [CrossRef]
- Kakiuchi-Kiyota, S.; Crabbs, T.A.; Arnold, L.L.; Pennington, K.L.; Cook, J.C.; Malarkey, D.E.; Cohen, S.M. Evaluation of expression profiles of hematopoietic stem cell, endothelial cell, and myeloid cell antigens in spontaneous and chemically induced hemangiosarcomas and hemangiomas in mice. Toxicol Pathol. 2013, 41, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Griffin, M.A.; Culp, W.T.N.; Rebhun, R.B. Canine and feline haemangiosarcoma. Vet. Rec. 2021, 189, e585. [Google Scholar] [CrossRef] [PubMed]
- Painter, C.A.; Jain, E.; Tomson, B.N.; Dunphy, M.; Stoddard, R.E.; Thomas, B.S.; Damon, A.L.; Shah, S.; Kim, D.; Gómez Tejeda Zañudo, J.; et al. The Angiosarcoma Project: Enabling genomic and clinical discoveries in a rare cancer through patient-partnered research. Nat. Med. 2020, 26, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Culp, W.T.; Drobatz, K.J.; Glassman, M.M.; Baez, J.L.; Aronson, L.R. Feline visceral hemangiosarcoma. J. Vet. Intern. Med. 2008, 22, 148–152. [Google Scholar] [CrossRef]
- Wong, K.; Ludwig, L.; Krijgsman, O.; Adams, D.J.; Wood, G.A.; van der Weyden, L. Comparison of the oncogenomic landscape of canine and feline hemangiosarcoma shows novel parallels with human angiosarcoma. Dis. Models Mech. 2021, 14, dmm049044. [Google Scholar] [CrossRef]
- D’Costa, S.; Yoon, B.I.; Kim, D.Y.; Motsinger-Reif, A.A.; Williams, M.; Kim, Y. Morphologic and molecular analysis of 39 spontaneous feline pulmonary carcinomas. Vet. Pathol. 2012, 49, 971–978. [Google Scholar] [CrossRef]
- Hahn, K.A.; McEntee, M. Prognosis factors for survival in cats after removal of a primary lung tumor: 21 cases (1979–1994). Vet. Surg. 1998, 27, 307–311. [Google Scholar] [CrossRef]
- Mehlhaff, C.J.; Mooney, S. Primary pulmonary neoplasia in the dog and cat. Vet. Clin. N. Am. Small Anim. Pract. 1985, 15, 1061–1067. [Google Scholar] [CrossRef]
- Goldfinch, N.; Argyle, D.J. Feline lung-digit syndrome: Unusual metastatic patterns of primary lung tumours in cats. J. Feline Med. Surg. 2012, 14, 202–208. [Google Scholar] [CrossRef]
- Husgafvel-Pursiainen, K.; Ridanpaa, M.; Anttila, S.; Vainio, H. p53 and ras gene mutations in lung cancer: Implications for smoking and occupational exposures. J. Occup. Environ. Med. 1995, 37, 69–75. [Google Scholar] [CrossRef]
- Suzuki, M.; Shiraishi, K.; Yoshida, A.; Shimada, Y.; Suzuki, K.; Asamura, H.; Furuta, K.; Kohno, T.; Tsuta, K. HER2 gene mutations in non-small cell lung carcinomas: Concurrence with Her2 gene amplification and Her2 protein expression and phosphorylation. Lung Cancer 2015, 87, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Grossman, D.A.; Hiti, A.L.; McNiel, E.A.; Ye, Y.; Alpaugh, M.L.; Barsky, S.H. Comparative oncological studies of feline bronchioloalveolar lung carcinoma, its derived cell line and xenograft. Cancer Res. 2002, 62, 3826–3833. [Google Scholar] [PubMed]
- Muscatello, L.V.; Oto, E.D.; Dignazzi, M.; Murphy, W.J.; Porcellato, I.; De Maria, R.; Raudsepp, T.; Foschini, M.P.; Sforna, M.; Benazzi, C.; et al. HER2 Overexpression and Amplification in Feline Pulmonary Carcinoma. Vet. Pathol. 2021, 58, 527–530. [Google Scholar] [CrossRef] [PubMed]
- Linderman, M.J.; Brodsky, E.M.; de Lorimier, L.P.; Clifford, C.A.; Post, G.S. Feline exocrine pancreatic carcinoma: A retrospective study of 34 cases. Vet. Comp. Oncol. 2013, 11, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Nicoletti, R.; Chun, R.; Curran, K.M.; de Lorimier, L.P.; Morges, M.A.; Rau, S.; Zwahlen, C.H.; Thamm, D.H. Postsurgical Outcome in Cats with Exocrine Pancreatic Carcinoma: Nine Cases (2007–2016). J. Am. Anim. Hosp. Assoc. 2018, 54, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Dedeaux, A.M.; Langohr, I.M.; Boudreaux, B.B. Long-term clinical control of feline pancreatic carcinoma with toceranib phosphate. Can. Vet. J. 2018, 59, 751–754. [Google Scholar]
- Forbes, S.A.; Beare, D.; Gunasekaran, P.; Leung, K.; Bindal, N.; Boutselakis, H.; Ding, M.; Bamford, S.; Cole, C.; Ward, S.; et al. COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015, 43, D805–D811. [Google Scholar] [CrossRef]
- Mayr, B.; Schaffner, G.; Reifinger, M.; Loupal, G. K-ras protooncogene mutations in feline pancreatic adenocarcinomas. Vet. Rec. 2003, 153, 468–469. [Google Scholar] [CrossRef]
- Crozier, C.; Wood, G.A.; Foster, R.A.; Stasi, S.; Liu, J.H.; Bartlett, J.M.; Coomber, B.L.; Sabine, V.S. KRAS Mutations in Canine and Feline Pancreatic Acinar Cell Carcinoma. J. Comp. Pathol. 2016, 155, 24–28. [Google Scholar] [CrossRef]
- Heldmann, E.; Anderson, M.C.; Wagner-Mann, C. Feline osteosarcoma: 145 cases (1990–1995). J. Am. Anim. Hosp. Assoc. 2000, 36, 518–521. [Google Scholar] [CrossRef]
- Dimopoulou, M.; Kirpensteijn, J.; Moens, H.; Kik, M. Histologic prognosticators in feline osteosarcoma: A comparison with phenotypically similar canine osteosarcoma. Vet. Surg. 2008, 37, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Bitetto, W.V.; Patnaik, A.; Schrader, S.C.; Mooney, S.C. Osteosarcoma in cats: 22 cases (1974–1984). J. Am. Vet. Med. Assoc. 1987, 190, 91–93. [Google Scholar] [PubMed]
- Nakano, Y.; Kagawa, Y.; Shimoyama, Y.; Yamagami, T.; Nomura, K.; Wakabayashi, H.; Sugiyama, Y.; Kobayashi, T. Outcome of appendicular or scapular osteosarcoma treated by limb amputation in cats: 67 cases (1997–2018). J. Am. Vet. Med. Assoc. 2021, 260, S24–S28. [Google Scholar] [CrossRef] [PubMed]


| Population | Benign Versus Malignant | Study |
|---|---|---|
| 59 tumours from 56 domestic cats in Tulsa, USA (1972 to 1973) | Overall, 83% of the tumours were malignant | [4] |
| 18,375 tumours from 51,322 domestic cats in Switzerland (1965–2008) | Overall, 80% of the tumours were malignant | [5] |
| 9683 cutaneous tumours from 9200 domestic cats in the UK (2006–2013) | Overall, 53% of the tumours were malignant | [6] |
| 685 tumours from domestic cats in Mexico City, Mexico (2006–2018) | Overall, 85% of the tumours were malignant | [7] |
| 475 tumours from 417 domestic cats in Portugal (2019) | Overall, 75% of the tumours were malignant | [8] |
| 1724 tumours from domestic cats in Portugal (2019–2020) | Overall, 75% of the tumours were malignant | [9] |
| Lymphoma Cell Lines: Genetic Investigations Performed and Results Obtained |
|---|
| 3201 cell line [26]:Examined for mutations in TP53; a non-synonymous mutation was found at codon 235 |
| 3201, FT-1, FL-74, KO-1, R96 cell lines [27]:Cytogenetic analysis and examined for mutations in TP53 and mRNA expression levels of MDM2; 3201, FL-74 and R96 showed centrosomal amplification and chromosomal instability, mutations in TP53 were found in all the cell lines (although only 3201 had a non-synonymous mutation at codon 235), and none of the cell lines showed elevated MDM2 mRNA levels |
| FL-7, FT-1, 2301, KO-1, R96 cell lines [28]:Examined for mutations in GADD45; no mutations were found |
| S87 cell line [29]:Cytogenetic analysis; showed centrosomal amplification and chromosomal instability |
| 3281, FT-1, MS4 cell lines [30]:Treated with a DNA methylation inhibitor; aberrant gene expression patterns observed |
| FT-1, MS4, KO-1 cell lines [31]:Genome-wide methylation profiling; showed thousands of CpG sites with gain of methylation at normally unmethylated CpG islands and loss of methylation at normally methylated non-CpG islands |
| Samples | c-KIT Mutations Reported | Study |
|---|---|---|
| 20 cutaneous MCTs from domestic cats 4 cutaneous MCTs from cheetahs |
| [112] |
| 16 intestinal MCTs from domestic cats |
| [99] |
| 24 cutaneous MCTs from domestic cats |
| [111] |
| 62 cutaneous, splenic or widespread MCTs from domestic cats |
| [103] |
| 10 splenic MCTs from domestic cats |
| [113] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ludwig, L.; Dobromylskyj, M.; Wood, G.A.; van der Weyden, L. Feline Oncogenomics: What Do We Know about the Genetics of Cancer in Domestic Cats? Vet. Sci. 2022, 9, 547. https://doi.org/10.3390/vetsci9100547
Ludwig L, Dobromylskyj M, Wood GA, van der Weyden L. Feline Oncogenomics: What Do We Know about the Genetics of Cancer in Domestic Cats? Veterinary Sciences. 2022; 9(10):547. https://doi.org/10.3390/vetsci9100547
Chicago/Turabian StyleLudwig, Latasha, Melanie Dobromylskyj, Geoffrey A. Wood, and Louise van der Weyden. 2022. "Feline Oncogenomics: What Do We Know about the Genetics of Cancer in Domestic Cats?" Veterinary Sciences 9, no. 10: 547. https://doi.org/10.3390/vetsci9100547
APA StyleLudwig, L., Dobromylskyj, M., Wood, G. A., & van der Weyden, L. (2022). Feline Oncogenomics: What Do We Know about the Genetics of Cancer in Domestic Cats? Veterinary Sciences, 9(10), 547. https://doi.org/10.3390/vetsci9100547







